Abstract
The Danube River is the second longest river in Europe, and its bacterial community composition has never been studied before over its entire length. In this study, bacterial community composition was determined by denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified portions of the bacterial 16S rRNA gene from a total of 98 stations on the Danube River (73 stations) and its major tributaries (25 stations), covering a distance of 2,581 km. Shifts in the bacterial community composition were related to changes in environmental conditions found by comparison with physicochemical parameters (e.g., temperature and concentration of nutrients) and the concentration of chlorophyll a (Chl a). In total, 43 distinct DGGE bands were detected. Sequencing of selected bands revealed that the phylotypes were associated with typical freshwater bacteria. Apparent bacterial richness in the Danube varied between 18 and 32 bands and correlated positively with the concentration of P-PO4 (r = 0.56) and negatively with Chl a (r = −0.52). An artificial neural network-based model explained 90% of the variation of apparent bacterial richness using the concentrations of N-NO2 and P-PO4 and the distance to the Black Sea as input parameters. Between the cities of Budapest and Belgrade, apparent bacterial richness was significantly lower than that of other regions of the river, and Chl a showed a pronounced peak. Generally, the bacterial community composition developed gradually; however, an abrupt and clear shift was detected in the section of the phytoplankton bloom. Large impoundments did not have a discernible effect on the bacterial community of the water column. In conclusion, the riverine bacterial community was largely influenced by intrinsic factors.
Prokaryotes play a major role in the biotic transformation of carbon and nutrients in aquatic ecosystems. Consequently, these organisms may alter the makeup of carbon and other chemicals during their transport in rivers. The sources, transport, and transformation of organic matter in rivers have been formalized conceptually (39, 40) and studied in situ (15). There is evidence that pelagic prokaryotic communities in rivers adapt to changes in the concentration and composition of organic carbon (20, 21) and nutrients (e.g., 3-5). Sekiguchi et al. (37) determined the bacterial community composition of the Changjiang River (China) as a reference for future changes caused by the construction of the Three Gorges dam. Their results showed that changes in the bacterial community (determined by denaturing gradient gel electrophoresis [DGGE] analysis and cloning) occurred gradually and that diversity decreased toward the river's delta near Shanghai.
In this study, we focus on the bacterial community composition of the Danube River over its entire length in order to determine the effects of human influences (cities and impoundments) versus those of large tributaries and changes in the geomorphology in different sections of the river. The Danube River is the second longest river in Europe, traveling between its sources in the Black Forest (Germany) and its delta in the Black Sea, a distance of 2,857 km. The Danube's catchment area is shared by 18 countries, covering 817 × 106 m2, and is home to roughly 83 × 106 people. We sampled the bacterial community composition of the Danube River and its major tributaries and side arms by DGGE analysis of PCR-amplified portions of the bacterial 16S rRNA gene at a total of 98 stations. Changes in apparent bacterial richness were related to physicochemical parameters (temperature and concentration of nutrients) and the concentration of chlorophyll a (Chl a). The data set was used to develop a mathematical model of apparent bacterial richness in the Danube River using artificial neural networks (ANNs). ANNs are best described as a data-driven modeling tool especially suited for complex real-world modeling tasks. The major advantages of ANNs are that the data do not need to fit a predefined model (e.g., the normal distribution), the linear and nonlinear relationships can be modeled simultaneously, and they are tolerant to noisy data due to their high parallelism. Previous studies demonstrate the usefulness of ANNs in microbial ecology (10, 32, 36, 42), and a detailed introduction to ANNs is given by Basheer and Hajmeer (2).
MATERIALS AND METHODS
Study sites.
During the Joint Danube Survey (13 August to 19 September 2001), a total of 98 sampling stations were occupied using the research vessel MLS Argus. Seventy-three stations were located on the Danube River, and 25 stations were located on its major tributaries and side arms from 0.1 to 1.4 km upstream of their confluences with the Danube River (Fig. 1) (a “keyhole markup language” [klm] file for use with “Google Earth” software provides exact positions of the sampling stations in the supplemental material). During the study period, the Danube River exhibited a typical summer situation with below-annual-average flow rates, and the flow rates in the tributaries varied between low and annual averages. Thus, the study area was characterized by relatively stable weather conditions without large rainfalls. The survey covered a total distance of 2,581 km, from Neu-Ulm (Germany) to the Danube's delta in the Black Sea. Water temperature (in °C), pH, and the dissolved oxygen saturation (DO%) were determined using submersible electrodes (WTW, Weilheim, Germany) placed at the same depths from which samples of nutrients and Chl a were taken (see below).
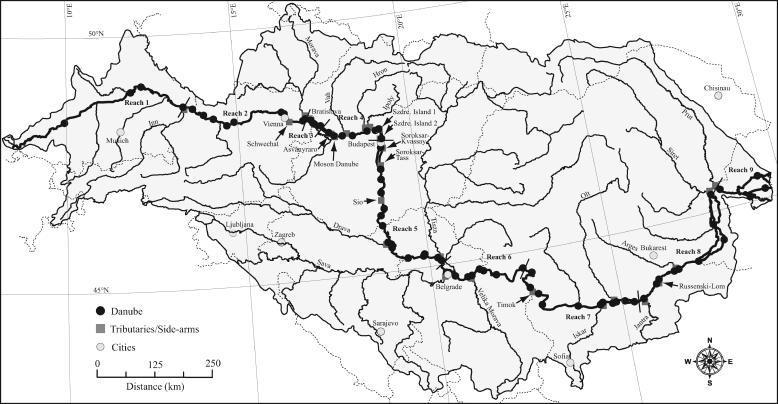
FIG. 1.
Geographic map of sampling stations occupied during this study. The shaded region represents the catchment area of the Danube River. The boundaries between the nine geomorphological reaches are indicated by bars crossing the Danube River.
Within the framework of the Joint Danube Survey, the longitudinal course of the Danube River was divided into nine geomorphological reaches (23). In order to aid data analysis, we have chosen to adopt this concept for the present study (Fig. 1). The definition of the reaches is governed largely by the geomorphological features of the river basin and anthropogenic influences such as damming. Reach 1 stretches between Neu-Ulm (Germany) and the Danube's confluence with the Inn River. Reach 2 begins at the confluence with the Inn River and stretches to the confluence with the Morava River. The Inn River contributes large amounts of water and sedimentary material of glacial origin, substantially reducing the Danube's water transparency. The Danube River within reaches 1 to 2 represents a typical alpine river. Reach 3 encompasses the Gabcikovo reservoir, caused by the construction of the Gabcikovo hydroelectric power plant. Increased sedimentation caused by the reduced flow velocities in the reservoir increases transparency. In Reach 4, from the Gabcikovo dam to just upstream of Budapest (Hungary), the Danube River starts to develop into a lowland river with reduced flow velocities and a widened horizontal profile. Reach 5 starts at Budapest and ends at the confluence with the Sava River, upstream of Belgrade (Serbia). In this reach, the lowland character of the river strengthens further. Reach 6 stretches between the confluence with the Sava River and the last of the Iron Gate dams. Within this reach, two large reservoirs directly follow each other and together constitute the biggest man-made impoundment of the Danube River. The stretch between the Iron Gate dam and the Danube's confluence with the Jantra River in Bulgaria constitutes reach 7. No further impoundments hinder the free flow of the Danube after the Iron Gate dam, and water column transparency is high. Reach 8 stretches between the Danube's confluence with the Jantra and the Prut. Within this reach, the Danube splits twice into two arms generating large islands, and water column transparency decreases. In reach 9, the Danube's delta into the Black Sea, the river splits into three major arms.
Determination of the concentration of nutrients and Chl a.
Samples for the determination of the concentrations of ammonium (N-NH4), nitrate (N-NO3), nitrite (N-NO2), phosphate (P-PO4), and Chl a were taken from the upper third of the water column in the navigation channel. The depth of the water column at the sampling locations varied between 0.5 and 15 m, and the water column was well mixed. Thus, the variation of these parameters over depth was negligible, and the results can be compared with apparent bacterial richness (see below). Samples were filtered through glass fiber filters (GF/F; Whatman) prior to analysis. Analysis was performed aboard ship immediately after sample collection, using spectrophotometry to determine the concentrations of N-NH4 according to ISO7150-1 (16) and N-NO3 as described by ISO7890-3 (17); ion chromatography was used to determine the concentration of N-NO2 (13), and the ascorbic acid method was used to determine the concentration of P-PO4 (13). Samples (0.5 to 2 liters) for the determination of Chl a concentration were filtered through glass fiber filters (GF/F; Whatman). Extraction, spectrometric determination of Chl a, and correction for phaeopigments were performed as described in ISO10260 (18).
Determination of bacterial community composition. (i) Sample collection.
Water samples (150 ml) were collected in sterilized glass bottles from a depth of 0.2 to 0.3 m in the navigation channel. Samples were filtered through 0.22-μm-pore-sized filters (47-mm diameter; Durapore GVWP; Millipore) to collect prokaryotic cells representing the total in situ prokaryotic community, within 0.5 h of sampling. The filters were stored frozen at −20°C until further analysis.
(ii) Nucleic acid extraction.
Extraction of the nucleic acids from the filters was performed using an UltraClean soil DNA kit (catalog no. 12800-100; MO BIO Laboratories). The filters were cut into small pieces and transferred into the extraction tubes using ethanol-flamed forceps and surgical scissors. The extraction procedure was performed as described by the manufacturer's instructions. The alternative lysis method for the extraction kit was employed, involving two heating steps to 70°C for 5 min in order to minimize shear damage to the nucleic acids. Nucleic acid extracts had a final volume of 50 μl in solution S5 (the solution contains no EDTA [the exact contents are the proprietary information of MO BIO Laboratories]) and were used directly as templates in PCRs.
(iii) PCR amplification.
A 586-bp-long fragment (from Escherichia coli, numbering positions 341 to 927) of the bacterial 16S rRNA gene was amplified from the nucleic acid extracts by PCR using the primer pair 341F (5′-CCT ACG GGA GGC AGC AG-3′ [31]) and 907R (5′-CCG TCA ATT CMT TTG AGT TT-3′ [35]). A 40-bp-long GC clamp (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′ [30]) was attached to the 5′ end of primer 341F to obtain PCR fragments suitable for DGGE analysis. The resulting PCR fragments had a length of 626 bp due to the GC clamp. We used 5 μl of the nucleic acid extracts as templates in PCRs. Each 50-μl PCR mixture contained 5 μl of 10× Taq buffer (100 mM Tris-HCl [pH 8.8], 500 mM KCl, 0.8% Nonidet P-40 [MBI Fermentas]), 4 μl of 25 mM MgCl2 (2 μM final concentration; MBI Fermentas), 6.25 μl of a 2 mM deoxynucleoside triphosphate mix (final concentration, 250 μM each; catalog no. R0241; MBI Fermentas), 0.5 μl of 100 μM primers 341F and 907R (final concentration, 1 μM; MWG-Biotech AG), and 0.25 μl of 5 U μl−1 Taq polymerase (catalog no. EP0401; MBI Fermentas). Cycling was performed in an iCycler thermal cycler (Bio-Rad) with an initial denaturation at 95°C for 1 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, and elongation at 72°C for 1 min. The final elongation step was performed at 72°C for 30 min in order to prevent the formation of artificial double bands in subsequent DGGE analyses (19). PCR fragments were cleaned and concentrated using a QIAquick PCR purification kit (QIAGEN) according to the manufacturer's instructions, resulting in a final volume of 28 μl in elution buffer (QIAGEN). Standard agarose gel electrophoresis was used to size and quantify the PCR fragments.
(iv) DGGE analysis.
DGGE analysis was performed on a DCode universal mutation detection system (Bio-Rad). The PCR products obtained from a single PCR of each sample were loaded on 6% polyacrylamide gels containing linear gradients of formamide and urea of 20 to 70%. Electrophoresis was performed at 100 V and 60°C for 16 h in 1× TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA [ethylenediaminetetraacetic acid], pH 8.3). The gels were stained with SYBR green I (1:10,000 dilution of stock solution; Molecular Probes) for 30 min before digitized gel images were obtained using a GelDOC 2000 (Bio-Rad) gel documentation system equipped with a charge-coupled-device camera.
(v) Analysis of DGGE banding patterns.
The images were analyzed for the number of bands per sample (presence versus absence), which served as a measure of apparent bacterial richness. Additionally, the results were transformed into a data matrix representing the presence and absence of bands by ones and zeros, respectively. The matrix was used to calculate Jaccard's dissimilarity index relative to the first sampled station in reach 1 according to the formula dissimilarity = 1 − (M/[M + U]), where M represents the number of bands present in both samples and U the total number of unique bands present in either one or the other of the compared samples. Jaccard's dissimilarity index ranges from 0 (identity) to 1 (no similarity). Detection frequencies of DGGE bands were calculated for each geomorphological reach by dividing the number of stations where a particular band was detected by the total number of stations in the reach. In order to identify prominent bands in the reaches, the detection frequencies for the bands were transformed according to the formula 1 = 0.5 < frequency ≤ 0.5 = 0. This matrix of prominent bands was used to visually display the relatedness of the reaches in a dendrogram based on Jaccard's dissimilarity index. The dendrogram was constructed with the program “neighbor” from the PHYLIP software package (9) using the unweighted pair group method with arithmetic averages.
Sequencing and phylogenetic analysis of selected DGGE bands.
In total, 78 bands from 33 different stations were excised from the DGGE gels. The gel slices were incubated in 200 μl of autoclaved Milli-Q water for 24 h at 4°C to extract the PCR fragments. Extracted PCR fragments were reamplified and separated by DGGE under identical conditions, as described above, using the original samples as standards. Reamplified PCR products that were correctly placed on the DGGE gels compared to the original samples were sequenced using primer 907R (35) by a commercial sequencing service (MWG Biotech, Germany). As a quality check, the nucleotide sequences were aligned with “ARB” (25) software against their closest relatives retrieved from GenBank by BLASTN searching (1). Taxonomic classification of the sequences was obtained by the Bayesian rRNA classification algorithm of the Ribosomal Database Project II, release 9.35 (6).
Modeling apparent bacterial richness using ANNs.
We used Mathematica 5.2 and the ANN application package (both from Wolfram Research) to implement feed-forward ANNs with one layer of hidden neurons and one output neuron. A bias term with a fixed value of one was included in the input and in the hidden layer. Prior to training, the data were scaled to a mean of zero and unity variance, and the network weights were initialized using the “LinearParameters” option in order to randomize the initial values of the nonlinear parameters within the range of the input data and to completely randomize the linear parameters. ANNs were trained by employing the Levenberg-Marquardt algorithm (29), and the progress of training was monitored using the root mean square error (RMSE) of the networks.
The data set used for modeling apparent bacterial richness consisted of the 72 stations along the Danube River (we were unable to obtain suitable PCR products for DGGE analysis from one of the stations), and the following nine input parameters were considered: the distance to the Black Sea (km), water temperature (°C), pH, and DO (%); N-NH4, N-NO3, N-NO2, and P-PO4 (all in mg liter−1); and Chl a (μg liter−1). Initially, the input parameters were used alone and in combination with up to two other parameters to train ANNs with one to five hidden neurons for 100 iterations. The initial values for the weights of the networks can have a profound influence on the outcome of training (14). Thus, 100 ANNs were initialized and trained as described for each set of input parameters and numbers of hidden neurons, respectively. In order to prevent overtraining (i.e., data being memorized by the ANN), we performed cross-validation (14). The data set was split into a training data set (80% of the data) and a validation data set (20% of the data). The training data set was used for adjusting the network parameters, and the validation data set was used only to validate the network at each iteration during training without interfering in parameter adjustment. Training was assumed to have converged when the sum of the RMSE of the training and the validation data sets reached a minimum. Finally, the best-performing ANNs for each set of input parameters and number of hidden neurons were determined by the smallest sum of the RMSE of the training and the validation data sets at the convergence of training. Comparisons between the results allowed us to identify suitable combinations of input parameters for modeling apparent bacterial richness.
These initial results indicated that the number of hidden neurons could still be increased to further improve network performance without overfitting the data, i.e., the combined RMSE of the training and the validation data sets had not yet reached a minimum when one to five hidden neurons were used. Thus, we increased the number of hidden neurons stepwise to a maximum of 11. For each set of input parameters and number of hidden neurons (6 to 11 hidden neurons), 1,000 ANNs were initialized, trained, and screened as described above. The best-performing set of input parameters and network structure were found by searching for the minimum of the combined RMSE of the training and the validation data sets between the sets of input parameters and numbers of hidden neurons.
Statistical analyses.
All statistical data analyses were performed with Mathematica 5.2 (Wolfram Research). Spearman rank correlation coefficients were used to determine the degree of correlation between parameters, and P values were corrected according to the Bonferroni method (where P was ≤0.05 per the number of parameters). Linear least-squares regression analysis was used to relate the observed values of apparent bacterial richness to the predictions of the ANN-based model. Differences in the slopes of linear regression analyses against each other or against a hypothetical slope of 1 were tested by calculating a t value according to the formula t = (|byx − Byx|)/(|Sbyx − SByx|), where byx and Byx represent the slopes of the regressions being compared, and Sbyx and SByx are the standard deviations of the slopes of the regressions. The P values are reported for the two-tailed t distribution. Analysis of variance (ANOVA) in combination with Scheffe's F multiple-comparison test was used to examine differences among the parameters of the geomorphological reaches. Student's unpaired t test was used to test differences in parameters between two sections of the river.
Nucleotide sequence accession numbers.
In total, 16 distinct, partial bacterial 16S rRNA gene sequences from this study were obtained and deposited in GenBank under the accession numbers DQ371837 to DQ371848 and DQ371850 to DQ371853.
RESULTS
Comparison among the Danube River, its tributaries, and side arms. (i) Physicochemical parameters.
In the Danube River, the temperature varied between 15.9 and 25.5°C, with the lowest and highest average values found in reaches 2 and 6, respectively (Table 1). A similar variation in temperature, from 15.5 to 26.7°C, was recorded in the tributaries and side arms (Table 2). The variation in pH in the Danube River, from 7.5 to 8.7, was comparable to the variation found in the tributaries and side arms, ranging from 7.3 to 8.6 (Table 2). The lowest average pH was recorded in reach 6, and the highest average values were found in reaches 1 and 5 (Table 1). DO ranged from 53 to 192% in the river, whereas in the tributaries and side arms, the variation was between 35 and 240% (Table 2). The average DO was lowest in reach 6 and highest in reach 5 (Table 1). Although DO was not measured at the same time of day, diel variations were negligible (data not shown) compared to the changes between reaches.
TABLE 1.
Parameters measured in the Danube Rivera
| Reach (n) | Temp (°C)
|
pH
|
DO (%)
|
N-NH4 (μg liter−1)
|
N-NO3 (mg liter−1)
|
N-NO2 (μg liter−1)
|
P-PO4 (μg liter−1)
|
Chl a (μg liter−1)
|
Bacterial richness (no. of bands)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | Avg | SD | |
| 1 (4) | 19.3 | 2.6 | 8.1 | 0.3 | 118 | 17 | 62 | 36 | 2.2 | 0.3 | 10 | 3 | 30 | 25 | 28.9 | 16.4 | 25.3 | 2.6 |
| 2 (9) | 18.8 | 0.8 | 8.0 | 0.1 | 102 | 6 | 19 | 11 | 1.3 | 0.1 | 11 | 3 | 26 | 22 | 10.0 | 2.8 | 26.0 | 1.6 |
| 3 (4) | 20.0 | 0.2 | 7.9 | 0.0 | 97 | 3 | 18 | 10 | 1.4 | 0.0 | 9 | 2 | 39 | 5 | 16.7 | 3.4 | 29.0 | 1.2 |
| 4 (8) | 21.3 | 0.5 | 8.0 | 0.1 | 104 | 8 | 16 | 9 | 1.3 | 0.1 | 7 | 2 | 38 | 6 | 23.8 | 10.1 | 24.7b | 2.1b |
| 5 (15) | 22.5 | 0.4 | 8.1 | 0.3 | 126 | 35 | 61 | 56 | 0.8 | 0.2 | 11 | 4 | 21 | 18 | 71.3 | 42.5 | 21.7 | 2.0 |
| 6 (9) | 23.2 | 0.9 | 7.5 | 0.1 | 67 | 12 | 164 | 34 | 0.8 | 0.1 | 23 | 9 | 60 | 8 | 6.7 | 4.4 | 28.2 | 2.3 |
| 7 (11) | 21.8 | 0.7 | 7.8 | 0.2 | 87 | 12 | 20 | 15 | 1.1 | 0.1 | 35 | 17 | 84 | 9 | 6.7 | 3.2 | 27.5 | 2.3 |
| 8 (9) | 20.3 | 0.4 | 7.8 | 0.1 | 85 | 11 | 26 | 18 | 1.1 | 0.1 | 14 | 5 | 93 | 27 | 12.6 | 5.2 | 29.6 | 2.4 |
| 9 (4) | 20.5 | 0.2 | 7.8 | 0.0 | 80 | 7 | 53 | 21 | 1.0 | 0.1 | 20 | 13 | 74 | 13 | 14.0 | 3.0 | 23.8 | 1.9 |
The table shows the number of samples (n), average values (Avg), and standard deviations (SD) for the parameters measured in the Danube River, calculated for each of the nine geomorphological reaches.
The number of samples for bacterial richness is seven (we failed to achieve a useable PCR product from one sample).
TABLE 2.
Parameters measured in the tributaries and side arms of the Danube Rivera
| Inflow/tributary | Reach | Label | Temp (°C) | pH | DO (%) | N-NH4 (μg liter−1) | N-NO3 (mg liter−1) | N-NO2 (μg liter−1) | P-PO4 (μg liter−1) |
|---|---|---|---|---|---|---|---|---|---|
| Inn | 2 | 1 | 15.5 | 7.9 | 106 | 20 | 0.5 | 15 | 35 |
| Schwechat | 2 | 2 | 23.6 | 8.6 | 125 | 40 | 1.9 | 27 | 130 |
| Morava | 2 | 3 | 23.2 | 8.3 | 100 | 10 | 1.3 | 6 | 140 |
| Asvanyraroc | 4 | 4 | 20.9 | 8.0 | 110 | 10 | 1.3 | 6 | 29 |
| Moson Danubec | 4 | 5 | 23.5 | 7.3 | 35 | 330 | 1.1 | 45 | 159 |
| Vah | 4 | 6 | 23.5 | 7.7 | 83 | 130 | 1.1 | 21 | 58 |
| Hron | 4 | 7 | 22.7 | 8.4 | 104 | 30 | 1.5 | 4 | 149 |
| Ipoly | 4 | 8 | 22.0 | 8.4 | 70 | 10 | 1.4 | 15 | 179 |
| Szentendre Island 1c | 4 | 9 | 21.7 | 8.1 | 113 | 20 | 1.3 | 6 | 55 |
| Szentendre Island 2c | 4 | 10 | 22.1 | 8.2 | 129 | 20 | 1.2 | 6 | 32 |
| Soroksar-Kvassayc | 5 | 11 | 22.9 | 8.5 | 146 | 20 | 0.9 | 12 | 16 |
| Soroksar-Tassc | 5 | 12 | 25.1 | 7.9 | 122 | 140 | 0.3 | 18 | 32 |
| Sio | 5 | 13 | 26.7 | 8.8 | 240 | 20 | 2.5 | 69 | 228 |
| Drava | 5 | 14 | 21.8 | 8.3 | 119 | 10 | 0.3 | 6 | 6 |
| Tisza | 5 | 15 | 24.4 | 7.4 | 59 | 230 | 0.5 | 21 | 65 |
| Sava | 6 | 16 | 24.7 | 7.8 | 81 | 70 | 0.6 | 6 | 98 |
| Velika Morava | 6 | 17 | 24.0 | 8.3 | 125 | 200 | 0.4 | 18 | 45 |
| Timok | 7 | 18 | 22.3 | NDb | 99 | 80 | 1.3 | 3 | 16 |
| Iskar | 7 | 19 | 21.8 | ND | 136 | 20 | 3.3 | 15 | 848 |
| Olt | 7 | 20 | 25.0 | ND | 91 | 30 | 0.3 | 30 | 94 |
| Jantra | 8 | 21 | 20.9 | 8.0 | 87 | 100 | 1.1 | 30 | 196 |
| Russenski-Lom | 8 | 22 | 18.1 | ND | 54 | 890 | 3.8 | 96 | 297 |
| Arges | 8 | 23 | 19.0 | ND | ND | 3240 | 0.1 | 36 | 238 |
| Siret | 8 | 24 | 18.1 | 7.7 | 63 | 100 | 1.1 | 39 | 65 |
| Prut | 8 | 25 | 19.6 | 7.8 | 83 | 50 | 1.1 | 21 | 68 |
Data indicate the geomorphological reach of confluence with the Danube River. Label numbers correspond to those shown in Fig. 2.
ND, not determined.
Oxbow lake or side arm.
N-NH4 varied between 10 and 190 μg liter−1 in the river and between 10 and 3,240 μg liter−1 in the tributaries and side arms (Table 2). The lowest and highest average concentrations of N-NH4 were found in reaches 4 and 6, respectively (Table 1). N-NO3 concentration varied between 0.5 and 2.7 mg liter−1 in the Danube River and between 0.1 and 3.8 mg liter−1 in the tributaries and side arms (Table 2). The lowest average concentration of N-NO3 was found in reaches 5 and 6, and the highest was recorded in reach 1 (Table 1). N-NO2 concentration ranged from 6 to 66 μg liter−1 in the river, with the lowest and highest average concentrations found in reaches 4 and 7, respectively (Table 1). The total variation of N-NO2 concentration in the tributaries and side arms was between 3 and 96 μg liter−1 (Table 2). P-PO4 concentration varied between 5 and 163 μg liter−1 in the river and between 6 and 848 μg liter−1 in the tributaries and side arms (Table 2). The lowest average concentration of P-PO4 was found in reach 5 and the highest in reach 8 (Table 1).
(ii) Concentration of Chl a and apparent bacterial richness.
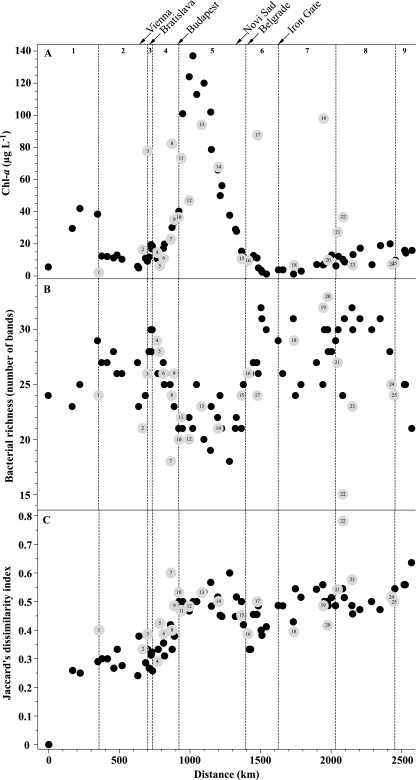
The concentration of Chl a in the Danube River varied by over 2 orders of magnitude, between 1.2 and 137 μg liter−1 and between 2.1 and 97.9 μg liter−1 in the tributaries and side arms, respectively (Fig. 2A). The average concentration of Chl a was highest in reach 5 (Table 1), where it displayed a pronounced peak (Fig. 2A). The lowest average concentrations of Chl a were found in reaches 6 and 7 (Table 1). The average concentration of Chl a in reach 5 was significantly higher than those in reaches 2 to 4 and 6 to 9 (ANOVA: P, <0.0001; F = 12.6; n = 73; Scheffe's F multiple-comparison test: reaches 2, 6, 7, and 8, P, <0.0001; reach 3, P = 0.0111; reach 4, P = 0.0023; reach 9, P = 0.0059).
FIG. 2.
Longitudinal development of (A) the concentration of Chl a, (B) the apparent bacterial richness as detected by DGGE analysis, and (C) Jaccard's dissimilarity index in the Danube River, its tributaries, and the side arms. Jaccard's dissimilarity index is based on the DGGE banding patterns and is calculated relative to the those of the first sampling station. Gray symbols indicate the tributaries and side arms, and numbered labels correspond to Table 2. Additionally, the figure shows the extent of the nine geomorphological reaches and the location of major cities and the Iron Gate dam.
Apparent bacterial richness in the Danube River ranged from 18 to 32 bands (Fig. 2B), with the lowest and highest average values in reaches 5 and 8, respectively (Table 1). The apparent level of bacterial richness in reach 5 was significantly lower than in reaches 2 to 3 and 6 to 8 (ANOVA: P, <0.0001; F = 15.3; n = 72; Scheffe's F multiple-comparison test: reach 2, P = 0.0070; reach 3, P = 0.0001; reaches 6 to 8, P, <0.0001). In the tributaries and side arms, apparent bacterial richness varied between 15 and 33 bands (Fig. 2B).
Bacterial community composition as determined by DGGE analysis.
In total, 43 distinct bands were detected on the DGGE gels (see Fig. S1 in the supplemental material). In order to detect the influences of the tributaries and side arms on the bacterial community of the river, the data were analyzed for bands that were not present immediately upstream of an inflow, were present in the inflow, and were present immediately downstream of the inflow (this analysis is not possible when two tributaries follow each other, e.g., the Hron and Ipoly tributaries). At most, five bands per inflow could be detected by fitting these criteria (constituting between 4 and 21% of the bands at the station immediately downstream of the inflow; see Fig. S1 in the supplemental material).
The riverine bacterial community developed gradually over the course of the river (Fig. 2C). In general, comparing the bacterial community of the first sampled station with subsequent communities showed that community relatedness decreased with increasing distance from the first station in the flow direction (Fig. 2C). This general pattern was disrupted for reaches 4 to 5 by an abrupt decrease in relatedness (increase in dissimilarity) in comparison to the first station, corresponding to decreases in apparent bacterial richness (Fig. 2B and C). Additionally, most of the bacterial communities in the tributaries and side arms were similar to those in the main stream in the vicinity of confluences (Fig. 2C). However, there also were exceptions like the bacterial communities of the rivers Inn (Fig. 2C, label 1), Hron (label 7), and Russenski-Lom (label 22), and to lesser degrees, rivers Timok (Fig. 2C, label 18), Olt (label 20), and Arges (label 23).
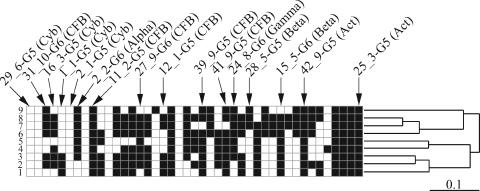
By looking at the banding patterns of prominent bands in the reaches, i.e., bands present in more than 50% of the stations in a reach, it becomes possible to compare the reaches with each other and to visualize their relatedness in a dendrogram (Fig. 3). Two groups consisting of reaches 1 to 5 and 6 to 9 could be identified. Within the first group, reaches 2 and 3 were most similar to each other, whereas in the second group, reaches 7 and 8 formed the tightest group (Fig. 3).
FIG. 3.
Banding patterns of the geomorphological reaches in the Danube River. Banding patterns of the geomorphological reaches displaying only prominent bands for the Danube River (black squares indicate bands found in more than 50% of the stations in a particular geomorphological reach [numbers 1 through 9]) and their relatedness based on Jaccard's dissimilarity index are shown. Additionally, the figure shows the position of sequenced bands and the abbreviations in brackets indicate the phylogenetic identity of the phylotypes (Cyb, Cyanobacteria; CFB, Cytophaga-Flavobacterium-Bacteroides; Alpha, Alphaproteobacteria; Beta, Betaproteobacteria; Gamma, Gammaproteobacteria; Act, Actinobacteria). A schematic representation of the DGGE banding patterns for each station obtained in this study can be found in Fig. S1 in the supplemental material.
Phylogenetic identification of selected DGGE bands.
The length of the partial 16S rRNA gene sequences obtained in this study varied between 507 and 526 bp (Table 3). Of the 16 unique bacterial 16S rRNA gene sequences obtained, 6 belong to the Cytophaga-Flavobacterium-Bacteroides group (phylotypes 11_2-G5, 12_1-G5, 27_9-G6, 31_10-G6, 39_9-G5, 41_9-G5), 4 belong to Cyanobacteria (1_1-G5, 16_3-G5, 2_1-G5, 29_6-G5), 2 belong to Actinobacteria (25_3-G5, 42_9-G5), 2 belong to Betaproteobacteria (15_5-G6, 28_5-G5), 1 belongs to Alphaproteobacteria (2_2-G6), and 1 belongs to Gammaproteobacteria (24_8-G6). The phylotypes 2_2-G6, 29_6-G5, 11_2-G5, 42_9-G5, and 16_3-G5 were obtained from samples from the Danube River and some from tributaries and side arms located between 599 and 1,391 km apart. By looking at the longitudinal distribution of the phylotypes, it is possible to identify rare phylotypes that were detected only in some stations, such as 29_6-G5, or in a limited region of the river, such as 2_1-G5 (see Fig. S1 in the supplemental material). Other phylotypes such as 11_2-G5, 42_9-G5, and 25_3-G5 were found in the majority of stations throughout the river (Fig. 3). The majority of the phylotypes showed a longitudinal distribution between these two extremes (Fig. 3; see Fig. S1 in the supplemental material).
TABLE 3.
Sequences of partial 16S rRNA genes obtained from the Danube River and its tributariesa
| Phylotype (sequence) | GenBank accession no. | Length (bp) | Classification (confidence level [%]) | Closest affiliation | Identity (%) |
|---|---|---|---|---|---|
| 1_1-G5 | DQ371841 | 507 | Order Deferribacterales (98) | Uncultured cyanobacterium clone SIMO-2301 (AY711667) | 99 |
| 2_1-G5 | DQ371847 | 507 | Class Cyanobacteria (97) | Uncultured cyanobacterium clone SIMO-2293 (AY711659) | 99 |
| Order Deferribacterales (96) | |||||
| 12_1-G5 | DQ371838 | 519 | Phylum Bacteroidetes (100) | CFBb bacterium strain WL5-12 (AF497890) | 92 |
| 11_2-G5 | DQ371837 | 517 | Genus Flavobacterium (100) | Flavobacterium sp. strain GOBB3-209 (AF321038) | 99 |
| 16_3-G5 | DQ371840 | 506 | Class Cyanobacteria (99) | Uncultured cyanobacterium clone SIMO-2301 (AY711667) | 99 |
| Order Deferribacterales (97) | |||||
| 25_3-G5 | DQ371843 | 509 | Order Actinomycetales (98) | Uncultured actinobacterium strain WL5-10 (AF497893) | 100 |
| 28_5-G5 | DQ371845 | 517 | Order Burkholderiales (100) | Uncultured betaproteobacterium clone Sta2-25 (AY562332) | 97 |
| 29_6-G5 | DQ371846 | 511 | Class Cyanobacteria (96) | Uncultured cyanobacterium DGGE gel band 2 (AY942896) | 97 |
| Order Deferribacterales (87) | |||||
| 39_9-G5 | DQ371851 | 518 | Genus Cyclobacterium (91) | Uncultured CFB group bacterium clone SFD1-9 (AF491677)b | 99 |
| 41_9-G5 | DQ371852 | 515 | Genus Cyclobacterium (95) | Bacteroidetes bacterium strain zo35 (AF531006) | 99 |
| 42_9-G5 | DQ371853 | 508 | Class Actinobacteria (97) | Uncultured actinobacterium clone R6 (AJ575502) | 99 |
| Order Actinomycetales (95) | |||||
| 2_2-G6 | DQ371848 | 500 | Phylum Proteobacteria (89) | Uncultured alphaproteobacterium clone DC8-48-1 (AY145598) | 99 |
| 15_5-G6 | DQ371839 | 526 | Order Burkholderiales (91) | Uncultured bacterium clone fl32-2 (AY544234) | 97 |
| 24_8-G6 | DQ371842 | 513 | Genus Acinetobacter (100) | Uncultured bacterium clone OS-26 (AB205957) | 100 |
| 27_9-G6 | DQ371844 | 520 | Order Sphingobacteriales (99) | Uncultured bacterium clone TLM09/TLMdgge12a (AF534433) | 90 |
| Family Crenotrichaceae (99) | |||||
| Genus Chitinophaga (99) | |||||
| 31_10-G6 | DQ371850 | 507 | Order Sphingobacteriales (97) | Uncultured Bacteroidetes bacterium clone LiUU-5-322 (AY509286) | 92 |
| Family Crenotrichaceae (97) | |||||
| Genus Chitinophaga (97) |
The table shows the GenBank accession number, length, classification according to the Bayesian classification algorithm of the Ribosomal Database Project II, closest affiliation, and percentage of nucleotide identity.
CFB, Cytophaga-Flavobacterium-Bacteroides group.
Covariation of physicochemical and biological parameters.
In the Danube River, a number of significant (Bonferroni corrected; P ≤ 0.005) relationships were found between the parameters, where −0.5 ≥ r ≥ 0.5 were detected (Table 4). DO, pH, and N-NO3 decreased, whereas N-NO2 and P-PO4 increased, with decreasing distance to the Black Sea. Temperature was negatively correlated with N-NO3, whereas pH was positively correlated with DO and Chl a. DO was negatively correlated with N-NO2 and P-PO4 and positively correlated with Chl a. Furthermore, N-NO2 correlated positively with P-PO4 and negatively with Chl a. P-PO4 was negatively correlated with Chl a and positively correlated with apparent bacterial richness. Moreover, Chl a correlated negatively with apparent bacterial richness.
TABLE 4.
Spearman rank correlation coefficients calculated for the parameters measured in the Danube Rivera
| Parameter | Distance to Black Sea | Temp | pH | DO | N-NH4 | N-NO3 | N-NO2 | P-PO4 | Chl a |
|---|---|---|---|---|---|---|---|---|---|
| Temp | −0.25 | ||||||||
| pH | 0.50* | −0.19 | |||||||
| DO | 0.63* | −0.15 | 0.92* | ||||||
| N-NH4 | −0.12 | 0.44* | −0.49* | −0.39* | |||||
| N-NO3 | 0.51* | −0.76* | 0.34* | 0.34* | −0.45* | ||||
| N-NO2 | −0.51* | 0.30 | −0.44* | −0.54* | 0.23 | −0.29 | |||
| P-PO4 | −0.75* | 0.02 | −0.47* | −0.62* | 0.06 | −0.05 | 0.55* | ||
| Chl a | 0.29 | 0.10 | 0.67* | 0.72* | −0.07 | −0.02 | −0.60* | −0.50* | |
| Bacterial richness | −0.26 | −0.25 | −0.34* | −0.48* | −0.06 | 0.19 | 0.33* | 0.56* | −0.52* |
Correlation coefficients (r) marked by an asterisk are statistically significant (Bonferroni corrected; P ≤ 0.005). Values in boldface indicate relevant correlation coefficients (−0.5 ≥ r ≥ 0.5).
Modeling apparent bacterial richness in the Danube River.
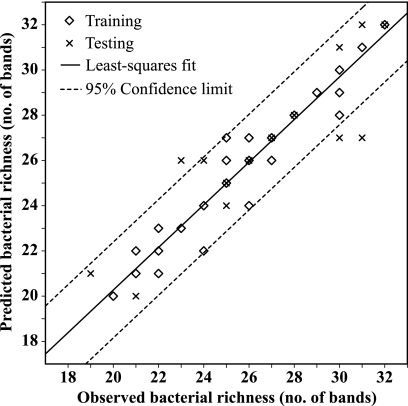
The best-performing ANN to model apparent bacterial richness used N-NO2, P-PO4, and the distance to the Black Sea as input parameters for an ANN with 10 hidden neurons. The ANN-based model explained 90% of the variation of apparent bacterial richness in the Danube River (Fig. 4). The slope of the linear least-squares regression (Fig. 4) of observed versus predicted values of apparent bacterial richness was not significantly different from 1 (t value = 0.75; P = 0.45). The slopes of linear least-squares regression analysis of observed versus predicted values of apparent bacterial richness performed with the training (y = 0.89 + 0.96x; n = 57) and test data set (y = 1.68 + 0.93x; n = 15) alone were not significantly different from 1 (training t value = 0.83, P = 0.40; testing t value = 0.29, P = 0.77) or from each other (t value = 0.08, P = 0.94).
FIG. 4.
Comparison between observed and predicted values of bacterial richness in the Danube River. Bacterial richness was determined by DGGE analysis, and the predictions were computed using the ANN developed in this study to model bacterial richness, as described in the text. The solid line represents a linear least-squares fit to all the data (training and test data), and the broken lines represent the boundaries of the 95% confidence region. The equation of the linear least-squares regression is y = 1.17 + 0.95x (n = 72; R2 = 0.90).
DISCUSSION
Discontinuity in the longitudinal development of the Danube River.
Based on physicochemical parameters, the Danube River can be divided into two stretches composed of reaches 1 to 5 and reaches 6 to 9 (Table 1). On one hand, DO and N-NO3 were significantly higher in reaches 1 to 5 than in reaches 6 to 9 (unpaired t test: DO, P, <0.0001; N-NO3, P = 0.0096). On the other hand, N-NO2 and P-PO4 were significantly lower in reaches 1 to 5 than in reaches 6 to 9 (unpaired t test: N-NO2 and P-PO4, P, <0.0001). The bacterial communities in reaches 1 to 5 and 6 to 9 formed two deeply branching groups (Fig. 3), reflecting this differentiation along the longitudinal dimension of the river.
The lowest apparent bacterial richness (Fig. 2B) and a clearly visible shift in bacterial community composition (Fig. 2C) were recorded in reach 5, where the highest Chl a concentration was measured (Fig. 2A). Chl a was significantly positively related to pH and DO (Table 4), and the highest average DO was found in reach 5 (Table 1). Thus, the increase in Chl a not only signaled an increase in phytoplankton biomass and/or Chl a content per cell but also indicated an increase in photosynthetic activity in the river. Phytoplankton species are well known to produce biologically available extracellular organic carbon during photosynthesis (11, 27). This photosynthetic extracellular release has been estimated to be ∼20% of the total photosynthetic carbon production (28). Thus, it is reasonable to assume that the high levels of photosynthetic activity in reach 5 in turn stimulated the growth of some members of the bacterial community that were able to use the sudden increase in organic carbon most efficiently. These fast-growing members of the community reduced the relative abundance of the less-adapted populations to a level below the detection limit of DGGE analysis, thus reducing apparent bacterial richness. A similar stimulation of bacterial growth and a concomitantly occurring decline in apparent bacterial richness have been reported previously (41).
Apparent bacterial richness correlated positively with P-PO4 (Table 4), thus indicating a potential phosphorus limitation to bacterioplankton. It is reasonable to assume that under phosphorus-limiting conditions, bacterial types with a highly efficient uptake machinery for phosphorus will have a competitive advantage over less efficient phosphorus-uptake bacterial types. Enhanced availability of phosphorus will allow more members of the community to increase in their relative abundance above the detection limit of DGGE analysis, thus increasing apparent bacterial richness.
Nitrite is an intermediate product of the oxidation of ammonium to nitrate, carried out exclusively by prokaryotes. Thus, N-NO2 indicates the presence and activity of nitrifying bacterial types. For example, Cébron et al. (4) showed that the Seine River (France) receives large amounts of nitrifiers due to the effluents of a wastewater treatment plant. Their results suggest that the pollution of the Seine River with N-NH4 downstream of the city of Paris facilitated the growth of these nitrifying bacterial types inoculated by the wastewater emissions. In our study, the highest average N-NH4 was found in reach 6 (Table 1), and this concentration level likely stimulated the growth of nitrifying bacterial types. The nitrifiers in turn substantially decreased N-NH4 (largest change between reaches 5 and 6; see Table 1) and increased N-NO2 downstream of reach 5. Thus, enhanced growth of nitrifying bacterial types in reaches 6 to 9 probably contributed to the shift in the riverine bacterial community between reaches 1 to 5 and 6 to 9.
Considerations concerning the interpretation of DGGE analysis.
PCR-based fingerprinting methods such as DGGE analysis have their limitations (7, 33, 38). However, a trend toward lower apparent bacterial richness is likely caused by a shift in evenness toward a few dominant phylotypes, reducing the relative abundance of other members of the community to below the detection limit of the method. This would imply that DGGE analysis detects abundant members of the bacterial community.
Samples for the characterization of the bacterial community were not prefiltered, and thus, our results might have been biased by the coamplification of plastid 16S rRNA genes (34). Nevertheless, all sequences retrieved from the DGGE gels were, without exception, of bacterial origin (Table 3). Additionally, the lowest apparent bacterial richness was recorded in the area with the highest Chl a concentration (Fig. 2A and B). Although we cannot completely exclude bias due to coamplified plastid 16S rRNA genes, our results suggest that their potential influence on our analysis was small.
Allochthonous and anthropogenic influences on the Danube River. (i) Tributaries and side arms.
The biggest tributaries of the Danube River are the rivers Inn (755 m3 s−1), Drava (450 m3 s−1), Tisza (720 m3 s−1), and Sava (2140 m3 s−1), together contributing ∼70% of the Danube's total water discharge. Thus, although some of the smaller tributaries differed substantially in their concentrations of nutrients (e.g., the Russenski-Lom and Arges) and Chl a (e.g., the Sio and Iskar) and apparent bacterial richness (e.g., the Hron and Russenski-Lom) (Tables 1 and 2; Fig. 2A and B), their influence on the Danube River was not detectable because of dilution effects. For this reason, we will focus the remainder of this discussion on the four biggest tributaries.
The temperature and Chl a concentration of the Inn River were substantially lower (Tables 1 and 2; Fig. 2A), and its bacterial community also differed compared to that in the Danube River (Fig. 2B and C). Thus, the signature of the Inn River is clearly detectable in the Danube downstream of confluence. The rivers Drava, Tisza, and Sava, however, were very similar to the Danube River in terms of Chl a and bacterial community composition (Fig. 2B and C; Table 1 and 2). Crump and Hobbie (8) studied the bacterial communities of two nonintersecting, temperate rivers. Those authors showed that the bacterial communities of the two rivers were nearly identical and changed synchronously over a period of 2.5 years. Their results suggest that intrinsic controls of prokaryotes were similar in the two rivers and that seasonal changes were driven by extrinsic factors such as climate. They further demonstrated that temperature was one of the best predictors for the temporal patterns in the bacterial community composition. In our study, the temperatures of the rivers Drava, Tisza, and Sava (Table 2) were similar to the Danube's temperature before confluence (before Drava, 22.4°C; before Tisza, 22.8°C; and before Sava, 23°C). Thus, the mechanisms suggested by Crump and Hobbie (8) might explain why the bacterial communities of the rivers Drava, Tisza, Sava, and the Danube were similar to each other around the points of confluence. Similarly, the large temperature difference between the Inn (Table 2) and the Danube River (before confluence, 21.4°C) might explain the differences in their bacterial communities. Although temperature differences or the lack thereof provide a plausible explanation for our results and are in principle supported by the study of Crump and Hobbie (8), further studies are needed to confirm this hypothesis for the Danube River and its biggest tributaries.
(ii) Gabcikovo and Iron Gate impoundments.
Intuitively, damming a river with high-flow velocities will have a bigger impact on the river ecosystem than an impoundment in a river with low-flow velocities. It is difficult to evaluate the impact of the two biggest man-made reservoirs in the Danube River due to a lack of comparison with the unaltered state. However, comparing parameters between the reservoirs and the areas adjacent to them should serve as a first indication of potential alterations.
The Gabcikovo reservoir had no detectable direct influence on the parameters measured in this study (Table 1; Scheffe's F test between reaches 2 and 4 resulted in a P value of >0.05 in every case). However, the dam increased underwater transparency in reach 4 by acting as a sediment trap due to the reduced flow velocities in the impoundment. It is misleading to compare reach 5 parameters with those of the Iron Gate reservoir due to the dramatic changes occurring in reach 5. However, a comparison between the Iron Gate reservoir and reach 7 showed that N-NH4 (unpaired t test, P, <0.0001) and N-NO3 (unpaired t test, P = 0.0008) were significantly different between these two stretches.
The Gabcikovo and the Iron Gate reservoirs had no discernible effect on the bacterial communities. Single prokaryotic cells are much smaller than the Kolmogorov microscale of turbulence (24). Thus, prokaryotes experience the surrounding water as a viscous medium and any effects on bacterial communities caused by a change in flow velocity, such as in a reservoir, will be indirect via cascading effects in the microbial food web (e.g., 26). Lindström and Bergström (22) investigated the influence of inlet bacteria on the community composition of bacterioplankton in two Swedish lakes differing in their hydraulic retention times. Their data suggest that the import of inlet bacteria has a substantial effect on the bacterial community composition of lakes with a short hydraulic retention time. River impoundments tend to have a short hydraulic retention time, and thus, the bacterial communities of the river and the reservoirs are expected to be similar.
(iii) Large cities.
The influence of urban areas on the bacterial community composition of rivers versus those of tributaries is difficult to evaluate. The input of tributaries can be determined by comparing the parameters before and after confluence; however, large cities also have a strong diffusive component (e.g., surface water runoff) and are for historical reasons often situated at river junctions, making the task even more difficult. Thus, although our data do not show abrupt changes in bacterial richness in the vicinity of urban areas (Fig. 2C), we have to acknowledge that the study design is not well suited to evaluate the impact of large cities on the riverine bacterial community. A more fine-grained sampling design in the vicinity of cities that includes seasonal changes will be necessary to achieve this.
Distribution of selected bacterial phylotypes.
In total, 16 distinct, partial 16S rRNA gene sequences were obtained (Table 3). Thus, we were able to phylogenetically characterize 37% of the bacterial community, as detected by DGGE analysis. In principal, some of the DGGE bands detected at stations far apart from each other might represent different phylotypes although they appear at the same position in the gel. Such a distance-related identity switch of DGGE bands would mask some of the changes in the bacterial community. We found no evidence for this; instead, we were able to obtain identical nucleotide sequences for phylotypes 2_2-G6, 29_6-G5, 11_2-G5, 42_9-G5, and 16_3-G5 (Table 3) at stations in the river and in some of the tributaries and side arms between 599 and 1,391 km apart. Although we cannot completely exclude the possibility of identical DGGE bands with different phylogenetic identities, our results suggest that this was not a major problem for our analysis.
All sequences obtained were closely related to typical freshwater bacteria (12, 43). The six phylotypes identified as belonging to the Cytophaga-Flavobacterium-Bacteroides group were detected frequently, with one phylotype being a prominent member of the community in all reaches (11_2-G5) (Fig. 3). The high detection frequency of these Cytophaga-Flavobacterium-Bacteroides phylotypes suggests that they were an important component of the bacterial community in the Danube River. The phylotypes 25_3-G5 and 42_9-G5, both belonging to Actinobacteria, were present in the majority of the sampling stations throughout the river (Fig. 3), apparently following an opportunistic lifestyle largely independent of environmental changes. Two of the cyanobacterial phylotypes (29_6-G5 and 2_1-G5; Fig. 3) were rarely detected, suggesting that they were of minor importance in the river. Additionally, none of the four identified cyanobacterial phylotypes was found in reach 5, where the highest Chl a concentration was detected (Fig. 2A and 3; see Fig. S1 in the supplemental material). Also, two phylotypes (12_1-G5 and 31_10-G6) with 92% and one (27_9-G6) with only 90% sequence identity to the closest related sequence in GenBank were obtained, possibly representing novel bacterial lineages.
Modeling apparent bacterial richness in the Danube River.
The ANN-based model of apparent bacterial richness developed in this study uses N-NO2, P-PO4, and the distance to the Black Sea as input parameters to an ANN with 10 hidden units. The ANN-based model explains 90% of the variation of apparent bacterial richness in the Danube River (Fig. 4) and is by far superior to a model based on stepwise multiple linear regression analysis (data not shown). The ANN-based model used a determining factor of apparent bacterial richness (P-PO4), an indicator for a process carried out by some members of the bacterial community (N-NO2), and a parameter representing the longitudinal dimension of the study area (distance to the Black Sea) to predict apparent bacterial richness with high accuracy (Fig. 4). The selection of these parameters was based on model performance. Thus, the ANN-based model of apparent bacterial richness supports our interpretation of the data given above.
Conclusions.
In general, the riverine bacterial community developed gradually over the course of the river. This pattern was disrupted in reach 5 by an abrupt decrease in apparent bacterial richness and a clear shift in bacterial community composition. Based on the longitudinal development of DO, N-NO3, N-NO2, and P-PO4 levels, the Danube River can be divided into an upper (reaches 1 to 5) and a lower (reaches 6 to 9) stretch. The bacterial communities in reaches 1 to 5 and 6 to 9 formed two deeply branching groups, reflecting this differentiation along the longitudinal dimension of the river. In particular, reach 5 marks the turning point with the highest average concentrations of Chl a and lowest average apparent bacterial richness. In reach 5, the Danube River increases its discharge by ∼45% within a distance of 200 km due to confluence with the rivers Drava, Tisza, and Sava. With the exception of the Inn River in reach 1, the bacterial communities of the biggest tributaries resembled the community of the Danube River. Thus, the shift in bacterial community composition between reaches 1 to 5 and 6 to 9 was not due to the import of new bacterial types; instead, the riverine bacterial community adapted to the changing environmental conditions. Large impoundments had no detectable effect on the bacterial community of the water column, most likely due to the short hydraulic retention time in these reservoirs. In conclusion, our data indicate that intrinsic factors such as the concentration of nutrients and phytoplankton production appeared to determine bacterial community composition. External factors such as large tributaries indirectly influenced the riverine bacterial community composition by altering the concentration of nutrients.
Supplementary Material
Acknowledgments
We thank Igor Liška and Péter Literáthy as the main organizers of the field campaign; Birgit Vogel, Carmen Hamchevici, and Aleksander Miletic for determining physicochemical parameters; Jozsef Nemeth, Boyan Boyanovski, and Ivan Traykov for determining Chl a values; and Erich Poetsch for sampling bacterial richness. We are grateful to Martina Burtscher and Georg Reischer for help in determining bacterial community composition and to Ulrich Schwarz for producing the map of the sampling stations. We thank two anonymous reviewers for improving the manuscript with their comments.
The Joint Danube Survey was carried out under the auspices of the International Commission for the Protection of the Danube River (ICPDR) and was cofinanced by the German Federal Environmental Agency and the Austrian Federal Ministry of Agriculture, Forestry, Environment and Water Management. The analysis of bacterial community composition was cofinanced by the Swiss Federal Institute for Environmental Science and Technology (ETH) and the International Association for Danube Research (IAD) and its Austrian Committee (AC-IAD).
Footnotes
Published ahead of print on 3 November 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Basheer, I. A., and M. Hajmeer. 2000. Artificial neural networks: fundamentals, computing, design, and application. J. Microbiol. Methods 43:3-31. [DOI] [PubMed] [Google Scholar]
- 3.Cébron, A., T. Berthe, and J. Garnier. 2003. Nitrification and nitrifying bacteria in the lower Seine River and estuary (France). Appl. Environ. Microbiol. 69:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cébron, A., M. Coci, J. Garnier, and H. J. Laanbroek. 2004. Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine River: impact of Paris wastewater effluents. Appl. Environ. Microbiol. 70:6726-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cébron, A., and J. Garnier. 2005. Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower Seine river (France). Water Res. 39:4979-4992. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 8.Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol. Oceanogr. 50:1718-1729. [Google Scholar]
- 9.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.65. Department of Genome Sciences, University of Washington, Seattle, WA. http://evolution.genetics.washington.edu/phylip.html.
- 10.Giacomini, M., C. Ruggiero, L. Calegari, and S. Bertone. 2000. Artificial neural network based identification of environmental bacteria by gas-chromatographic and electrophoretic data. J. Microbiol. Methods 43:45-54. [DOI] [PubMed] [Google Scholar]
- 11.Giroldo, D., and A. A. H. Vieira. 2005. Polymeric and free sugars released by three phytoplankton species from a freshwater tropical eutrophic reservoir. J. Plankton Res. 27:695-705. [Google Scholar]
- 12.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg, A. E. (ed.). 1985. Standard methods for the examination of water and wastewater, 16th ed. American Public Health Association, American Water Works Association, Water Pollution Control Federation, Washington, DC.
- 14.Haykin, S. 1999. Neural networks, 2nd ed. Prentice Hall, Upper Saddle River, NJ.
- 15.Hedges, J. I., E. Mayorga, E. Tsamakis, M. E. McClain, A. Aufdenkampe, P. Quay, J. E. Richey, R. Benner, S. Opsahl, B. Black, T. Pimentel, J. Quintanilla, and L. Maurice. 2000. Organic matter in Bolivian tributaries of the Amazon River: a comparison to the lower mainstream. Limnol. Oceanogr. 45:1449-1466. [Google Scholar]
- 16.International Organization for Standardization. 1984. ISO7150-1. Determination of ammonium—part 1: manual spectrometric method. p. 1-7. http://www.iso.org.
- 17.International Organization for Standardization 1988. ISO7890-3. Determination of nitrate—part 3: spectrometric method using sulfosalicyclic acid. p. 1-4. http://www.iso.org.
- 18.International Organization for Standardization 1992. ISO10260. Spectrometric determination of the chlorophyll-a concentration. p. 1-6. http://www.iso.org.
- 19.Janse, I., J. Bok, and G. Zwart. 2004. A simple remedy against artificial double bands in denaturing gradient gel electrophoresis. J. Microbiol. Methods 57:279-281. [DOI] [PubMed] [Google Scholar]
- 20.Judd, K. E., B. C. Crump, and G. W. Kling. 2006. Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87:2068-2079. [DOI] [PubMed] [Google Scholar]
- 21.Kirchman, D. L., A. I. Dittel, S. E. G. Findlay, and D. Fischer. 2004. Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquat. Microb. Ecol. 35:243-257. [Google Scholar]
- 22.Lindström, E. S., and A.-K. Bergström. 2004. Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol. Oceanogr. 49:125-136. [Google Scholar]
- 23.Literáthy, P., V. Koller-Kreimel, and I. Liska. 2002. Joint Danube survey. Technical report of the International Commission for the Protection of the Danube River. International Commission for the Protection of the Danube River (ICPDR). http://www.icpdr.org/icpdr-pages/jds.htm.
- 24.Logan, B. E., and D. L. Kirchman. 1991. Uptake of dissolved organics by marine bacteria as a function of fluid motion. Mar. Biol. 111:175-181. [Google Scholar]
- 25.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maar, M., L. Arin, R. Simó, M.-M. Sala, F. Peters, and C. Marrasé. 2002. Combined effects of nutrients and small-scale turbulence in a microcosm experiment. II. Dynamics of organic matter and phosphorus. Mar. Ecol. Prog. Ser. 29:63-72. [Google Scholar]
- 27.Marañón, E., P. Cermeño, and E. Fernández. 2004. Significance and mechanisms of photosynthetic production of dissolved organic carbon in a coastal eutrophic ecosystem. Limnol. Oceanogr. 49:1652-1666. [Google Scholar]
- 28.Marañón, E., P. Cermeño, and V. Pérez. 2005. Continuity in the photosynthetic production of dissolved organic carbon from eutrophic to oligotrophic waters. Mar. Ecol. Prog. Ser. 299:7-17. [Google Scholar]
- 29.Marquardt, D. 1963. An algorithm for least-squares estimation of nonlinear parameters. SIAM J. Appl. Math. 11:431-441. [Google Scholar]
- 30.Muyzer, G., S. Hottenträger, A. Teske, and C. Wawer. 1996. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 1-23. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer, Dordrecht, The Netherlands.
- 31.Muyzer, G., A. Teske, and C. O. Wirsen. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 32.Noble, P. A., K. D. Bidle, and M. Fletcher. 1997. Natural microbial community compositions compared by a back-propagating neural network and cluster analysis of 5S rRNA. Appl. Environ. Microbiol. 63:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappé, M. S., M. T. Suzuki, K. L. Vergin, and S. J. Giovannoni. 1998. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl. Environ. Microbiol. 64:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 36.Schryver, J. C., C. C. Brandt, S. M. Pfiffner, A. V. Palumbo, A. D. Peacock, D. C. White, J. P. McKinley, and P. E. Long. 2006. Application of nonlinear analysis methods for identifying relationships between microbial community structure and groundwater geochemistry. Microb. Ecol. 51:177-188. [DOI] [PubMed] [Google Scholar]
- 37.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorp, J. H., and M. D. DeLong. 2002. Dominance of autochthonous autotrophic carbon in food webs of heterotrophic rivers. Oikos 96:543-550. [Google Scholar]
- 40.Vannote, R. L., G. W. Minshall, K. W. Cummins, J. R. Sedell, and C. E. Cushing. 1980. The river continuum concept. Can. J. Fish. Aquat. Sci. 37:130-137. [Google Scholar]
- 41.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2005. Linking bacterial richness with viral abundance and prokaryotic activity. Limnol. Oceanogr. 50:968-977. [Google Scholar]
- 42.Winter, C., A. Smit, T. Szoeke-Dénes, G. J. Herndl, and M. G. Weinbauer. 2005. Modelling viral impact on bacterioplankton in the North Sea using artificial neural networks. Environ. Microbiol. 7:881-893. [DOI] [PubMed] [Google Scholar]
- 43.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.