Summary
Ezrin/Radixin/moesin (ERM) family members provide a regulated link between the cortical actin cytoskeleton and the plasma membrane to govern membrane structure and organization. Here we report the crystal structure of intact insect moesin, revealing that its essential yet previously uncharacterized α-helical domain forms extensive interactions with conserved surfaces of the core FERM domain. These interdomain contacts provide a functional explanation for how PIP2 binding and tyrosine phosphorylation of ezrin lead to activation, and also provide an understanding of previously enigmatic loss-of-function missense mutations in the tumor suppressor merlin. Sequence conservation and biochemical results indicate that this structure represents a complete model for the closed state of all ERM-merlin proteins, wherein the central α-helical domain is an active participant in an extensive set of inhibitory interactions that can be unmasked, in a rheostat like manner, by coincident regulatory factors that help determine cell polarity and membrane structure.
Keywords: Structure, actin, ERM, merlin, coiled-coil
Introduction
The plasma membrane is organized into functional regions, with the distinct apical and basolateral domains of polarized epithelial cells providing a well-studied example. To assemble, maintain and regulate the composition and structure of these domains, eukaryotes have evolved proteins that link the underlying cytoskeleton to specific membrane proteins. Among the best understood class are the ezrin/radixin/moesin (ERM) family which provide a conformationally regulated linkage from the cortical actin cytoskeleton to the plasma membrane, especially in structures like microvilli 1; 2.
ERM proteins consist of three principal domains (Fig. 1a). The best characterized of these are the band four-point-one/ezrin/radixin/moesin (FERM) domain 3 and the C-terminal tail domain. The N-terminal ∼300 residue FERM domain consists of three lobes, designated F1, F2 and F3, that are tightly associated in a cloverleaf type structure 4. The FERM domain of ERM proteins binds directly to integral membrane proteins, such as CD43, CD44 or ICAM1-3, through their positively-charged juxtamembrane regions 5; 6; 7; 8; 9, or indirectly through the PDZ-containing scaffolding proteins EBP50/NHERF and E3KARP 10; 11; 12. The C-terminal tail domain (often referred to as the C-terminal tail or C-terminal ERM Association Domain or CERMAD) spans the last ∼100 residues and contains an F-actin binding site in the last 30 residues 13; 14; 15. This domain interacts with the FERM domain as an extended, meandering polypeptide beginning with a β-strand associated with β5 in F3 followed by four helices, the first two of which bind lobe F2 and second two lobe F3 (Fig. 2a) 4. The FERM-tail complex represents a dormant form of the protein in which membrane protein and active binding sites are masked.
Figure 1.
Domain structure of Sfmoesin and primary sequence of the ERM-merlin α-helical domain. (a) Domain structure of Sfmoesin. Residue numbers at the domain boundaries are indicated. (b) Alignment of ERM-Merlin α-helical domains. The sequence for αC is folded back (runs right to left) to indicate its register with the αB helix. Helical regions are indicated by a yellow coil, and the β1 strand of the C-terminal tail by a red arrow. The a and d positions of the coiled-coil heptad repeat 16 are shown with orange and cyan backgrounds. These positions interact with the other helix of the coil, as shown. Residues that are disordered in both Sfmoesin structures are shown with lower case italics. Sequence numbering corresponds to that of Sfmoesin, and the asterisks indicate invariant or highly conserved residues. Sites sensitive to trypsin digestion in the radixin α-helical domain (black arrows), and positions in human merlin associated with cancer (purple arrows) are indicated. Sequences used are as follows: human merlin (HsMerlin), SwissProt accession no. P35240; human radixin (HsRadixin), P35241; human ezrin (HsEzrin), P15311; human moesin (HsMoesin), P26038; and D. melanogaster moesin (DmMoesin), GenBank accession no. NP_996392.
Figure 2.
Comparison of dormant human and Sfmoesin structures. (a) The human FERM-C-terminal domain complex (PDB code 1EF1). The three lobes of the ERM domain (F1, F2 and F3) are colored cyan and the C-terminal domain red. The β1 strand of the C-terminal domain is contributed by a crystal-packing interaction. (b) The 2.1 Å Sfmoesin structure. The α-helical domain (yellow) folds into three extended helices (αA, αB and αC), each of which containing elements that pack against the FERM domain. The αB and αC helices form an anti-parallel coiled-coil. (c) In the 3.0 Å structure, 67 more residues of the ∼70Å αB/αC coiled-coil are revealed.
Linking the FERM and C-terminal domains is an essential but structurally uncharacterized domain of ∼190 residues, referred to as the α-helical domain, the most conserved feature of which is a heptad repeat characteristic of α-helical coiled-coils 16 (Fig. 1b). This region has been proposed to form an extended helical tether in activated ERM proteins linking the membrane binding determinants of the FERM domain to the actin-binding determinant in the C-terminal tail 17. Although crystal structures have been reported for activated FERM domains as well as for the inactive FERM domain complexed with the C-terminal tail of human moesin, they have at most revealed only a small portion of the important and enigmatic α-helical domain.
Equally enigmatic is the neurofibromatosis 2 (NF2) tumor suppressor protein merlin, which is closely related to ERM proteins and shares all of the above features except for actin binding 18; 19. Mutations in merlin that lead to loss of tumor suppression are often disruptive, either truncating the protein or interfering with the proper fold of the protein. Some missense mutations of merlin associated with NF2 have been mapped to the interface of the FERM and C-terminal tail domain 4, suggesting that their association is critical for tumor suppressor activity. However, many more mutations remain unexplained given the current available models.
A particularly interesting aspect of ERM proteins is that they can exist in at least two conformational states 1, an active open form with the FERM and C-terminal tail domain dissociated, and a dormant closed form similar to that described for the human moesin FERM-C-terminal tail domain complex 4. Dissociation of the FERM and C-terminal domains unmasks binding sites for other proteins. EBP50 binds to a region of the FERM domain that overlaps with the C-terminal tail, 20; 21, and the ICAM-2 receptor binds to the radixin FERM domain at a site analogous to the first β-strand in the C-terminal tail 22. This site is also analogous to where integrin tails anchor to the talin FERM domain 23, and to where a regulatory intramolecular linker region interacts with the FERM domain of focal adhesion kinase 24. Evidence suggests these states are primarily regulated by the phosphorylation of a threonine in the C-terminal domain (equivalent to Thr558 in moesin) and/or the binding of PIP2 1. Thr558 lies buried in the FERM-C-terminal tail interface, and its phosphorylation is expected to favor domain dissociation. PIP2 binds to a site between lobes F1 and F3 and has been proposed to confer subtle conformational changes that favor dissociation of the interface, for this site is not obviously masked in the dormant protein 25. Although merlin is expected to have the same domain architecture as ERM proteins 4; 26, it is activated differently via phosphorylation at other sites and the self-associated, closed form is thought to be responsible for its tumor suppressor activity 27.
In this manuscript, we report the structure of the full-length, dormant monomeric moesin endogenous to ovarian Spodoptera frugiperda cells (Sfmoesin), the ERM protein expected to play an essential role in establishing polarity in insect embryos 28. The central α-helical domain is seen to contain three helices, including a striking 70 Å anti-parallel coiled-coil, that form extensive, unanticipated and functionally important interactions with the FERM domain. We argue that this structure provides a complete and relevant model for the dormant state of ERM proteins and for what is thought to be the relevant tumor suppressor state of merlin.
Results
Overall structure
Sfmoesin was purified with yields of 0.5 mg/L from ovarian insect cells, and is a stable monomer as determined by gel filtration chromatography. The cDNA sequence we determined corresponds to a protein of 575 residues, having 83% identity with Drosophila moesin and ∼60% identity with human ERM proteins. The most variability occurs within residues 315-500, which spans most of the α-helical domain and the beginning of the C-terminal tail (Fig. 1). Within this region, Sfmoesin and human ERM proteins have < 25% sequence identity. Residues 299-461 have significant coiled-coil probability 29, with hydrophobic residues dominating at positions a and d of the heptad repeat (Fig. 1b).
We determined two closely related crystal structures of Sfmoesin at 2.1 and 3.0 Å resolution (Table I; Fig. 2), with the lower resolution structure revealing a much larger portion of the α-helical domain. In addition to the expected tri-lobed FERM domain and C-terminal tail interaction, the α-helical domain is revealed to consist of three extended helices (αA, B and C). The first helix folds back under lobe F1, as has been observed in several prior structures of the open form of ERM-merlin proteins (Fig. 3a), and is followed by helices that form the outgoing (αB) and incoming (αC) segments of a 70 Å antiparallel coiled-coil. The N-terminus of αB (residues 328-350) is not paired with αC, and instead uses the hydrophobic residues of its heptad repeat to interact with a highly-conserved surface on the F1 lobe (Fig. 1b, 4). Comparison of the moesin coiled-coil with those from other proteins demonstrates that it has standard geometry, including the N-terminal portion of αB that interacts with F1 (Fig. 3).
Table I.
Crystallographic data and refinement statistics
| A (Initial Dataset) | B (High-resolution) | C (Low-resolution) | |
|---|---|---|---|
| X-ray Source: | ALS 8.3.1 | APS 17-ID | APS 19-BM |
| Wavelength (Å) | 1.116 | 1.000 | 1.033 |
| Resolution (Å) | 3.5 | 2.1 | 3.0 |
| Space group | R32 | R32 | R32 |
| Cell constants (Å) | a=b=124.1, c=285.0 | a=b=123.7, c=283.2 | a=b=126.9, c=272.5 |
| Unique reflections | 10518 | 43611 | 17202 |
| Redundancy | 7.5 (7.6) a | 4.9 (3.5) a | 5.4 (5.3) a |
| Rsym (%)b | 22.9 (68.1) | 7.9 (33.8) | 14.4 (43.7) |
| Completeness (%) | 96.8 (98.1) | 88.5 (71.0) | 99.3 (99.7) |
| <I>/<σI> | 7.7 (2.4) | 19.7 (3.3) | 10.9 (3.3) |
| Refinement statistics | |||
| Resolution (Å) | 50 – 2.1 | 50 – 3.0 | |
| Total reflections | 43,611 (2506)c | 17202 (1258)c | |
| Protein atoms | 4004 | 4507 | |
| Non-protein atoms | 438 | 64 | |
| R.m.s.d. bond lengths (Å) | 0.010 | 0.007 | |
| R.m.s.d. bond angles (°) | 1.11 | 0.93 | |
| Est. coordinate error (Å) | 0.06 | 0.17 | |
| Average B-factor (Å2) | 24.5 | 35.9 | |
| Ramachandran plot | |||
| most favored (%) | 96.4 | 95.4 | |
| disallowed (%) | 0.2 | 0.2 | |
| Rworkd | 17.9 (16.8) | 18.1 (26.9) | |
| Rfreee | 21.5 (22.9) | 24.9 (33.5) | |
| Rfinalf | 17.6 (16.3) | 18.4 (24.7) | |
Numbers in parentheses correspond to the highest resolution shell of data; data set A: 2.74 – 2.6 Å; data set B: 2.18-2.10 Å; data set C: 3.11-3.00 Å.
Rsym = ΣhklΣi |I(hkl)i - I(hkl)|/ Σhkl I(hkl)i, where I(hkl) is the mean intensity of i reflections after rejections. A–1.0 I/σI cutoff was applied to data set C.
Numbers in parentheses correspond to the highest resolution shell of data; data set B: 2.15-2.10 Å; data set C: 3.08-3.00 Å.
Rwork = Σhkl||Fobs(hkl)| -|Fcalc(hkl)||/ Σhkl |Fobs(hkl)|; no I/σ cutoff was used during refinement.
5% of the truncated data set was excluded from refinement to calculate Rfree.
Final R-factor after last several rounds of refinement when all reflections were used.
Figure 3.
Comparison of the Sfmoesin α-helical domain with active ERM domain structures and other coiled-coil domains. (a) The structures of “activated” merlin, radixin, and moesin (PDB entries 1ISN, 1J19, 1E5W, respectively) superimposed on the 2.1 Å structure of Sfmoesin, showing that the αA helix does not greatly alter its orientation upon activation. (b) The Rad50 coiled-coil (PDB entry 1L8D) superimposed on that of the 3.0 Å structure of Sfmoesin in the same orientation as panel (a). In this structure, Rad50 is dimerized through a hook-like structure at the turn of the coiled-coil 48. (c) Superposition of the methyl-accepting chemotaxis protein coiled-coil (PDB entry 2CH7) 49. This coiled-coil is half of an antiparallel four-helix bundle. Note that the N-terminal segment of Sfmoesin αB, which does not participate in coiled-coil with αA, maintains proper coiled-coil geometry. Superpositions are the optimal fits reported by a search of the PDB using the DALI server 50.
Figure 4.
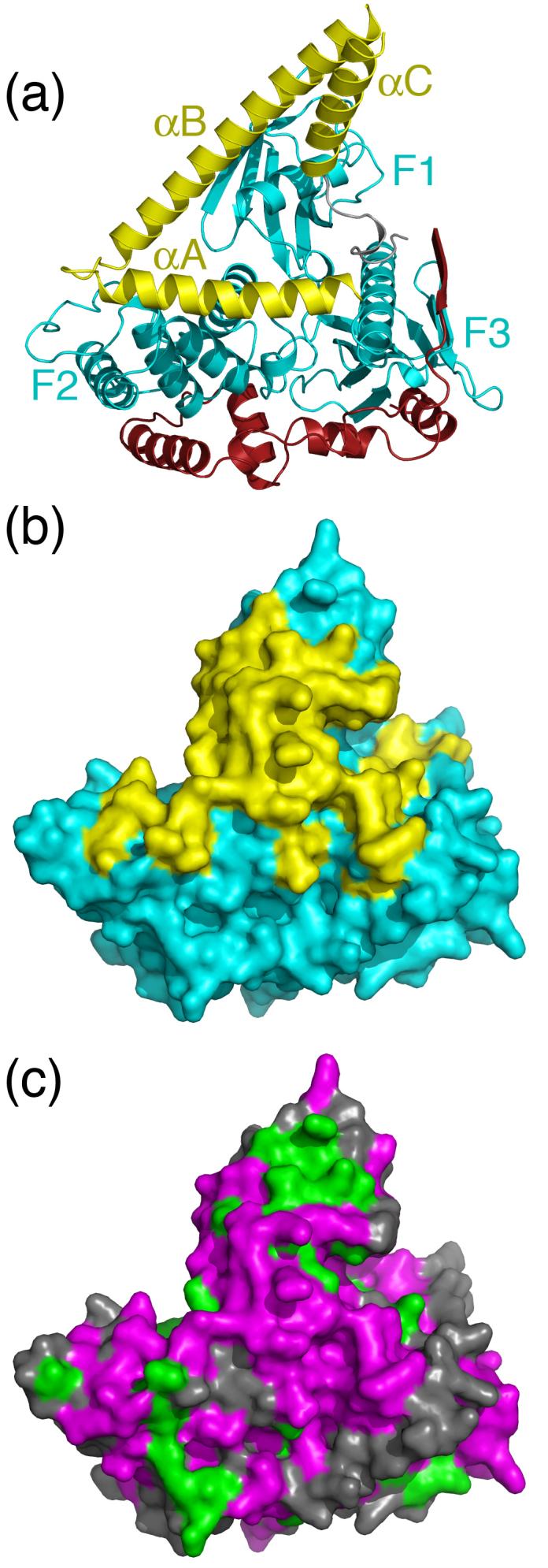
Extent and sequence conservation of the surfaces buried by the α-helical domain and linker region. (a) The Sfmoesin FERM domain. The view is rotated by ∼180° around a vertical axis from that in Fig. 2. (b) Molecular surface of the FERM domain. Yellow regions are those in contact with the α-helical domain and linker region (∼1800 Å2 of buried accessible surface area). (c) Conservation of the FERM domain. Magenta regions correspond to residues that are either identical or conservatively substituted (e.g. Asp/Glu, Arg/Lys, Ser/Thr) in all ERM-merlin proteins. Green regions correspond to residues conserved only in the ERM family.
Although the αB-αC loop of the α-helical domain is not observed, there is sufficient room in the crystal lattice for the missing residues to form a helical hairpin, and there is also no possibility for the formation of a crystalline dimer mediated by the coiled-coil (Fig. 5b). An examination of B-factors shows that the FERM domain on the whole is very well ordered, as are the segments of the α-helical domain that anchor its αA, αB and αC helices to the F1 lobe (Fig. 5b). However, B-factors for residues in the coiled-coil increase steadily as their distance from the F1 lobe increases, rising to > 140 Å2 at the termini. Nevertheless, helical electron density for the backbone of these residues is unambiguous. C-terminal to the αC helix (ending with Thr460), a “linker region” threads through the cleft formed between lobes F1 and F3, with residues 461 through 465 forming a short 310-helix and residues 465 through 472 adopting the conformation of a type II polyproline helix. The visible density ends at 472, and begins again at Glu486, the first residue of a β-strand associated with lobe F3 that defines the start of the C-terminal tail domain. The intervening disordered loop is enriched with acidic residues in ERM proteins (Fig. 1b).
Figure 5.
Crystal packing interactions and mobility of the α-helical domain. (a) Stereo view along the crystallographic 2-fold axis of a D3 center in the crystals of the 3.0 Å structure. Each asymmetric unit in the cluster is colored uniquely. In this structure, differences in lattice packing tilt the αB/αC coiled-coil region by 7°, allowing it to form crystal contacts with a 2-fold related coiled-coil via the side chains of Gln426 and Leu430 (ball and stick models). Only residues 400-409 are missing in its helical turn. This structure eliminates the possibility of a dimer mediated by the coiled-coil in the crystalline lattice. (b) Temperature factors as a function of residue position in the 2.1 and 3.0 Å structures of Sfmoesin. The regions of the α-helical domain that contact the FERM domains are as stable as the FERM domain, while those at greater distances from the ERM domain have gradually increasing mobility. The change in crystal contacts between the two structures orders more of the αB/αC coiled-coil in the low resolution structure, but also disorders part of the αA-αB loop (residues 312-325).
Interactions of the α-helical domain with the FERM Domain
Unexpectedly, the central α-helical domain of moesin forms multiple, extensive contacts with highly conserved surfaces of the FERM domain (Fig. 4). One surface, formed by both the F1 and F2 lobes, interacts with the αA helix, and two other faces of F1 stabilize and give direction to the αB/αC coiled-coil: one face interacts with the N-terminal portion of αB and the other provides a specific docking site for the residues at and after the C-terminal end of helix αC. These two interaction surfaces will be referred to as the “launching” and “landing” pads for the antiparallel coiled-coil, respectively. The contacts that the α-helical domain and the subsequent linker region (Fig. 1) form with the FERM domain bury 3,800 Å2 of total accessible surface area.
The interactions of αA are bipartite (Fig. 6a). The highly conserved N-terminus of αA (residues 298-313) forms extensive hydrophobic interactions with a continuous surface of F1 and F2, wherein the side chain of Ile94 (in F2) is buried in a hydrophobic pocket formed by Val302, Met305, Lys306 and Ala 309 (in αA), while the side chain of Met305 (αA) is buried in a pocket formed by Arg40 and Trp43 (in F1) and His465 (linker region). In the second interaction region, the C-terminus of αA and the subsequent αA-αB loop form primarily electrostatic contacts with F2. The basic side chains of Arg184, Lys316, Arg320, Arg332 (all invariant or highly conserved) cluster together and are in close proximity to the acidic side chains of Glu96, Glu185 and Glu326 (also highly conserved). The backbone of the αA-αB loop is disordered in the 3.0 Å structure (Fig. 5b), and has relatively high B-factors in the 2.1 Å structure, suggesting marginal stability of the interactions.
Figure 6.
Stereo views of interdomain contacts of the Sfmoesin α-helical domain and linker region. (a) The αA helix (yellow) interacts with the F1 and F2 lobes (cyan) in a bipartite fashion. Carbon atoms are shown with the same color as the backbone, oxygens red, nitrogens blue and sulfurs green. Specific hydrogen bonds are shown as black dashed lines. Residues colored with white carbons indicate side chains that are disordered or exist in multiple conformations in the 2.1 Å crystal structure. (b) The launching pad involves the hydrophobic face of αB and a highly conserved surface of the F1 lobe. Met347 is analogous to the site of tyrosine phosphorylation in ezrin 35;36, which leads to activation of ezrin in response to growth factor stimulation. Mutation of the residue analogous to Leu344 to proline in merlin is associated with NF2 39. Both modifications in the context of the Sfmoesin structure likely disrupt this interface. (c) The landing pad is formed by the end of the αC helix and the linker region. Contacts between the linker region and the F1 lobe are dominated by backbone-side chain interactions, possibly explaining the lack of strong sequence conservation in this region of the α-helical domain. Mutation of the residue analogous to Trp43 and residues in the F1 αA helix in merlin are also associated with NF2 39; 51. These changes likely disrupt the observed interdomain contacts.
The “launching pad” interaction for the αB/αC coiled-coil is formed by the hydrophobic face of the αB helix just prior to the coiled-coil, which abuts the αA helix of the F1 lobe (Fig. 6b). The highly conserved residues that make up this surface, Tyr340, Leu344 and Met347 occupy the “d-a-d” positions of the coiled-coil heptad repeat. Specific hydrogen bonds are also formed between the side chain of Gln337 (αB) and the backbone of Glu15 (F1), the side-chain of Arg343 with backbone carbonyls of residues 35 and 36, and the Tyr340 hydroxyl with the side chain of Asp13.
The “landing pad” interactions involve both the end of the αC helix and the linker region (Fig. 6c). The amino group of Lys35, conserved as either Lys or Arg among ERM-merlin proteins, caps the end of the αC helix. Lys27 and Asp31, which form an invariant salt bridge within F1, coordinate the buried hydroxyl of Thr461. His465 forms a hydrogen bond with the side chain of Asp301 in the first turn of the αA helix, joining the two ends of the α-helical domain. The side chain of His466 packs in a conserved hydrophobic pocket formed by Lys27, Phe30, Val42 and Leu61. Subsequently, the side chain of Val467 packs against F3, and Glu469 forms two hydrogen bonds with the backbone nitrogens of residues 61 and 62. Invariant moesin residues Glu289 and Arg293 (F3) together form a salt bridge as well as three hydrogen bonds with the backbone of residues 465, 466 and 468.
Comparisons with previous ERM structures
Prior structures of truncated/activated ERM proteins have included only the first 49, 27 and 14 residues of the α-helical domain (Fig. 3a). The intact Sfmoesin αA helix stays more tightly associated with the FERM domain than the others, possibly because they are missing subsequent residues that, as in Sfmoesin, would help restrain the C-terminal end of this helix.
Overlay of Sfmoesin with the radixin·IP3 complex demonstrates that the IP3 binding site is sterically blocked by the linker region (Fig. 7). Moreover, the disordered loop joining the linker region to the C-terminal tail is intensely acidic in ERM proteins (Fig. 1b). Thus the linker and subsequent loop dramatically lower the electrostatic potential of this region (Fig. 7b,c), which is believed to associate with negatively charged membranes. Because the 2 h soak of IP3 into the Sfmoesin crystals used for the low resolution data set showed no electron density for this ligand, IP3 alone under the crystal harvesting conditions is insufficient to displace these structures, rendering them an effective mask for PIP2 binding. This appears to conflict with a recent report that PIP2 binding is a prerequisite for C-terminal domain phosphorylation in ezrin 30. However, IP3 may be a poor mimic for PIP2 in the context of a phospholipid bilayer.
Figure 7.
The known intermolecular binding sites of ERM proteins are masked in the dormant structure. (a) The ICAM-2 (orange cartoon), IP3 (ball and sticks), and EBP50 (green coil) ligands mapped onto the structure of Sfmoesin (from PDB entries 1J19, 1GC6 and 1SGH, respectively). (b) Solvent accessible surface of the FERM domain of Sfmoesin with the α-helical domain and C-terminal domain superimposed. The surface is oriented as in panel (a) and colored by its electrostatic potential, contoured from −6.0 (red) to 6.0 kT/e- (blue). The intensely basic surface likely helps moesin to bind negatively charged lipid bilayers, such as those that contain PIP2 (see panel a). (c) The electrostatic surface of dormant Sfmoesin. The association of the α-helical domain and the linker region ablates the positive charge of the surface and masks the PIP2 site. The ICAM-2 peptide binding site is masked by the β1-strand of the C-terminal domain. The electrostatic calculation does not take into account the negatively charged, disordered loop (residues 473-485) that connects the linker to the C-terminal domain.
Finally, the Sfmoesin structure confirms the structure and register of the β1 strand in the C-terminal tail (Figs. 1,2b, 7a,b), which was domain-swapped in the human moesin FERM/C-terminal domain complex and masks the ICAM-2 peptide binding site observed in the radixin-ICAM-2 complex 22. The intact path of this strand to the remainder of the C-terminal domain is therefore fully defined in Sfmoesin. Sequence comparison suggests that all ERM-merlin proteins will share an analogous β-strand interaction with the F3 domain (Fig. 1b).
Discussion
A model relevant to the entire ERM-merlin family
The structure of the endogenous, intact Sfmoesin reported here provides a very important advance in the study of ERM-merlin proteins as it defines the complete dormant state of these proteins. For merlin, it has been postulated that the analogous closed conformation is responsible for tumor suppressor activity 27. In this conformation, the Sfmoesin structure demonstrates that there are extensive interactions of the central α-helical domain with the FERM domain, indicating that the α-helical domain plays a surprisingly active role in masking additional ligand binding and activation sites.
Conservation in other ERM-merlin proteins of the arrangement of the Sfmoesin αA, αB and αC helices in the α-helical domain is strongly supported by sequence conservation and biochemical studies. Unlike the αA helix, the coiled-coil portion of αB and αC (but not the αA-αB or αB-αC loops) has an almost perfect heptad repeat. Our structure also easily accommodates the seven residue insertion found in human radixin, ezrin and merlin proteins within or near the αA-αB loop (Fig. 1b). In addition, limited proteolysis of the recombinant isolated α-helical domain of human radixin show rapid cleavage at residue 330 followed by cleavage at residue 351 to yield a relatively stable fragment spanning 352-469 17. The first cleavage site corresponds to the highly mobile αA-αB loop (Fig. 4b) and the second matches the position within αB just before the start of the coiled-coil segment. The stable proteolytic fragment is thus a reasonable match with the αB/αC coiled-coil region of the Sfmoesin structure (Fig. 1b). This correlation leads to the rather remarkable inference that, even in the absence of contacts with the FERM domain, there are features intrinsic to the α-helical domain that partition it into the three distinct helices observed in the Sfmoesin structure.
Surfaces of the FERM domain that contact the α-helical domain are remarkably well-conserved (Fig. 4), supporting a biological role for these interactions. Interdomain contacts include the docking of αA onto the FERM domain via Met305, Lys306 and Ala 309 (Fig 6a), as observed in prior crystal structures of activated ERM-merlin proteins (Fig. 3a). The N-terminus of αB from the α-helical domain interacts with a nearly invariant surface of the F1 lobe with no previously ascribed function. The helical domain αC helix is followed by a stop signal (“T/STPxxxxxxE”) conserved among moesin proteins that also contains two residues (the second Thr and the ultimate Glu) that form specific interactions with lobe F1 (Fig. 6c). Although ezrin, radixin and merlin have a polyproline stretch in this region (Fig. 1b), this is nevertheless compatible with the salient features of the Sfmoesin linker region. Specifically, the most important interactions made by the “spacer” residues (marked “x” above) as they wind toward the IP3 binding site are made primarily by their main chain atoms (Fig. 6c). In addition, the backbone conformation of the linker is superimposable with a polyproline helix. Finally, it seems likely that the conserved, highly acidic region joining the linker region and the C-terminal domain (Fig. 1b) in ERM proteins could play a functional role in hindering PIP2 and membrane interactions until the α-helical and/or C-terminal domains become displaced.
The α-helical domain in activated ERM-merlin proteins
Biochemical studies of ERM-merlin proteins and the Sfmoesin structure now support the idea that the α-helical domain can adopt a variety of physiologically relevant conformations, ranging from the relatively condensed, inactive structure that we observe in Sfmoesin, to a relatively protected one in which the domain is dissociated from the FERM domain yet forms 2-3 helices that retain the observed αB/αC coiled-coil interaction, to a fully extended helix. The existence of the latter two states is supported by limited proteolytic digestion and biophysical studies of the isolated α-helical domain of radixin, and the activated structure of moesin17; 31.
For full activation of ERM proteins, large conformational changes are needed to expose various regulatory sites, including that for PIP2 which is masked by the linker region (Fig. 7). This direct model for activation is in contrast with a previous model wherein it was speculated that PIP2 binding led to activation via relatively subtle conformational changes 25. Unraveling the αB/αC coiled-coil observed in the dormant state also appears to be a prerequisite for gaining access to the buried hydrophobic residues that compose the A-kinase anchoring sites reported for ezrin and merlin 32; 33. Consistent with this, binding of merlin to RIβ only occurs in forms of merlin that mimic the “open” state (e.g. C-terminally truncated). Thus, the putative “switchblade-like” opening of the αB/αC coiled-coil into an extended helix may represent yet another functionally relevant unmasking event. The interconversion of helical and non-helical regions required for the proposed switchblade-like opening of the helical domain is well-precedented, perhaps most dramatically as seen in the pH triggered conformational change seen in hemaglutinin-mediated membrane fusion of the influenza virus 34. Conversion of the αB/αC coiled-coil to a fully extended state upon activation is also supported by biophysical studies on the isolated helical domain of radixin in which this region appears to form an unusual monomeric 240 Å long helix 17. This seems possible, given the strong helical potential of the αA-αB and αB-αC loops (Fig. 1b) and the fact that the 50 residues of the α-helical domain present in an activated moesin FERM structure continue as an uninterrupted helix that leaves the FERM domain in the direction of αA 31, rather than breaking at the αA-αB turn as is observed in Sfmoesin (Fig. 3a). Conversion to this extended form is likely to be stabilized by ligands that favor binding to a single amphipathic helix, such as AKAPs.
Until now there has been no concrete model for how Tyr353 phosphorylation of ezrin contributes to its activation 35; 36. The structurally equivalent residue in Sfmoesin (Met347) is buried at the end of the launching pad of αB (Fig. 6b) and it is reasonable to assume that in ezrin Tyr353 interacts similarly. Phosphorylation of Tyr353 in activated ezrin would be expected to strongly favor release of the α-helical domain from the FERM domain.
Finally, our studies provide insight into the structures of the dormant ERM homodimers that when compared to dormant monomers have indistinguishable patterns of limited proteolysis 37. We propose that in these dimers, the αB-αC turn does not exist, and instead two molecules associate via a fully-extended αB and αC helix that forms an intermolecular antiparallel coiled-coil and allows the C-terminal tail domain of each chain to interact with the FERM domain of the other chain. In this way exactly the same launching and landing pad contacts would be maintained. Consistent with this model, the αB-αC turn consists of exactly seven residues (Fig. 1b), so that the heptad repeat would be perfectly maintained in such a domain swapped dimer. While the physiological relevance of dormant homodimers is unknown, this structural arrangement could be present in the ezrin-merlin (and other) heterodimers that likely do exist in vivo 38.
An explanation for enigmatic mutations in the tumor suppressor merlin
Many of the missense mutations associated with NF2 27; 39 defied explanation when mapped onto prior atomic structures of ERM-merlin proteins. When mapped on the tertiary structure of Sfmoesin (Fig. 1b), the structural impact of many of these merlin mutations becomes clear, lending further support to the biological relevance of the α-helical domain contacts. Of the non-truncating mutations of merlin that are known to be pathogenic in cells, 16 are found in the F1 lobe, 9 in F2, 6 in F3, 10 in the α-helical domain and 7 in the C-terminal domain. Of those that are purely missense (not insertions or deletions), 10 are in F1, 4 in F2, 4 in F3, 10 in the α-helical domain, and 6 in the C-terminal tail. The preponderance of disruptions in the F1 lobe and the α-helical domain implies an important role for these domains in maintaining merlin in a stable, closed, active form. The F1 lobe αA helix, straddled by the αB and αC helices of the α-helical domain (Fig. 6), is particularly rich in NF2-associated disruptions. Also, the F1 lobe αA-β3 loop, with residues interacting with both the αA helix and the landing pad, is also relatively rich in mutations. Mutations identified in the α-helical domain of merlin are dispersed throughout, but some are found in the highly conserved N-terminus of αA: at Leu323 (L339F in merlin), a residue that helps to stabilize the αA-αB loop (Fig. 6a), and at Leu344 (L360P), a buried αB residue in the launching pad interface (Fig. 6b). This remarkable correlation between sites of disease-causing merlin mutations and sites involved in stabilizing interactions between the α-helical and FERM domains supports the conclusion that the closed form of merlin is required for its tumor suppressor activity. The structure reported here finally provides a solid structural foundation for studies to unravel the mechanisms of merlin action.
An active regulatory role for the α-helical domain
The crystal structure of the FERM-C-terminal domain complex of human moesin showed that the extensive FERM-C-terminal domain complex are held together by five largely independent interacting parts 4. We can now extend this model to include the launching and landing pads of the central α-helical domain, that together with the coiled-coil interactions provide sixth, seventh and eighth points of interaction with the FERM domain that can be independently modulated to influence the net affinity of the masking interactions. Interestingly, these interaction points with the FERM domain are, apparently, purposefully of lower affinity than what they could be. For example, ICAM and EBP50 peptides have conserved sequence features not present in the ERM sequences, and they bind to their respective sites on the FERM domain much more tightly in trans (nM affinities) than does the tail domain in cis. Such distributed binding involving many purposefully medium or low affinity interactions creates a net high affinity interaction via the chelate effect 40, but which can in principle have its affinity fine tuned to many different levels by a wide variety of effectors. In addition to known effectors already discussed above, the interactions of the α-helical domain allow speculation that another modulator of activation could be the binding of SH3 domains to the polyproline sequences that fill the F1-F3 cleft in ezrin, radixin and merlin. In accordance with this idea, the focal adhesion kinase FERM domain, although distantly related to ERM proteins and lacking an α-helical domain, has a regulatory linker region C-terminal to the FERM domain that occupies the cleft between its F1 and F3 lobes. The linker forms a β-strand interaction, analogous to but opposite in orientation to the β1 strand of the Sfmoesin C-terminal domain, and a polyproline helix, bound to F3, that can be sequestered by the SH3 domain of Src 24.
We conclude that the central α-helical domain is not just a passive structural feature that is important for facilitating effective cytoskeletal-membrane linkage once the activated proteins are generated. Instead, it is actively involved in masking, burying ∼2000 Å2 of FERM domain surface area, about 75% of that buried by the C-terminal tail interaction. Together, the central and C-terminal tail domains bury 4700 Å2 which is 25% of the accessible surface area of the FERM domain and directly mask all known sites for activating ligands or modifications (i.e. the PIP2 binding site and phosphorylation sites Thr558 or ezrin Tyr353) or docking targets (EBP50/NHERF, ICAM and actin). As alluded to in 4 and extended here from five to eight points of interaction, we propose that ERM proteins are subject to a ‘rheostat mechanism’ for a graded regulation of activation. Multiple regulatory pathways that impinge on the ERM proteins could be combined and/or integrated to allow for different levels of release of the coiled-coil domain, depending on their sites of interaction, and hence unmasking of ligand binding sites. Finally, our studies have now yielded an important image that fully describes the structural components involved in the closed form of ERM-merlin proteins. Such information will be exceptionally useful in the design and interpretation of a new generation of experiments aimed at unraveling the activation mechanisms of ERM proteins and to better understand the tumor suppression mechanism of merlin.
Materials and Methods
Purification
The endogenous moesin protein from Sf ovarian cells was purified following the same protocol used to purify G protein-coupled receptor kinase 2 (GRK2) 41. Except in the first purification, wherein Sfmoesin was mistakenly purified instead of GRK2, Sfmoesin-containing fractions were identified by Western analysis using a rabbit polyclonal antibody (a gift from D. Kiehart, Duke University) at 1:40,000 dilution. The final purification step was over two tandem S200 gel filtration columns (Amersham Pharmacia Biotech) equilibrated in 20 mM HEPES (pH 7.8-8.0), 200 mM NaCl, 2 mM DTT. The protein eluted as a protein with expected molecular weight of 74 kDa based on comparison with Bio-Rad Gel Filtration standards, and the resulting homogeneous moesin was concentrated to 4.75 mg/ml for crystallization. The total yield was 0.12 to 0.45 mg/L Sf cell culture.
Crystallization
Sfmoesin crystals grew by hanging drop vapor diffusion using the same conditions used for GRK2 42, with 100 mM HEPES pH 7.8 as the well solution buffer. For harvesting, cryoprotectant solution (25 % PEG400, 15 % PEG 8K, 50 mM phosphoserine pH 7.5, 800 mM NaCl, 800 mM urea, 1 mM DTT, 22 mM HEPES pH 8.0 and 22 mM HEPES pH 7.8) was added 1 μL at a time into the hanging drop, and subsequently crystals were transferred into 100% cryoprotectant solution. The crystals were then flash frozen on nylon loops (Hampton Research) in liquid nitrogen.
Diffraction Data collection
A preliminary 3.5 Å data set (Data Set A), collected at the Advanced Light Source (ALS) on a Quantum 210 CCD detector (ADSC) at beam line 8.3.1, was used for the initial structure determination. A higher resolution data set (Data Set B) was later collected at the Advanced Photon Source (APS) on a Quantum 4 CCD detector (ADSC) at beam line 17-ID. Lastly, a 3.0 Å data set (Data Set C) was collected at APS beam line 19-BM from a crystal soaked in harvesting solution supplemented with 1 mM IP3 for 2 hours. Data were indexed, integrated and scaled using HKL2000 43. The crystal soaked in IP3 had significantly different cell constants (>1% change), suggesting a conformational change. Unit cell parameters and data collection statistics are summarized in Table I.
Identification of Moesin by Mass Spectrometry
Sfmoesin was purified using the same scheme used for GRK2, had a similar apparent molecular weight (74 kDa for Sfmoesin, 80 kDa for GRK2) and crystallized under conditions identical to GRK2. However, molecular replacement using the structure of GRK2 failed to yield a solution. To verify the identity of the crystallized protein, a drop of the protein solution (roughly 5 μg) was taken from the hanging drops used for crystallization, run on a 10 % SDS-PAGE gel, and then subjected to in gel trypsin digestion followed by MALDI mass spectrometry at the Institute for Cellular and Molecular Biology Protein Core Facility (UT Austin). Four abundant peptide peaks were generated, and their resulting masses were best matched to D. melanogaster moesin (residues 29-QLFDQVVK-36, 42-EVWFFGLQYTDSK-54, 195- IAQDLEMYGVNYFEIR-210, 239-IGFPWSEIR-247), suggesting that the crystallized protein was in fact the moesin protein endogenous to S. furgiperda, which had not previously been sequenced.
Structure determination and Refinement
Phases were determined by straightforward molecular replacement using the structure of the FERM/C-terminal tail complex of human moesin (PDB code: 1EF1) as a search model, reflections from Data Set A and the program PHASER from the CCP4 suite 44. The resulting model was refined with simulated annealing in CNS 45 and then REFMAC5 46. The sequence of D. melanogaster moesin was used for initial model building. The high resolution of Data Set B permitted “sequencing by electron density” in the most well-resolved regions, and refinement was continued in REFMAC5. Upon sequencing of the Sfmoesin cDNA (see below), the refinement was completed (Table I). At an R factor of 17.9%, and Rfree of 21.5% the use of Rfree was discontinued so that all reflections could be used during the last few rounds of refinement. The final model for Data Set B contains residues 3-364, 442-470 and 486-575 (out of 575 total) and has Rfinal =16.3%. Difference Fourier analysis between Data Sets B and C revealed no significant electron density that might correspond to IP3, which was soaked in to the crystals used for Data Set C. However, these maps revealed that significantly more of the α-helical domain was ordered in Data Set C, and so an atomic model was refined using this data set as well. The final model from Data Set C contains residues 3-320, 326-399, 410-472, and 486-575. The different unit cell constants for Data Set C (Table I) presumably derive from the two hour soak in harvesting solution (as compared to several minutes for Data Set B). In the 3.0 Å structure, changes in crystal contacts disorder the αA-αB loop (residues 312-325) and lead to the formation of a new crystal contact stabilizing the αB/αC coiled-coil (Fig. 5a). In both structures, the backbone of residue 252 is in a disallowed region of the Ramachandran plot (Table I). This residue, conserved as Asp or Glu in all ERM-merlin proteins, is in the i+1 position of a type II′ β-turn (normally occupied by glycine), has good electron density, and the analogous residue in other ERM structures has the same conformation. The backbone nitrogen of Asp252 forms a hydrogen bond with residue 486, the first visible residue of β1 in the C-terminal domain. Coordinates and intensities corresponding to Data Sets B and C are deposited with the Protein Data Bank under the accession numbers 2I1J and 2I1K, respectively.
Sequencing of Sfmoesin
To determine the sequence of Sfmoesin, a portion of the highly conserved FERM domain was amplified using PCR with degenerate oligonucleotide primers followed by 5' and 3' RACE to complete the flanking sequences. Total RNA was isolated from Sf9 cell cultures using Trizol (Invitrogen). First strand synthesis was performed using random hexamer primers and Multiscribe (Applied Biosystems) reverse transcriptase. Using this template DNA, PCR reactions were performed using degenerate primers previously described for cloning of D. melanogaster moesin 47 as well as degenerate primers based on Sfmoesin protein sequence determined from mass spectrometry. Once the sequence of the FERM domain was obtained, 5' and 3' RACE were performed to obtain flanking sequences using the ‘GeneRacer’ system (Invitrogen) following the manufacturer's instructions. The sequence is available as Genbank entry ######.
Acknowledgments
We thank D. Lodowski (UT Austin), Ingo Focken and Jochen Huber (Sanofi-Aventis Deutschland GmbH) for technical assistance. Support was provided by American Heart Association Scientist Development Grant 0235273N, NIH Grant HL071818, American Cancer Society Research Scholar Grant 04-185-01 (to JJGT), NIH grant NS034783 (to RGF), and NIH grant GM36652 (to APB). The Advanced Light Source (ALS) is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory. Beam line 17-ID of the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) at the Advanced Photon Source (Argonne, IL) is supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Illinois Institute of Technology (IIT), executed through the IIT Center for Synchrotron Radiation Research and Instrumentation. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source, was supported by the U. S. Department of Energy, Office of Biological and Environmental Research, under Contract No. W-31-109-ENG-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nature Reviews Cell and Molecular Biology. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 2.Polesello C, Payre F. Small is beautiful: what flies tell us about ERM protein function in development. Trends Cell Biol. 2004;14:294–302. doi: 10.1016/j.tcb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, Chasis JA, Conboy JG, Gascard P, Takakuwa Y, Huang SC, Benz EJ, Jr., Bretscher A, Fehon RG, Gusella JF, Ramesh V, Solomon F, Marchesi VT, Tsukita S, Tsukita S, Hoover KB, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–2. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 4.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–70. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 5.Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–80. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885–95. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5- bisphosphate. J Biol Chem. 1998;273:21893–900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- 8.Helander TS, Carpen O, Turunen O, Kovanen PE, Vaheri A, Timonen T. ICAM-2 redistributed by ezrin as a target for killer cells. Nature. 1996;382:265–8. doi: 10.1038/382265a0. [DOI] [PubMed] [Google Scholar]
- 9.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reczek D, Berryman M, Bretscher A. Identification of EBP50: A PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–79. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino- terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–8. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- 12.Murthy A, Gonzalez-Agosti C, Cordero E, Pinney D, Candia C, Solomon F, Gusella J, Ramesh V. NHE-RF, a regulatory cofactor for Na(+)- H+ exchange, is a common interactor for merlin and ERM (MERM) proteins. J Biol Chem. 1998;273:1273–6. doi: 10.1074/jbc.273.3.1273. [DOI] [PubMed] [Google Scholar]
- 13.Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–75. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turunen O, Wahlstrom T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–53. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestonjamasp K, Amieva MR, Strassel CP, Nauseef WM, Furthmayr H, Luna EJ. Moesin, ezrin, and p205 are actin-binding proteins associated with neutrophil plasma membranes. Mol Biol Cell. 1995;6:247–59. doi: 10.1091/mbc.6.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen C, Parry DAD. 〈-Helical coiled coils - a widespread motif in proteins. Trends Biochem Sci. 1986;11:245–248. [Google Scholar]
- 17.Hoeflich KP, Tsukita S, Hicks L, Kay CM, Ikura M. Insights into a single rod-like helix in activated radixin required for membrane-cytoskeletal cross-linking. Biochemistry. 2003;42:11634–41. doi: 10.1021/bi0350497. [DOI] [PubMed] [Google Scholar]
- 18.Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;75:826. doi: 10.1016/0092-8674(93)90501-g. [DOI] [PubMed] [Google Scholar]
- 19.Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–21. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- 20.Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117:1547–52. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- 21.Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–89. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Hamada K, Shimizu T, Yonemura S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. Embo J. 2003;22:502–14. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 24.Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the FERM domain of focal adhesion kinase. J Biol Chem. 2006;281:252–9. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- 25.Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. Embo J. 2000;19:4449–62. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu T, Seto A, Maita N, Hamada K, Tsukita S, Hakoshima T. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J Biol Chem. 2002;277:10332–6. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- 27.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis--the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–77. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 28.Miller KG. A role for moesin in polarity. Trends Cell Biol. 2003;13:165–8. doi: 10.1016/s0962-8924(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 29.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 30.Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol. 2004;164:653–9. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards SD, Keep NH. The 2.7 A crystal structure of the activated FERM domain of moesin: an analysis of structural changes on activation. Biochemistry. 2001;40:7061–8. doi: 10.1021/bi010419h. [DOI] [PubMed] [Google Scholar]
- 32.Dransfield DT, Bradford AJ, Smith J, Martin M, Roy C, Mangeat PH, Goldenring JR. Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. Embo J. 1997;16:35–43. doi: 10.1093/emboj/16.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gronholm M, Vossebein L, Carlson CR, Kuja-Panula J, Teesalu T, Alfthan K, Vaheri A, Rauvala H, Herberg FW, Tasken K, Carpen O. Merlin links to the cAMP neuronal signaling pathway by anchoring the RIbeta subunit of protein kinase A. J Biol Chem. 2003;278:41167–72. doi: 10.1074/jbc.M306149200. [DOI] [PubMed] [Google Scholar]
- 34.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 35.Krieg J, Hunter T. Identification of the two major epidermal growth factor-induced tyrosine phosphorylation sites in the microvillar core protein ezrin. J Biol Chem. 1992;267:19258–65. [PubMed] [Google Scholar]
- 36.Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–30. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bretscher A, Gary R, Berryman M. Soluble ezrin purified from placenta exists as stable monomers and elongated dimers with masked C-terminal ezrin-radixin-moesin association domains. Biochemistry. 1995;34:16830–7. doi: 10.1021/bi00051a034. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen R, Reczek D, Bretscher A. Hierarchy of merlin and ezrin N-and C-terminal domain interactions in homo- and heterotypic associations and their relationship to binding of scaffolding proteins EBP50 and E3KARP. J Biol Chem. 2001;276:7621–9. doi: 10.1074/jbc.M006708200. [DOI] [PubMed] [Google Scholar]
- 39.Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Human Mutation. 2006 doi: 10.1002/humu.20393. in press. [DOI] [PubMed] [Google Scholar]
- 40.Fersht A. Structure and mechanism in protein science. W. H. Freeman and Company; 1999. pp. 345–346. [Google Scholar]
- 41.Lodowski DT, Barnhill JF, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Purification, crystallization and preliminary X-ray diffraction studies of a complex between G protein-coupled receptor kinase 2 and Gβ1γ2. Acta Crystallogr D Biol Crystallogr. 2003;59:936–9. doi: 10.1107/s0907444903002622. [DOI] [PubMed] [Google Scholar]
- 42.Lodowski DT, Barnhill JF, Pyskadlo RM, Ghirlando R, Sterne-Marr R, Tesmer JJ. The role of Gβγ and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry. 2005;44:6958–6970. doi: 10.1021/bi050119q. [DOI] [PubMed] [Google Scholar]
- 43.Otwinoski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 12):1622–4. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 45.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, GrosseKunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.McCartney BM, Fehon RG. Distinct cellular and subcellular patterns of expression imply distinct functions for the Drosophila homologues of moesin and the neurofibromatosis 2 tumor suppressor, merlin. J Cell Biol. 1996;133:843–52. doi: 10.1083/jcb.133.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–6. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 49.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–7. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 50.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–80. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 51.Turunen O, Sainio M, Jaaskelainen J, Carpen O, Vaheri A. Structure-function relationships in the ezrin family and the effect of tumor-associated point mutations in neurofibromatosis 2 protein. Biochim Biophys Acta. 1998;1387:1–16. doi: 10.1016/s0167-4838(98)00103-4. [DOI] [PubMed] [Google Scholar]








