Abstract
Mutations in the gene ELOVL4 have been shown to cause stargardt-like macular dystrophy. ELOVL4 is part of a family of fatty acid elongases and is yet to have a specific elongase activity assigned to it. We generated Elovl4 Y270X mutant mice and characterized the homozygous mutant as well as homozygous Elovl4 knockout mice in order to better understand the function or role of Elovl4. We found that mice lacking a functional Elovl4 protein died perinatally. The cause of death appears to be from dehydration due to faulty permeability barrier formation in the skin. Further biochemical analysis revealed a significant reduction in free fatty acids longer than C26 in homozygous mutant and knockout mouse skin. These results implicate the importance of these long chain fatty acids in skin barrier development. Furthermore, we suggest that Elovl4 is likely involved in the elongation of C26 and longer fatty acids.
Keywords: ELOVL4, fatty acids, skin, STGD3, mouse model.
1. Introduction
Using a positional cloning approach, ELOVL4 was identified as the causal disease gene responsible for stargardt-like macular dystrophy (STGD3, MIM 600110), an autosomal dominant form of juvenile macular degeneration characterized by decreased visual acuity, macular atrophy, and extensive flecks 1, 2. The disease-causing gene ELOVL4 encodes a protein with sequence and structural similarities to the ELO family of proteins3, 4. The ELO family of proteins is involved in the elongation of long chain fatty acids, suggesting that ELOVL4 may have a similar function (hence the name, ELOVL), and is highly expressed in rod and cone photoreceptor cells4-6. Sequence analysis of human ELOVL4 cDNA predicts a protein of 314 amino acids that share homology with members of the yeast Elo (elongation of long chain fatty acid) family and the human ELO1 homolog (HELO1) 4, 7. HELO1 and the ELO family members possess biochemical features that suggest their participation in reduction reactions occurring during fatty acid elongation 8, 9.
Mutational analysis of the ELOVL4 gene in five large STGD-like macular dystrophy pedigrees revealed a 5 base-pair deletion, resulting in a frame-shift and the introduction of a stop codon, 51 codons from the end of the coding region 4. Subsequently, two single base-pair deletions, 789delT and 794delT, in ELOVL4 were identified in an independent large Utah pedigree, confirming the role of the ELOVL4 gene in a subset of dominant macular dystrophies 10. Both the 5 base-pair deletion and the two single base-pair deletions are predicted to result in a similar truncated ELOVL4 protein. A third mutation in ELOVL4, 270stop, which should also generate a truncated ELOVL4 protein, has been identified in a Dutch family with dominant STGD 11. In addition, a recent report suggest that variants in ELOVL4 may predispose individuals to AMD12.
The mutant ELOVL4 misfolds and sequesters the wild type ELOVL4 into a aggregated form in cell cultures, suggesting a dominant negative mechanism by which mutant ELOVL4 causes macular degeneration in humans13-17. We have further shown lipofuscin accumulation, electroretinogram abnormality, and RPE and photoreceptor degeneration in mutant ELOVL4 transgenic mice18. Mice heterozygous for the Elovl4 5base-pair deletion developed progressive photoreceptor degeneration, while mice haploinsufficient for Elovl4 developed no noticeable retinal degeneration 19-21. This evidence suggests that mutant ELOVL4 exerts a dominant negative effect, resulting in retinal degeneration.
Mice lacking a functional copy of Elovl4 die perinatally 19, 21, 22. Mice harboring two 5 base-pair mutant copies of ELOVL4 have been shown to have reduced levels of N-acyl-very long chain fatty acids (VLCFAs) and epidermal free fatty acids greater than 26 carbons in chain length and lack w-0-acylceramide and acylglucosylceramide 22. The alterations in lipid content have been suggested to result in a skin permeability barrier defect shown to occur in these mice and lead to perinatal lethality, but the effect of having two mutant copies of Elovl4 cannot be ruled out as a possible cause of this lethality.22.
We have generated Elovl4 270x mutation knockin mice (Elovl4270x/270x) and have found that homozygote mice with two mutant copies of Elovl4 die perinatally, just like the 5 base-pair mutant and knockout mice described previously. We then did a comparative study with our Elovl4 knockout (Elovl4-/-) and Elovl4270x/270x mice. Both have similar skin phenotypes, failing to develop a functional stratum corneum (SC) permeability barrier. Skin total lipids of from both groups were compared and mice lacking a functional copy of ELOVL4 are unable to produce adequate quantities of free fatty acids greater than C26. Heterozygous animals have lipid levels between the wild-type and homozygous mice. Elovl4-/- and Elovl4270x/270x mice dehydrate at a rate of about 3% of their body weight per hour after birth and die within 2-3 hours. Lacking a functional copy of Elovl4 leads to reduced VLCFA levels affecting the skins ability to hold water and ultimately resulting in dehydration and death shortly after birth.
2. Methods
Generation and characterization of Elovl4 270x knockin mouse
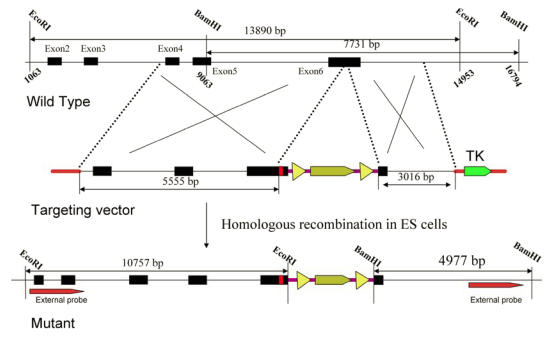
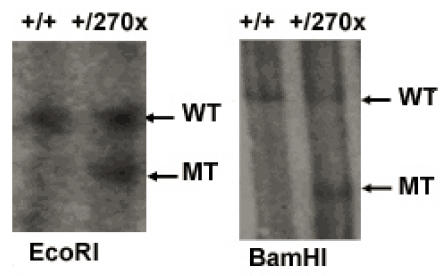
The targeting vector was derived from a 129/SVJ bacterial artificial chromosome clone that includes exon 6 from the Elovl4 gene. The construct consisted of Y270X mutation and HA tagged Elovl4 fragment, followed by a loxP-flanked neomycin resistance (neo) cassette, a 5.5-kb 5_ homology arm, and a 3-kb 3_ homology arm. The herpes simplex virus thymidine kinase (TK) was used as a counter selection marker (Fig. 1). The targeting vector was electroporated into 129/SVJ embryonic stem (ES) cells after linearized with NotI. Clones in which homologous recombination resulted in targeted replacement of exon 6 were identified by Southern blot analysis of genomic DNA digested with EcoRI and BamHI, using external probes, and then confirmed by PCR (Fig. 2). A karyotyped positive clone was injected and generated chimeras, which transmitted the recombinant locus. Genotyping of Elovl4 mutant mice was performed by PCR on genomic earclips DNA using oligonucleotides Elovl4-KI-gt-L(tgtctctctacaccgactgc)/Elovl4-KI-gt-R(tattcacgccgtttgatgag) to produce 174-bp and 201-bp bands from the wild-type and targeted allele, respectively (Fig. 3). A 280-bp neo cassette fragment was amplified from mutant mice using Pgk-neo-Int.F (cttgggtggagaggctattc) and Pgk-neo-Int.R (aggtgagatgacaggagatc) (Fig. 3). Elovl4 mutant mice were maintained in a C57BL/6 background.
Figure 1.
Schematic diagram of the Elovl4 locus and targeting construct. The construct contains 5.5 kb and 3 kb of the 5' and 3' exon6 genomic sequence. A HA tag was inserted before the induced Y270X mutation. A floxed PGKNEO (Neo), positive selection marker, was also inserted into exon6. There is a HSV-TK counter selection cassette in the 5' end of the vector. The wild type band expected from EcoRI digestion is 13.9 kb, mutant band is 10.8 kb. The wild type and mutant bands expected from BamHI digestion are 7.7 and 5.0 kb, respectively.
Figure 2.
Southern blot analysis of genomic DNA after digestion with EcoRI, BamHI and hybridization with 5', 3' external probes.
Figure 3.
PCR genotyping of mouse genomic DNA from Elovl4+/+ and Elovl4+/270x mice. (A) A single 174-bp fragment appears in the case of wild-type mice, and a 201-bp fragment appears in the case of heterozygous mice along with the wild-type fragment. (B) A 280-bp neo cassette fragment appears in the case of mutant mice.
Skin permeability assay
Embryonic day 19 (E19) mice were dehydrated for 1 min each in 25, 50, 75 and 100% methanol/PBS, prior to rehydration in PBS. The mice were stained for 5 min in 0.1% toluidine blue /PBS and then destained in a large volume of PBS prior to being photographed using a Nikon digital camera.
Dehydration assay
Dehydration was measured by weighing each mouse every 30 minutes for 6 hours. The mice are kept at 37 degrees C. Water loss is determined by weight loss in percent of initial birth weight. Water is presumed to be the sole contributor to the weight change. Littermates are compared to each other and statistical analysis is performed using SPSS.
Tissue preparation
E19 embryos are fixed in 4% paraformaldehyde/0.1M PBS overnight at 4 degrees C. The embryos are then cryoprotected in a graded series of sucrose/0.1M PBS (10-30%). Embryos are subsequently frozen in OCT compound and cryosectioned at 12µm in thickness.
Histological analysis
Sectioned slides will be stained with hematoxylin and eosin (H&E) and methyl green. H&E sections are stained for 3 minutes with hematoxylin followed by another 2 minute incubation in eosin and finally dehydrated in a series of alcohol rinses. Methyl green (0.5%) is used to stain sections for 5 minutes and then rinsed. Sections are viewed and photographed with a Zeiss Axiovert 200 microscope.
Mouse tissue lipid extraction
Mouse skin is removed from the torso of E19 mice and frozen on dry ice. Tissues are weighed and divided into approximately 0.2g portions. The Folch method for total lipid extraction of tissues is used for the extraction process. This is followed by a silicic acid column separation of lipid classes from Folch washed lower phase lipid extracts. Final purification & identification of lipids from the silicic acid column fractions is done by thin layer chromatography on washed silica gel G plates 0.25u, 20 x 20 cm and capillary GC. Two capillary columns differing in polarity are used to correctly identify and quantitate the fatty acid methyl esters (FAME). The columns that are routinely used are: 50 meter x .2u ID x .11u film OV-1 and a 100 meter x.2u ID x .15u film SP-2560 capillary column. Standards of known fatty acid composition and reagent blanks are analyzed with each set of samples. Individual lipids are expressed as a percentage of the total lipid analyzed.
GC/MS
FAME were generated as described above and subjected to GC/MS analysis. The sample injection volume is 1ul with a 1:35split at injection. Analysis is performed on an Agilent 6890/5973 GC/MS operating in negative chemical ionization (NCI) mode using ammonia as reagent gas (source pressure, 1.6 Torr; flow, 2 ml/min). Separation of the fatty acid-PFB esters was accomplished in a 61minute analysis using an AT-Silar-100 capillary column (30m x 0.25mm x 0.2 μm), part # 12632 Alltech Assoc. Inc. Each fatty acid is matched to the labeled internal standard of closest chain length, retention time, and concentration. Individual lipids are expressed as a percentage of the total lipid analyzed.
3. Results
Skin permeability
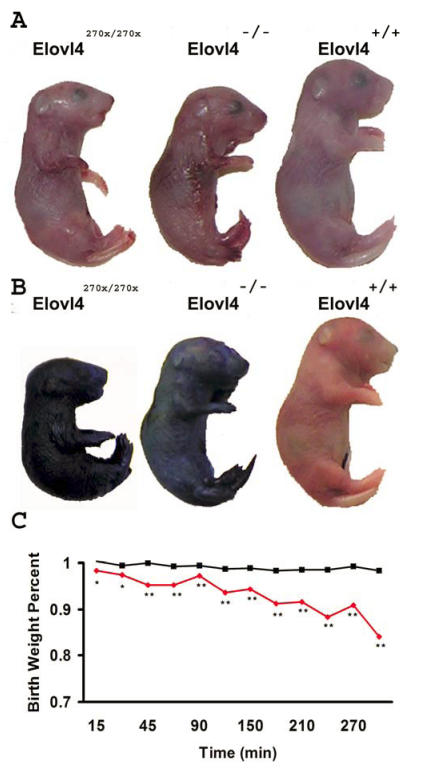
Toluidine blue normally does not penetrate skin with a fully developed stratum corneum 23. We examined mice at E19 when the stratum corneum has developed. Elovl4270x/270x and Elovl4-/- mice have a thin and shiny skin (Figure 4A) while wild type and heterozygous mice did not absorb the toluidine blue; Elovl4270x/270x and Elovl4-/- mice both absorbed the dye and were stained blue accordingly (Figure 4B). Mice lacking a functional copy of ELOVL4 are unable to develop a proper skin permeability barrier and are therefore susceptible to dehydration in a desiccating environment and infection.
Figure 4.
Permeability barrier failure at E19. (A) Elovl4270x/270x and Elovl4-/- mice have shiny semi-transparent skin compared to wild type pups. (B) Elovl4270x/270x and Elovl4-/- mice stain blue in a permeability assay demonstrating their lack of a functional permeability barrier. (C) Average birth weight for Elovl4270x/270x and Elovl4-/- pups (n=10) are shone in red and wild type with Elovl4+/270x and Elovl4+/- (n=33) are in black. Elovl4270x/270x and Elovl4-/- pups lose a significant amount of water compared to mice with a functional barrier. (one-tailed p-values *< 0.05 **<0.001)
Dehydration
Because Elovl4270x/270x and Elovl4-/- mice are perinatal lethal we removed the pups from pregnant females at E19. Each pup was weighed after being cleaned with a moistened kimmwipe. The pups were weighed every 15 minutes for the first hour and then every 30 minutes up to 5 hours (Figure 4C). Mice lacking a functional copy of Elovl4 lost about 3% of their body weights within the first half hour, a significant change compared to mice with at least one copy of Elovl4. By 5 hours after birth Elovl4270x/270x and Elovl4-/- mice lost 15% of their initial body weight while wild-type and heterozygous mice lost less than 2% of their body weight during this period. This loss in body weight is attributed to the evaporation of water through the skin.
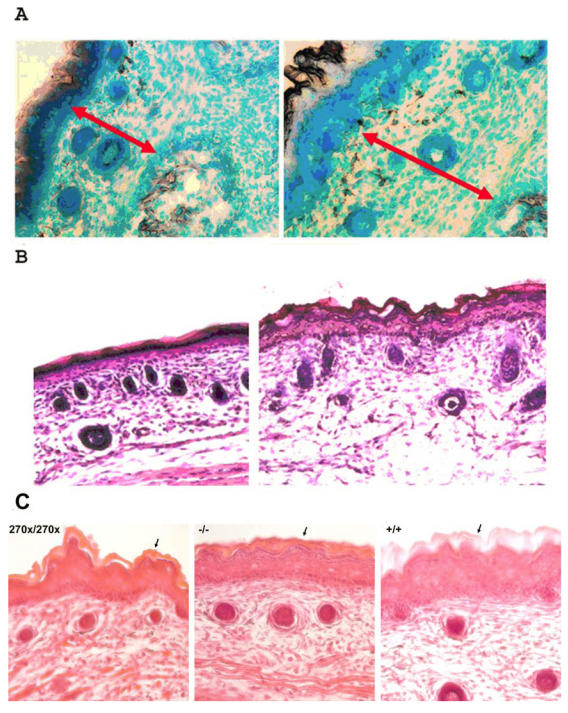
Skin Histology
Care was taken to collect sections from the torso and face at nearly identical locations in the mice. Images show the medial dorsal area of the sections from the torso (Figure 5A&B) and apical nasal sections (Figure 5C). No noticeable skin differences were seen in heterozygous animals compared to wild type, but mice lacking a functional copy of Elovl4 had several changes. Both Elovl4-/- and Elovl4270x/270x mice have dermis and epidermis layers that are about 70% of normal dermis and epidermis and appear to be compacted (Figure 5A). The outer stratum corneum of the epidermis from Elovl4-/- and Elovl4270x/270x is thinner and more fragile compared to normal mice (Figure 5B&C).
Figure 5.
Histological changes occur in E19 mouse skin. (A)Methyl green stain. (Left) Mice lacking a functional Elovl4 have a thin and fragile stratum corneum and have a much thinner epidermis and dermis. (Right) Wild type skin has a well developed, thick stratum corneum. Red arrows are drawn across the dermis to show the size difference more clearly. (B) H&E stain. Once again mice lacking a functional Elovl4 (left) have a very thin stratum corneum, epidermis and dermis. Hair cells appear to be normal. (C) H&E stain showing Elovl4270x/270x, Elovl4-/- and wild-type mouse skin. Arrows indicate the SC.
Lipid differences in Elovl4-/- and Elovl4270x/270x skin
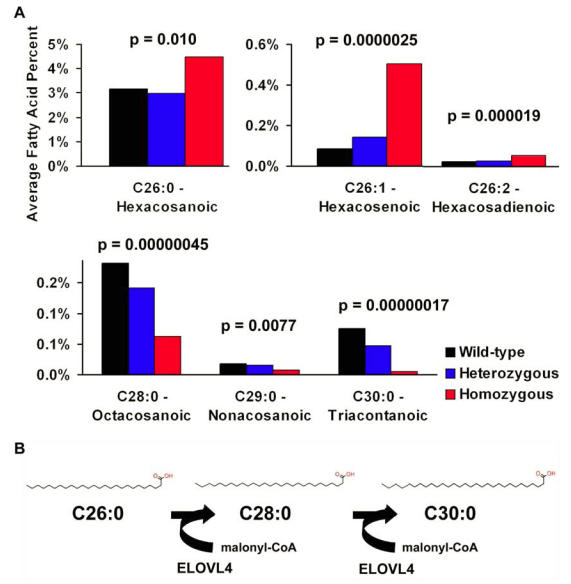
We compared skin lipid profiles from Elovl4+/+, Elovl4+/-, Elovl4 -/-, Elovl4 +/270x and Elovl4270x/270x mice (Tables 1 and 2). Subtle differences were noted in omega 3 and omega 6 lipids, but none were significant. Significant changes in lipid content were observed in VLCFA's. All 26 carbon fatty acids analyzed, C26:0, C26:1, and C26:2, were significantly elevated in mice lacking a functional copy of Elovl4 and correspondingly C28:0, C29:0, and C30:0 fatty acids were reduced (p<0.01). C30:0 was barely detectable in Elovl4 -/- mice and not detected at all in Elovl4270x/270x mice.
Table 1.
Fatty acids measured from E19 skin samples. Significant changes using linear regression on combined Elovl4 270x and knockout are in bold. Heterozygous and Homozygous represent combined values of both Elovl4 270x and knockout mice. (* decreasing and # increasing compared to wild-type)
| Elovl4+/+ | Elovl4+/270x | Elovl4270x/270x | Elovl4+/- | Elovl4-/- | Heterozygous | Homozygous | |
|---|---|---|---|---|---|---|---|
| C8:0 - Octanoic | 0.1326% | 0.4273% | 0.3018% | 0.0042% | 0.0044% | 0.2157% | 0.1531% |
| C9:0 - Nonanoic | 0.7218% | 2.4780% | 1.9750% | 0.0134% | 0.0175% | 1.2457% | 0.9963% |
| C10:0 - Capric | 0.2219% | 0.7687% | 0.6058% | 0.0033% | 0.0044% | 0.3860% | 0.3051% |
| C11:0 - Undecanoic | 0.0596% | 0.1698% | 0.1671% | 0.0051% | 0.0085% | 0.0875% | 0.0878% |
| C12:0 - Lauric | 0.5006% | 1.5230% | 1.3846% | 0.0533% | 0.0847% | 0.7881% | 0.7347% |
| C13:0 - Tridecanoic | 0.0552% | 0.1247% | 0.1050% | 0.0214% | 0.0268% | 0.0731% | 0.0659% |
| C14:0 - Myristic | 1.2141% | 1.2878% | 1.1034% | 1.1025% | 1.0868% | 1.1952% | 1.0951% |
| C15:0 - Pentadecanoic | 0.4841% | 0.4167% | 0.4651% | 0.4655% | 0.4073% | 0.4411% | 0.4362% |
| C16:0 - Palmitic | 19.8759% | 17.3620% | 14.4374% | 22.6083% | 21.9211% | 19.9851% | 18.1792% |
| C17:0 - Heptadecanoic | 0.4221% | 0.3611% | 0.3879% | 0.4419% | 0.4203% | 0.4015% | 0.4041% |
| C18:0 - Stearic | 10.9095% | 11.9813% | 10.3545% | 10.3991% | 9.1910% | 11.1902% | 9.7728% |
| C20:0 - Arachidic | 0.4324% | 0.4672% | 0.4800% | 0.3888% | 0.3732% | 0.4280% | 0.4266% |
| C21:0 - Heneicosanoic* | 0.0677% | 0.0584% | 0.0600% | 0.0598% | 0.0481% | 0.0591% | 0.0540% |
| C22:0 - Behenic | 1.3721% | 1.1230% | 1.2751% | 1.3003% | 1.1393% | 1.2116% | 1.2072% |
| C23:0 - Tricosanoic | 0.4071% | 0.2416% | 0.3257% | 0.4066% | 0.3112% | 0.3241% | 0.3185% |
| C24:0 - Lignoceric | 6.8596% | 4.6244% | 7.6147% | 7.2884% | 6.9943% | 5.9564% | 7.3045% |
| C25:0 - Pentacosanoic | 0.9133% | 0.4115% | 0.7714% | 1.2422% | 1.1816% | 0.8268% | 0.9765% |
| C26:0 - Hexacosanoic# | 3.1668% | 1.8104% | 4.1685% | 4.1543% | 4.8060% | 2.9823% | 4.4873% |
| C28:0 - Octacosanoic* | 0.1820% | 0.0903% | 0.0557% | 0.1929% | 0.0704% | 0.1416% | 0.0630% |
| C29:0 - Nonacosanoic* | 0.0180% | 0.0106% | 0.0086% | 0.0194% | 0.0055% | 0.0150% | 0.0070% |
| C30:0 - Triacontanoic* | 0.0754% | 0.0292% | 0.0000% | 0.0655% | 0.0105% | 0.0473% | 0.0052% |
| C10:1 - Caproleic | 0.0018% | 0.0106% | 0.0043% | 0.0000% | 0.0000% | 0.0053% | 0.0021% |
| C11:1 - Undecaenoic | 0.0694% | 0.2124% | 0.1801% | 0.0061% | 0.0075% | 0.1093% | 0.0938% |
| C12:1 - Dodecaenoic | 0.0059% | 0.0080% | 0.0129% | 0.0013% | 0.0028% | 0.0046% | 0.0078% |
| C13:1 - Tridecaenoic | 0.0027% | 0.0080% | 0.0086% | 0.0000% | 0.0000% | 0.0040% | 0.0043% |
| C15:1 - Pentadecaenoic | 0.0084% | 0.0292% | 0.0214% | 0.0010% | 0.0019% | 0.0151% | 0.0116% |
| C16:1(n-9) | 0.5706% | 0.5762% | 0.4370% | 0.5887% | 0.5609% | 0.5824% | 0.4989% |
| C17:1 - Heptadecaenoic | 0.5139% | 0.1779% | 0.2507% | 0.2972% | 0.7126% | 0.2376% | 0.4816% |
| C18:1(n-9) - Oleic | 16.0053% | 14.9932% | 15.6582% | 15.7484% | 14.9762% | 15.3708% | 15.3172% |
| C20:1(n-9) - Eicosenoic | 0.1894% | 0.2230% | 0.2400% | 0.1567% | 0.1667% | 0.1898% | 0.2034% |
Table 2.
Fatty acids measured from E19 skin samples continued. Significant changes using linear regression on combined Elovl4 270x and knockout are in bold. Heterozygous and Homozygous represent combined values of both Elovl4 270x and knockout mice. (# increasing compared to wild-type)
| Elovl4+/+ | Elovl4+/270x | Elovl4270x/270x | Elovl4+/- | Elovl4-/- | Heterozygous | Homozygous | |
|---|---|---|---|---|---|---|---|
| C20:3(n-9) - Mead | 0.1953% | 0.1671% | 0.1071% | 0.2297% | 0.2196% | 0.1984% | 0.1634% |
| C22:1(n-9) - Erucic | 0.1021% | 0.1009% | 0.1218% | 0.1522% | 0.1127% | 0.1266% | 0.1173% |
| C24:1(n-9) - Nervonic | 1.0265% | 1.0195% | 1.1293% | 1.0112% | 0.9972% | 1.0153% | 1.0632% |
| C25:1# | 0.0210% | 0.0106% | 0.0343% | 0.0457% | 0.0671% | 0.0282% | 0.0507% |
| C26:1 - Hexacosenoic# | 0.0835% | 0.0584% | 0.3708% | 0.2243% | 0.6395% | 0.1414% | 0.5051% |
| C14:1 - Myristoleic | 0.0454% | 0.0372% | 0.0364% | 0.0660% | 0.0649% | 0.0516% | 0.0507% |
| C16:1(n-7) - Palmitoleic | 3.5607% | 2.2245% | 2.4490% | 4.8816% | 4.5576% | 3.5531% | 3.5033% |
| C16:1(n-5) | 0.0050% | 0.0000% | 0.0000% | 0.0078% | 0.0104% | 0.0039% | 0.0052% |
| C18:1(n-7) - Vaccenic | 3.0881% | 2.9465% | 2.7320% | 3.1959% | 3.0467% | 3.0712% | 2.8893% |
| C18:1(n-5) | 0.0000% | 0.0000% | 0.0000% | 0.0000% | 0.0000% | 0.0000% | 0.0000% |
| C20:3(n-7) | 0.1179% | 0.1195% | 0.1328% | 0.1050% | 0.1024% | 0.1123% | 0.1176% |
| C14:2 - Myristolenic | 0.0096% | 0.0212% | 0.0193% | 0.0019% | 0.0028% | 0.0116% | 0.0110% |
| C16:2 - Palmitolenic | 0.1756% | 0.1087% | 0.2421% | 0.1873% | 0.2207% | 0.1480% | 0.2314% |
| C18:2(n-6) - Linoleic | 10.1454% | 10.6251% | 11.9408% | 8.9727% | 9.2647% | 9.7989% | 10.6028% |
| C18:2(N-6) - Conj - Rumenic | 0.1839% | 0.3613% | 0.3493% | 0.0268% | 0.0262% | 0.1941% | 0.1878% |
| C18:3(n-6) - Gamma Linolenic | 0.1628% | 0.1805% | 0.1371% | 0.1566% | 0.1618% | 0.1686% | 0.1495% |
| C20:2(n-6) - Eicosadienoic | 3.5252% | 0.5044% | 0.5250% | 6.7875% | 7.8270% | 3.6460% | 4.1760% |
| C20:3(n-6) - Dihomo-g-linolenic | 0.6143% | 0.7512% | 0.6150% | 0.5521% | 0.5801% | 0.6516% | 0.5976% |
| C20:4(n-6) - Arachidonic | 4.6884% | 9.4628% | 8.7692% | 0.3853% | 0.3928% | 4.9240% | 4.5810% |
| C22:2(n-6) - Docosadienoic | 0.2134% | 0.0717% | 0.0686% | 0.3878% | 0.4528% | 0.2297% | 0.2607% |
| C22:4(n-6) - Adrenic | 1.3861% | 1.6590% | 1.3005% | 1.4364% | 1.5560% | 1.5477% | 1.4283% |
| C22:5(n-6) - Docosapentaenoic | 0.2672% | 0.5785% | 0.4564% | 0.0533% | 0.0550% | 0.3159% | 0.2557% |
| C24:2 | 0.3384% | 0.3611% | 0.3792% | 0.3045% | 0.3049% | 0.3328% | 0.3421% |
| C26:2 - Hexacosadienoic# | 0.0249% | 0.0239% | 0.0600% | 0.0283% | 0.0484% | 0.0261% | 0.0542% |
| C18:3(n-3) - Alpha Linolenic | 0.2979% | 0.2867% | 0.2228% | 0.2503% | 0.2115% | 0.2685% | 0.2171% |
| C20:3(n-3) | 0.0953% | 0.1485% | 0.0257% | 0.0126% | 0.0160% | 0.0805% | 0.0208% |
| C20:5(n-3) - Eicosapentaenoic | 0.1046% | 0.1805% | 0.1521% | 0.1145% | 0.1460% | 0.1475% | 0.1491% |
| C22:3(n-3) - Docosatrienoic | 1.2824% | 0.0026% | 0.0043% | 0.0023% | 0.0028% | 0.0024% | 0.0036% |
| C22:5(n-3) | 0.6352% | 0.7829% | 0.6792% | 0.5535% | 0.6417% | 0.6682% | 0.6604% |
| C22:6(n-3) - Docosahexanoic | 1.5897% | 3.9786% | 2.8194% | 2.6529% | 3.3451% | 3.3158% | 3.0823% |
We also examined whether or not these fatty acids were dependent on Elovl4 expression. Using linear regression modeling we found that the 26 carbon fatty acids increased while C28:0, C29:0, and C30:0 decreased as Elovl4 expression decreased (Figure 6A). Without normal expression of Elovl4, C26 fatty acids build up and longer fatty acids decrease or almost disappear entirely. Taken together with the individual statistics, this data suggests that Elovl4 is responsible for elongating both saturated and unsaturated 26 carbon fatty acids. (Figure 6B)
Figure 6.
Fatty acid alterations suggest Elovl4 may be elongating C26 and C28 fatty acids. (A) Saturated and unsaturated C26 fatty acids in the skin are greatly increased while longer chain fatty acids are decreased in Elovl4270x/270x and Elovl4-/- mice. The longest measured C30:0 is almost non-existent. (B) ELOVL4 may function as an elongase for VLCFAs, beginning with C26 as diagrammed above.
4. Discussion
We have addressed the functional relevance of Elovl4 by utilizing two mouse models, Elovl4-/- and Elovl4270x/270x mice. Both models were perinatal lethal in the homozygous condition. Elovl4+/- mice develop no significant phenotype while Elovl4+/270x have yet to be studied by our group, however a 5bp deletion mutant was studied and found to develop a retinal degeneration-like phenotype with age.20, 21 By investigating the perinatal lethality we uncovered a possible fatty acid elongase activity for Elovl4 and found that Elovl4 is necessary for skin barrier development and mouse survival.
Mice develop a protective skin barrier during the last few days of embryonic development. The most crucial layer for this protection is the stratum corneum (SC), the uppermost layer of the epidermis. The SC filled with ordered stack of cornified envelops (proteins) and lipid bilayers. The lipid bilayers are made up of a 1:1:1 ratio of cholesterol, free fatty acids, and ceramides. Altering any one of these pools could result in a faulty barrier.
Our examinations show that free fatty acids longer than C26 are dramatically reduced in Elovl4-/- and Elovl4270x/270x mouse skin while the total fatty acid pool remains the same. A study of human atopic dermatitis, a skin disease resulting in impaired permeability barrier function, showed that free fatty acids ≥C26 are significantly reduced in affected patient's skin.24 Longer chain fatty acids are more hydrophobic and therefore have a greater ability to prevent water loss that shorter chain fatty acids. Missing nearly entire classes of VLCFAs could disrupt the highly ordered state of the SC and allow for water penetration.
Free VLCFAs are important for ceramide synthesis as well, so missing these fatty acids could affect more than one lipid bilayer component of the SC. In fact another group has shown that w-0-acylceramides are non-existent in mutant Elovl4 mice.22 Ceramides are made up of sphingosine and fatty acids, the former requiring free fatty acids itself. Altering the lipid chain length of two classes of the “mortar” making up the SC could be detrimental to the integrity of the permeability barrier.
By using both a knockout and a knockin model we are able to conclude that it is the lack of the Elovl4 protein that results in these lipid and skin barrier changes and not a gain of function by the mutant protein. In fact, just missing one functional copy of Elovl4 is sufficient to significantly reduce lipid levels, though not to a detrimental level. Mice with only one copy of Elovl4 are able survive normally. Our studies in conjunction with the previously reported Elovl4 5bp deletion study demonstrate that Elovl4 likely functions in elongating C26 to C28 and possibly beyond. Unsaturated and odd chain fatty acids may also use Elovl4 for similar chain lengths such as C26:1-C28:1 or C27-C29.
The lack of these VLCFAs results in a skin barrier defect developing into an atopic dermatitis- like phenotype. Dehydration assays show that Elovl4270x/270x and Elovl4-/- mice lose about 3% of their initial body weight an hour compared to less than 0.3% an hour lost by wild type and heterozygous mice. Nearly all of the Elovl4270x/270x and Elovl4-/- mice died within 2 hours of birth and all died within 4 hours of birth. Altering the temperature at which pups were kept did not appear to affect this rate of water loss. (Data not shown)
Elovl4 is expressed in other areas of the body. The brain in particular has high Elovl4 expression and is also a site where loss of a gene would be cause for concern. We examined the brain, specifically the hippocampus and cerebellum where Elovl4 expression is highest, and found no histological abnormalities. We also examined the brain for fatty acid differences in Elovl4-/- mice and found no significant changes. Other ELO family proteins are expressed in the brain and could share redundant function with Elovl4 suggesting a possible reason for the lack of fatty acid changes in Elovl4-/- mice.22 Histological and a biochemical alterations found in the skin seem to be the main cause of perinatal lethality in both Elovl4270x/270x and Elovl4-/- mice.
In conclusion, Elovl4 appears to act as a fatty acid elongase responsible for the generation of fatty acids of chain length greater than C26. This elongase activity is especially important in the skin, where free fatty acids and ceramides are an integral part of the SC, the skins permeability barrier. Removing the longest VLCFAs results in a smaller, weaker SC that loses its ability to prevent water loss in a desiccating environment. Elovl4270x/270x and Elovl4-/- mice consequently dehydrate very quickly and die as a result. In an accompanying paper 25, Li et al report the depletion of certain VLCFA containing ceramides in Elovl4 deficient mice that also have impaired SC barrier function and are neonatal lethal. Together these findings demonstrate Elovl4 is essential for lipid biosynthesis, skin permeability barrier function, and neonatal survival.
Acknowledgments
This research was supported by grants from Knights Templar Eye Research Foundation (ZY), and the following grants (to KZ): NIH (R01EY14428, R01EY14448); the Ruth and Milton Steinbach Fund, Ronald McDonald House Charities, the Macular Vision Research Foundation. We would like to thank Ann Moser and the Peroxisomal Diseases Laboratory at the Kennedy Krieger Institute, Johns Hopkins University, Baltimore MD for fatty acid measurements.
References
- 1.Donoso LA, Frost AT, Stone EM, Weleber RG, MacDonald IM, Hageman GS, Cibis GW, Ritter R 3rd, Edwards AO. Autosomal dominant Stargardt-like macular dystrophy: founder effect and reassessment of genetic heterogeneity. Arch Ophthalmol. 2001;119(4):564–570. doi: 10.1001/archopht.119.4.564. [DOI] [PubMed] [Google Scholar]
- 2.Stone EM, Nichols BE, Kimura AE, Weingeist TA, Drack A, Sheffield VC. Clinical features of a Stargardt-like dominant progressive macular dystrophy with genetic linkage to chromosome 6q. Arch Ophthalmol. 1994;112(6):765–772. doi: 10.1001/archopht.1994.01090180063036. [DOI] [PubMed] [Google Scholar]
- 3.Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272(28):17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 4.Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27(1):89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- 5.Mandal MN, Ambasudhan R, Wong PW, Gage PJ, Sieving PA, Ayyagari R. Characterization of mouse orthologue of ELOVL4: genomic organization and spatial and temporal expression. Genomics. 2004;83(4):626–635. doi: 10.1016/j.ygeno.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Zhang XM, Yang Z, Karan G, Hashimoto T, Baehr W, Yang XJ, Zhang K. Elovl4 mRNA distribution in the developing mouse retina and phylogenetic conservation of Elovl4 genes. Mol Vis. 2003;9:301–307. [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards AO, Donoso LA, Ritter R 3rd. A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest Ophthalmol Vis Sci. 2001;42(11):2652–2663. [PubMed] [Google Scholar]
- 8.Cinti DL, Cook L, Nagi MN, Suneja SK. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog Lipid Res. 1992;31(1):1–51. doi: 10.1016/0163-7827(92)90014-a. [DOI] [PubMed] [Google Scholar]
- 9.Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol. 2000;149(3):707–718. doi: 10.1083/jcb.149.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein PS, Tammur J, Singh N, Hutchinson A, Dixon M, Pappas CM, Zabriskie NA, Zhang K, Petrukhin K, Leppert M et al. Diverse macular dystrophy phenotype caused by a novel complex mutation in the ELOVL4 gene. Invest Ophthalmol Vis Sci. 2001;42(13):3331–3336. [PubMed] [Google Scholar]
- 11.Maugeri A, Meire F, Hoyng CB, Vink C, Van Regemorter N, Karan G, Yang Z, Cremers FP, Zhang K. A novel mutation in the ELOVL4 gene causes autosomal dominant Stargardt-like macular dystrophy. Invest Ophthalmol Vis Sci. 2004;45(12):4263–4267. doi: 10.1167/iovs.04-0078. [DOI] [PubMed] [Google Scholar]
- 12.Conley YP, Jakobsdottir J, Weeks DE, Mah T, Ferrell RE, Gorin MB. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005;14(14):1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- 13.Karan G, Yang Z, Zhang K. Expression of wild type and mutant ELOVL4 in cell culture: subcellular localization and cell viability. Mol Vis. 2004;10:248–253. [PubMed] [Google Scholar]
- 14.Ambasudhan R, Wang X, Jablonski MM, Thompson DA, Lagali PS, Wong PW, Sieving PA, Ayyagari R. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004;83(4):615–625. doi: 10.1016/j.ygeno.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Karan G, Yang Z, Howes K, Zhao Y, Chen Y, Cameron DJ, Lin Y, Pearson E, Zhang K. Loss of ER retention and sequestration of the wild-type ELOVL4 by Stargardt disease dominant negative mutants. Mol Vis. 2005;11:657–664. [PubMed] [Google Scholar]
- 16.Vasireddy V, Wang XF, Huang J, Vijaysarathy C, Jablonski M, Petty HR, Sieving PA, Ayyagari R. 5-Bp-Deletion Mutant ELOVL4 Protein Causes Altered Localization of the Wild Type ELOVL4 Protein in COS-7 Cells- Probable Mechanism Underlying Stargardt's-Like Macular Degeneration. Invest Ophthalmol Vis Sci. 2005;46(5):1643. [Google Scholar]
- 17.Grayson C, Molday RS. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J Biol Chem. 2005;280(37):32521–32530. doi: 10.1074/jbc.M503411200. [DOI] [PubMed] [Google Scholar]
- 18.Karan G, Lillo C, Yang Z, Cameron DJ, Locke KG, Zhao Y, Thirumalaichary S, Li C, Birch DG, Vollmer-Snarr HR et al. Lipofuscin accumulation, abnormal electrophysiology, and photoreceptor degeneration in mutant ELOVL4 transgenic mice: A model for macular degeneration. Proc Natl Acad Sci U S A. 2005;102(11):4164–4169. doi: 10.1073/pnas.0407698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raz-Prag D, Ayyagari R, Fariss RN, Mandal MN, Vasireddy V, Majchrzak S, Webber AL, Bush RA, Salem N Jr, Petrukhin K et al. Haploinsufficiency is not the key mechanism of pathogenesis in a heterozygous Elovl4 knockout mouse model of STGD3 disease. Invest Ophthalmol Vis Sci. 2006;47(8):3603–3611. doi: 10.1167/iovs.05-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasireddy V, Jablonski MM, Mandal MN, Raz-Prag D, Wang XF, Nizol L, Iannaccone A, Musch DC, Bush RA, Salem N Jr et al. Elovl4 5-bp-deletion knock-in mice develop progressive photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2006;47(10):4558–4568. doi: 10.1167/iovs.06-0353. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Chen Y, Cameron DJ, Wang C, Karan G, Yang Z, Zhao Y, Pearson E, Chen H, Deng C et al. Elovl4 haploinsufficiency does not induce early onset retinal degeneration in mice. Vision Res. 2007 doi: 10.1016/j.visres.2006.10.023. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasireddy V, Uchida Y, Salem N, Kim SY, Mandal MN, Reddy GB, Bodepudi R, Alderson NL, Brown JC, Hama H et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (>=C28) and the unique {omega}-O-acylceramides in skin leading to neonatal death. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddl480. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125(8):1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- 24.Macheleidt O, Kaiser HW, Sandhoff K. Deficiency of epidermal protein-bound omega-hydroxyceramides in atopic dermatitis. J Invest Dermatol. 2002;119(1):166–173. doi: 10.1046/j.1523-1747.2002.01833.x. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Sandhoff R, Kono M, Zerfas P, Hoffmann V, Ding BCH, Proia RL, Deng CX. Depletion of ceramides with very long-chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int J Biol Sci. 2007;3:120–128. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]