Abstract
Zearalenone is a mycotoxin with estrogenic effects on mammals that is produced by several species of Fusarium. We found that zearalenone and its derivatives inhibit the growth of filamentous fungi on solid media at concentrations of ≤10 μg/ml. The fungitoxic effect declined in the order zearalenone > α-zearalenol > β-zearalenol. The mycoparasitic fungus Gliocladium roseum produces a zearalenone-specific lactonase which catalyzes the hydrolysis of zearalenone, followed by a spontaneous decarboxylation. The growth of G. roseum was not inhibited by zearalenone, and the lactonase may protect G. roseum from the toxic effects of this mycotoxin. We inactivated zes2, the gene encoding zearalenone lactonase in G. roseum, by inserting a hygromycin resistance cassette into the coding sequence of the gene by means of Agrobacterium tumefaciens-mediated genetic transformation. The zes2 disruption mutants could not hydrolyze the lactone bond of zearalenone and were more sensitive to zearalenone. These data are consistent with a hypothesis that resorcylic acid lactones exemplified by zearalenone act to reduce growth competition by preventing competing fungi from colonizing substrates occupied by zearalenone producers and suggest that they may play a role in fungal defense against mycoparasites.
Zearalenone [6-(10-hydroxy-6-oxo-trans-1-undecenyil)-resorcylic acid lactone] is a mycotoxin produced by several species of Fusarium, most notably Fusarium graminearum and Fusarium culmorum. Zearalenone and its derivatives exert estrogenic and anabolic effects on mammals. Carryover of zearalenone from infected grain to feedstuff causes reproductive problems in pigs, sheep, and other farm animals, including precocious sexual development, vulva enlargement, pseudopregnancy, loss of embryos, and reduced litter size (10, 14, 24). When digested with grain-based food, zearalenone may cause hyperestrogenism in children (33). Apart from its estrogenicity, zearalenone is genotoxic in mice and is a suspected carcinogen (5, 15, 30). The intake of zearalenone by some human populations is close to the estimated tolerable limits (9), and the amount of zearalenone allowed in grain, feedstuff, and food is regulated in several countries (1). Current ecological and economic constraints on grain production encourage minimum tillage practices and reduced fungicide application, so infection of cereal plants with Fusarium spp. is increasing and contamination with mycotoxins is becoming a more serious threat to grain production.
One promising strategy for reducing zearalenone contamination is enzymatic degradation. The only commercial product on the market claimed to enzymatically detoxify zearalenone is the feed additive Mycofix Plus (Biomin GmbH, Herzogenburg, Austria). There are no published laboratory data demonstrating the hydrolysis of zearalenone by Mycofix Plus or by its active component, and such activity was not detected by us (21). The recently discovered yeast species Trichosporon mycotoxinivorans (27) degrades zearalenone and is expected to provide future versions of Mycofix Plus with genuine zearalenone-hydrolyzing activity. Another fungus with proven zearalenone-degrading activity is Gliocladium roseum (8), and a zearalenone lactonase gene has been cloned from this mycoparasite by two research groups (19, 34; E. H. Crane, J. T. Gilliam, P. Karlovsky, and J. R. Maddox, 3 October 2002, World Trade Organization patent application 02/076205; P. Karlovsky, E. H. Crane, J. T. Gilliam, and J. R. Maddox, U.S. patent application 20030073239). The sequences published independently differ in three amino acids (P. Karlovsky, unpublished data). The biological function of zearalenone esterase, as well as the function of zearalenone itself, is unknown.
The objectives of this work were (i) to demonstrate the growth inhibition of filamentous fungi by zearalenone and (ii) to evaluate the effect of disruption of the zes2 gene of G. roseum on the ability of the fungus to hydrolyze zearalenone and the resistance of G. roseum to zearalenone. Our working hypothesis is that zearalenone acts as an agent of interference competition (active protection of substrate from colonization by competitors) and that the zearalenone-specific lactonase of G. roseum protects the fungus from the fungitoxic effect of the mycotoxin. We suggest a biological function for zearalenone production and an ecological role for this fungal metabolite in a nonagricultural setting.
MATERIALS AND METHODS
Microbial strains.
G. roseum DSM62726 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Agrobacterium tumefaciens AGL1 (23) was used for fungal transformation. Escherichia coli DH5α (16) was used as the host for the construction and maintenance of plasmid vectors.
The fungal and protoctist species used in the investigation of the effect of zearalenone on growth rate and colony morphology were Epicoccum purpurascens AvT206, Cladosporium herbarum AvT264, and Alternaria alternata AvT64 (provided by A. von Tiedemann, Goettingen University, Goettingen, Germany); Sporotrichium thermophile H98 and Thermoascus aurantiacus H103 (provided by Jean Wagner, Hohenheim University, Stuttgart, Germany); Phytophthora nicotianae DSM1829 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany); Stagonospora avenae H121, Stagonospora nodorum Sn80, and F. graminearum HOH587 (provided by Hanno Wolf, Hohenheim University, Stuttgart, Germany); Septoria avenae f. sp. tritici ATCC 26371 (American Type Culture Collection, Manassas, VA); and Phytophthora parasitica P6623 (provided by Arthur de Cock, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands).
Culture conditions and media.
GM7 medium (20) was prepared by dissolving 3 g l-asparagine, 1 g KH2PO4, 0.5 g MgSO4 · 7H2O, 50 mg CaCl2, 10 mg FeCl3 · 7H2O, 1 mg thiamine, and 20 g glucose in 1 liter of distilled water; the pH was adjusted to 5.6. Media were sterilized by autoclaving and solidified with 1.5% agar. A. tumefaciens was grown in LB medium (31) supplemented with kanamycin, rifampin, and carbenicillin at 50, 50, and 25 μg/ml, respectively. Induction medium (IM) for A. tumefaciens was prepared as described by de Groot et al. (7). Malt extract agar (MEA) and Czapek-Dox agar were purchased from Difco (Detroit, Mich.) and prepared according to the manufacturer's instructions. Synthetic nutrient-poor agar was prepared as described by Gerlach and Nirenberg (12).
Chemicals.
Zearalenone was obtained from Fermentek Ltd. (Jerusalem, Israel) in a purity of 99%. α- and β-zearalenol were purchased from Sigma-Aldrich (Steinheim, Germany). Stock solutions of zearalenone and its derivatives were made in ethanol at a concentration of 10 mg/ml and kept at −20°C. All other chemicals were of “pro analysis” quality (analytical grade).
General DNA isolation and manipulation methods.
Total DNA of G. roseum was isolated by using a modified cetyltrimethylammonium bromide protocol (4). Plasmid DNA isolation, restriction enzyme digestion, ligation, PCR, and DNA electrophoresis in agarose gels were performed according to standard protocols (31).
Construction of the zes2 disruption vector.
The cassette used for the disruption of zes2 was constructed by inserting a hygromycin resistance gene into the coding sequence of zes2. Flanking regions consisting of 960 bp (upstream sequence and 3′ part of the zes2 coding region) and 1,064 bp (5′ part of the zes2 coding region and the downstream sequence) were amplified with primers (Table 1) derived from the genomic DNA of G. roseum (GenBank accession no. AB076037) and containing recognition sequences for SacI/HindIII (upstream and 3′ regions) and KpnI/XbaI (5′ and downstream regions). PCR amplification was performed with 35 cycles of 96°C for 30 s, 61°C for 30 s, and 72°C for 50 s with a final extension at 72°C for 5 min.
TABLE 1.
PCR primers used in this study
| Template | Name | Sequence (5′-3′)a | Product size (bp) |
|---|---|---|---|
| zes2 coding sequence | OL500 | TACATACATATGCGCATTCGCAGCACAA | 820 |
| OL501 | AAGAGAAAGATCTTCAAAGATACTTCTGCGTA | ||
| zes2 3′ and upstream sequences | OL505 | AAGACAGAGCTCAACCAACCAGCCAGAAGTTAGA | 960 |
| OL506 | AAGACAAAGCTTGGTCCAGTAGCTTTGTTGGCA | ||
| zes2 5′ and downstream sequences | OL507 | AAGACAGGTACCCAAACACCGCTGTGCTCGA | 1,064 |
| OL508 | AAGACATCTAGAGGGCTGGTCTCCCGTAC | ||
| hph | HygBu | AAAAGTTCGACAGCGTCTCC | 930 |
| HygBd | CGGCGAGTACTTCTACACAGC |
Underlined recognition sequences for restriction enzymes: OL505, SacI; OL506, HindIII; OL507, KpnI; OL508, XbaI.
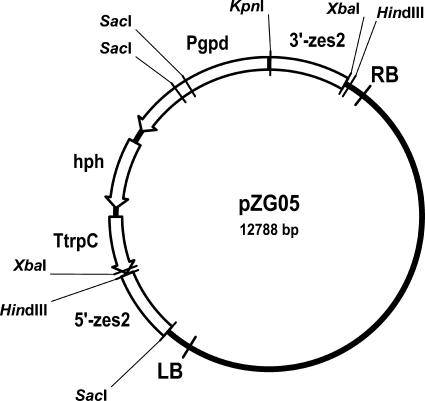
The hygromycin B resistance cassette contained the hygromycin phosphotransferase gene from E. coli (hph) under the control of the gpdA promoter and the trpC terminator from A. nidulans isolated as a 4-kb KpnI-HindIII fragment from binary vector pPK2 (6). The resistance cassette was fused with the 1,064-bp KpnI-XbaI PCR fragment carrying the 5′ part and downstream region of zes2 inserted between the HindIII and XbaI sites of pUC57 (GenBank accession no. Y14837). The resistance cassette connected to the 3′ and downstream regions of zes2 was isolated as a 5-kb HindIII/XbaI fragment and joined in a tri-fragment ligation with the SacI/HindIII-digested PCR product of the amplified upstream and 3′ parts of zes2 and SacI/XbaI-digested binary vector pPK2 to yield pZG05 (Fig. 1). The length of the T-DNA, from the left border to the right border, was ∼6.5 kb. The vector was introduced into A. tumefaciens AGL1 by electroporation (26) with a pulse of 2.5 kV in a 2-mm cuvette, which was bridged with a resistance of 400 Ω and a capacitance of 25 μF.
FIG. 1.
Physical map of zes2 disruption vector pZG05. Pgpd, promoter region from gpd gene; TtrpC, termination region from trpC gene (both from Aspergillus nidulans); LB and RB, left and right boundaries of T-DNA, respectively.
Southern hybridization.
Disruption of zes2 in the genome of G. roseum after A. tumefaciens-mediated transformation was evaluated by Southern hybridization with a PCR-generated, digoxigenin-labeled zes2 fragment as the probe. The probe was amplified from the genomic DNA of G. roseum by using dUTP conjugated with digoxigenin (Roche Diagnostics GmbH, Penzberg, Germany) and primers OL500 and OL501 (Table 1), generating a product of 820 bp. Six micrograms of HindIII-digested genomic DNA from putative transformants was resolved in a 0.8% agarose gel and blotted on Hybond N+ nylon membrane (Amersham Biosciences Europe GmbH, Freiburg, Germany). Hybridization and detection of the digoxigenin-labeled probe with streptavidin-phosphatase complex and chemiluminescence substrate CDP-star was performed according to the manufacturer's (Roche Diagnostics GmbH) instructions. Alternatively, chemifluorescence of dephosphorylated dimethylacridinone phosphate (Molecular Probes, Eugene, OR) was used to localize the probe, and fluorescent signals were recorded with a laser scanner (Typhoon 8600; Amersham Biosciences Europe GmbH).
A. tumefaciens-mediated transformation of G. roseum.
A. tumefaciens carrying the pZG05 disruption vector was grown with shaking (220 rpm) at 28°C overnight. Cells were washed with IM, transferred to fresh IM containing 0.2 mM acetosyringone, and grown to an optical density at 660 nm of 0.15. The culture was shaken at 28°C until its optical density doubled. One hundred microliters of A. tumefaciens culture and a suspension of 1 × 106 G. roseum spores in 100 μl water were mixed and spread on a cellophane membrane overlying the IM agar supplemented with 0.2 mM acetosyringone. After coincubation for 48 h, the membranes were transferred to agar plates with GM7 medium containing 250 μg/ml hygromycin B to select transformants and 200 μM cefotaxime to prevent the growth of A. tumefaciens.
HPLC-mass spectrometric analysis of zearalenone and its conversion products.
Reverse-phase high-performance liquid chromatography (HPLC) with a diode array and mass spectrometric detectors was used. The column was a Polaris C18-A (5 μm, 150 by 2 mm; Varian, Darmstadt, Germany), the mobile phase was 55% methanol-45% water-5% acetonitrile, the flow rate was 0.2 ml/min, the column temperature was 40°C, detection was done with a diode array detector (absorbance at 200 to 800 nm), and mass spectrometry was done by electrospray in positive mode (scan for a mass/charge ratio of 250 to 500). Preparation of samples was done by extracting culture supernatants with ethyl acetate and removing the solvent on a rotary evaporator. The residue was dissolved in the mobile phase.
Examination of the effect of zearalenone and its derivatives on fungal growth.
MEA was autoclaved, cooled to 60°C, and amended with zearalenone and its derivatives by adding appropriate amounts of stock solutions in methanol. The concentration of methanol was adjusted to 5% in all media, including controls. We have not observed any effect of methanol at this concentration on the growth of G. roseum. For qualitative assays, fungal strains were inoculated on solidified media by placing agar blocks overgrown with mycelium onto the agar close to the margin of the petri dish. Inoculated plates were incubated for 7 days at 20°C under a long-day light regimen (16 h light, 8 h dark). For quantitative assays, round, mycelium-covered agar blocks cut with a cork borer (6 mm diameter) out of agar plates were placed in the middle of petri dishes with solid medium. Plates were incubated at 24°C in the dark, and colony diameter was measured every 48 h. Two values measured in perpendicular directions were recorded at each time point.
RESULTS
Inhibition of fungal growth by zearalenone.
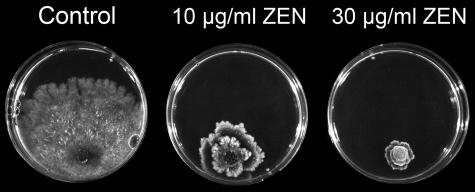
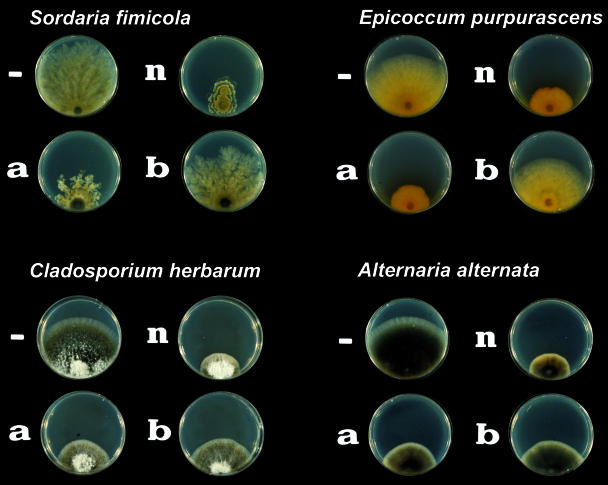
Zearalenone strongly inhibits the growth of Sordaria fimicola (Fig. 2). An inhibitory effect is detectable at a zearalenone concentration as low as 2 μg/ml. An additional 10 species of filamentous fungi and fungus-like protoctists were inoculated onto MEA plates with zearalenone. With the exception of G. roseum, the growth of all of the species tested was inhibited by zearalenone at ≥10 μg/ml. Growth of G. roseum was not inhibited by zearalenone on agar media. The zearalenone derivative α-zearalenol had growth inhibitory effects on S. fimicola similar to those of zearalenone, but β-zearalenone was less fungitoxic (Fig. 3). Growth in medium with zearalenone and zearalenols altered the morphology of S. fimicola colonies and pigment production by E. purpurascens (Fig. 3). Similarly, growth inhibition of C. herbarum by zearalenone derivatives declined in the order zearalenone > α-zearalenol > β-zearalenol (Fig. 3). In liquid media, even the growth (dry weight) of the zearalenone producer F. graminearum was slightly inhibited by zearalenone at 20 μg/ml (data not shown).
FIG. 2.
Inhibition of S. fimicola growth by zearalenone. S. fimicola was inoculated into petri dishes filled with MEA alone (control) or with MEA amended with zearalenone (ZEN) at 10 μg/ml and 30 μg/ml. Zearalenone was added to agar medium cooled to 60°C as a methanol stock solution. The final concentration of methanol was set to 5% in all three media. Inoculated plates were incubated at 20°C for 7 days under a 16-h light, 8-h dark cycle.
FIG. 3.
Effects of zearalenone derivatives on fungi. The fungal species specified were inoculated onto petri dishes filled with MEA alone (−) and medium amended with zearalenone (n), α-zearalenol (a), and β-zearalenol (b) at 20 μg/ml. Mycotoxins were added to agar medium cooled to 60°C as a methanol stock solution. The final concentration of methanol was set to 5% in all media. Inoculated plates were incubated at 20°C for 7 days under a 16-h light, 8-h dark cycle.
Disruption of zearalenone lactonase production in G. roseum by insertion of a resistance cassette into the zes2 gene.
G. roseum spores (106) were subjected to transformation with A. tumefaciens. They were plated on GM7 plates supplemented with 250 μg/ml hygromycin B, and 23 Hygr colonies were selected for further analysis.
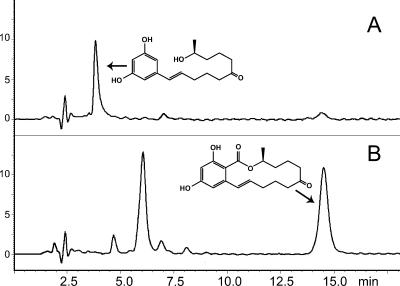
A time series of HPLC chromatograms was generated for each sample and assigned to one of two classes. The first class (Fig. 4 A) contained chromatograms of the wild-type strain and 11 putative transformants. Zearalenone was rapidly replaced with a conversion product eluting from the column at 3.8 min. The UV spectrum of the product possessed three maxima with a relative distance similar to that of the maxima of zearalenone and a hypsochromic shift of 20 nm. The mass of the molecular ion [M + 1] corresponding to this product was 293, which was 26 units less than the molecular mass of zearalenone. These data confirm the identity of the product as a hydrolysis product of zearalenone (acquisition of 18 mass units) which underwent spontaneous decarboxylation (loss of 44 mass units). We concluded that these transformants retained a zearalenone lactonase activity identical to that of the wild type, having the hygromycin resistance cassette integrated ectopically.
FIG. 4.
HPLC analyses of the products of zearalenone degradation by G. roseum. Products of the transformation of zearalenone by wild-type G. roseum (A) and a zes2 disruption mutant (B) were separated by HPLC and detected by A236. The structure of zearalenone is drawn adjacent to the zearalenone peak in chromatogram B, and the structure of the hydrolyzed and decarboxylated degradation product of zearalenone (8) is drawn adjacent to the corresponding peak of chromatogram A. A large peak at the elution time of approximately 6 min corresponds to a putative unknown transformation product of zearalenone.
The second class contained chromatograms of 12 samples (Fig. 4B). The conversion of zearalenone was much slower, leaving a substantial part of the exogenous zearalenone unchanged after 2 weeks of incubation. The conversion product eluted from the column with a retention time of 6 min, which indicates that it is more polar than zearalenone (retention time, 14.6 min). The absorption spectrum of the product(s) corresponding to this peak was indistinguishable from the UV spectrum of zearalenone, indicating that the aromatic ring and its functional groups remained unchanged. The mass of the putative molecular ion [M + 1] was 16 units higher than the mass of the molecular ion of zearalenone, suggesting that the product was an oxidized derivative of zearalenone.
Analysis of the zes2 locus in G. roseum transformants.
Hygromycin-resistant colonies were checked by PCR for the presence of the hygromycin phosphotransferase gene (primers HygBu and HygBd, Table 1) and the absence of the intact zes2 coding region. For the latter purpose, primers within the coding sequence of zes2 flanking the hygromycin cassette in pZG05 were used (OL500 and OL501, Table 1). Among 23 putative transformants examined by PCR, 11 contained both the hygromycin cassette and the intact zes2 sequence, indicating ectopic integration of the vector. These transformants metabolized zearalenone in the same manner as the wild-type strain (Fig. 4A). Twelve hygromycin-resistant strains lost the amplification product originating from the intact zes2 gene, indicating that they had undergone a double recombination event. All of these transformants exhibited type B zearalenone metabolism (Fig. 4).
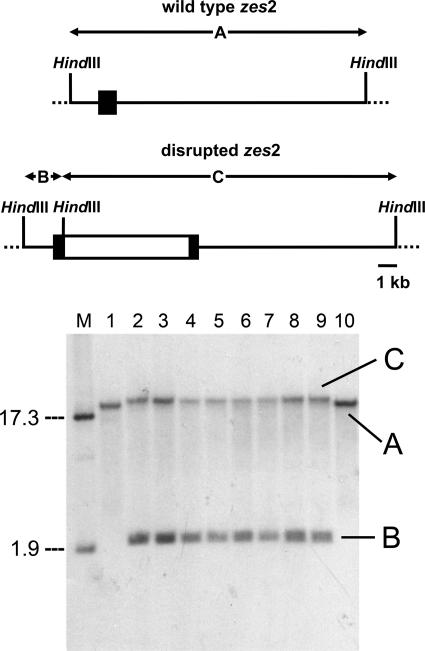
Eight potential mutants with zes2 disrupted were examined by Southern blotting and hybridization. Genomic DNA was digested with HindIII (Fig. 5), which does not cut zes2 but does have a single recognition sequence in the hygromycin resistance cassette. The sum of fragments from the zes2 disruption mutants equals the sum of the zes2 fragment from the wild-type strain plus the length of the hygromycin resistance cassette. The size of the fragment detected in the wild-type strains was estimated to be 15.6 kb, and the putative transformants produced fragments of 17.5 kb and 2.1 kb. The difference (4.0 kb) is approximately the size of the resistance cassette (4 kb), which is consistent with the hypothesis that the transformants identified were true gene replacements.
FIG. 5.
Southern hybridization analysis of zes2 disruption mutants of G. roseum. The upper panel shows a scheme of the insertional mutagenesis of zes2. A Southern blot is presented in the lower panel. Genomic DNA was digested with HindIII, separated on a 0.8% agarose gel, and hybridized with an 820-bp fragment of zes2. Lane M, size marker (in kilobases); lanes 1 and 10, untransformed wild-type strain; lanes 2 to 9, eight putative zes2 disruption mutants.
Effect of zearalenone on the growth of zes2 disruption mutants.
The growth of the zes2 mutant was retarded by zearalenone at 20 μg/ml, while the wild type was unaffected (Table 2). Ectopic transformants behaved indistinguishably from the wild type (data not shown). Colonies of wild-type G. roseum grown on agar plates containing Czapek-Dox medium with zearalenone at 20 μg/ml exhibited a fluffy phenotype indistinguishable from that of colonies grown without zearalenone, while the zes2 mutants grew as compact colonies with a reduced amount of aerial mycelium.
TABLE 2.
Effect of zearalenone on growth of G. roseum zes2 mutants
| G. roseum strain | Growth rate (mm/day)a
|
|
|---|---|---|
| Czapek-Dox medium | Czapek-Dox medium with 20 μg zearalenone/ml | |
| Wild type | 1.37 ± 0.70 | 1.46 ± 0.080 |
| zes2-1 | 1.44 ± 0.11 | 0.78 ± 0.044 |
| zes2-2 | 1.55 ± 0.14 | 0.58 ± 0.53 |
| zes2-3 | 1.54 ± 0.088 | 0.67 ± 0.087 |
Agar plates were incubated at 24°C in darkness for 12 days. Radial growth was recorded in two perpendicular directions, and means ± standard deviations of six measurements are given.
DISCUSSION
The observation that G. roseum has a lactonase activity that can hydrolyze zearalenone and the high substrate specificity of the enzyme (37) both suggest that zearalenone plays a role in the interaction of this mycoparasite with Fusarium species. Our finding of inhibition of the growth of many filamentous fungi by zearalenone supports the hypothesis that zearalenone helps Fusarium spp. reduce or inhibit the growth of many fungi. Decreased growth by the zes2 disruption mutants of G. roseum in the presence of zearalenone further supports this hypothesis. The protection of a substrate colonized by zearalenone-producing Fusarium spp. against competitors appears to be limited to fungi, since zearalenone does not affect the growth of bacteria (3) although it induces the SOS repair system in some bacterial species (13).
In spite of their economic importance and their impact on the health of humans and farm animals, the biological function of many mycotoxins remains unknown. Zearalenone was the first mycotoxin of Fusarium spp. for which a biological function was hypothesized. Nelson et al. suggested in the 1970s that zearalenone controls sexual development in Gibberella zeae (29, 38, 39, 40). This hypothesis persists (22) even though there are several lines of evidence that contradict it, including a lack of correlation between zearalenone synthesis and perithecium production (11, 36) and the fact that F. culmorum makes large amounts of zearalenone but does not reproduce sexually. (F. culmorum strains that have lost zearalenone synthesis would outgrow zearalenone producers if zearalenone did not increase their fitness by a mechanism unrelated to sexual reproduction.) Our results provide another argument to retire the sex hormone hypothesis: if zearalenone acts as an agent of interference competition, it is unlikely to control sexual reproduction. This does not, however, exclude the possibility that zearalenone exerts subtle effects on the physiology of its producers.
The existence of a functional gene encoding zearalenone-specific lactonase in G. roseum and the inducibility of this gene by zearalenone (37) indicate that the interaction of the mycoparasite with fungi producing zearalenone or similar resorcylic lactones is frequent enough to exert selection pressure for the maintenance of detoxifying lactonase. This enzyme may be one reason why Gliocladium spp. are among the most frequently found and effective fungal antagonists of F. culmorum in wheat (35). To address the ecological role of zearalenone in detail, the effect of zearalenone and its degradation on fungal fitness needs to be evaluated in mixed cultures and microcosms. The zes2 mutants described here and known non-zearalenone-producing mutants of F. graminearum (11, 22) should facilitate these studies. The results will be relevant for the assessment of the ecological impact of a large-scale release of genetically modified crops expressing zearalenone lactonase (17, 18).
Many filamentous fungi and yeasts convert zearalenone to a mixture of α- and β-zearalenol (2, 25, 28, 32, 41). β-Zearalenone production may be regarded as detoxification because this derivative has much less of an effect on fungal growth than zearalenone does (Fig. 3). Neither wild-type G. roseum nor its zes2 mutants reduced zearalenone to zearalenols in detectable amounts.
Acknowledgments
We thank Susanne Frick, Leibniz Institute of Plant Biochemistry (Halle/Saale, Germany), for providing A. tumefaciens AGL1; Hanno Wolf for providing S. fimicola; John-Bryan Speakman (BASF) and Helgard H. Nirenberg (BBA, Berlin, Germany) for taxonomic characterization of fungal isolates; Ursula Hettwer for help with HPLC; A. von Tiedemann, H. Wolf, A. de Cock, and J. Wagner for providing microbial strains; and Claudia Nordmann for technical assistance.
J. Utermark was supported by a grant from Niedersachsen Israel Funds.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Anonymous. 2005. Commission regulation (EC) no. 856/2005 amending regulation (EC) no. 466/2001 as regards to Fusarium toxins. Off. J. Eur. Union 143:3-8. [Google Scholar]
- 2.Boeswald, C., G. Engelhardt, H. Voegl, and P. R. Wallnoefer. 1995. Metabolism of the Fusarium mycotoxins zearalenone and deoxynivalenol by yeast strains of technological relevance. Nat. Toxins 3:138-144. [DOI] [PubMed] [Google Scholar]
- 3.Boutibonnes, P. 1978. A preliminary report about antibacterial properties of the mycotoxin zearalenone. Pharmacology 4:491-492. [Google Scholar]
- 4.Brandfass, C., and P. Karlovsky. 2006. Simultaneous detection of Fusarium culmorum and F. graminearum in plant material by duplex PCR with melting curve analysis. BMC Microbiol. 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coe, J. E., K. G. Ishak, J. M. Ward, and M. J. Ross. 1992. Tamoxifen prevents induction of hepatic neoplasia by zeranol, an estrogenic food contaminant. Proc. Natl. Acad. Sci. USA 89:1085-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covert, S., P. Kapoor, M. Lee, A. Briley, and C. J. Nairn. 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105:259-264. [Google Scholar]
- 7.de Groot, M. J. A., P. Bundock, P. J. J. Hooykaas, and A. G. M. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 8.El-Sharkawy, S., and Y. J. Abul-Hajj. 1988. Microbial cleavage of zearalenone. Xenobiotica 18:365-371. [DOI] [PubMed] [Google Scholar]
- 9.Eriksen, G. S., and J. Alexander. 1998. Nordic Council of Ministers TemaNord no. 502. Nordic Council of Ministers, Copenhagen, Denmark.
- 10.Farnworth, E. R., and H. L. Trenholm. 1983. The metabolism of the mycotoxin zearalenone and its effects on the reproductive tracts of young male and female pigs. Can. J. Anim. Sci. 63:967-975. [Google Scholar]
- 11.Gaffoor, I., D. W. Brown, R. Plattner, R. H. Proctor, W. Qi, and F. Trail. 2005. Functional analysis of the polyketide synthase genes in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Eukaryot. Cell 4:1926-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlach, W., and H. I. Nirenberg. 1982. The genus Fusarium—a pictorial atlas. Mitt. Biol. Bundesanst. Land-Forstwirtsch. Berlin-Dahlem 209:1-406. [Google Scholar]
- 13.Ghedira-Chekir, L., K. Maaroufi, A. Zakhama, F. Ellouz, S. Dhouib, E. E. Creppy, and H. Bacha. 1998. Induction of a SOS repair system in lysogenic bacteria by zearalenone and its prevention by vitamin E. Chem. Biol. Interact. 113:15-25. [DOI] [PubMed] [Google Scholar]
- 14.Green, M. L., M. A. Diekman, J. R. Malayer, A. B. Scheidt, and G. G. Long. 1990. Effect of prepubertal consumption of zearalenone on puberty and subsequent reproduction of gilts. J. Anim. Sci. 68:171-178. [DOI] [PubMed] [Google Scholar]
- 15.Grosse, Y., L. Chekir-Ghedira, A. Huc, S. Obrecht-Pflumio, G. Dirheimer, H. Bacha, and A. Pfohl-Leszkowicz. 1997. Retinol, ascorbic acid and α-tocopherol prevent DNA adduct formation in mice treated with the mycotoxins ochratoxin A and zearalenone. Cancer Lett. 114:225-229. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, J. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Higa, A., M. Kimura, K. Mimori, T. Ochiai-Fukuda, T. Tokai, N. Takahashi-Ando, T. Nishiuchi, T. Igawa, M. Fujimura, H. Hamamoto, R. Usami, and I. Yamaguchi. 2003. Expression in cereal plants of genes that inactivate Fusarium mycotoxins. Biosci. Biotechnol. Biochem. 67:914-918. [DOI] [PubMed] [Google Scholar]
- 18.Higa-Nishiyama, A., N. Takahashi-Ando, T. Shimizu, T. Kudo, I. Yamaguchi, and M. Kimura. 2005. A model transgenic cereal plant with detoxification activity for the estrogenic mycotoxin zearalenone. Transgenic Res. 14:713-717. [DOI] [PubMed] [Google Scholar]
- 19.Kakeya, H., N. Takahashi-Ando, M. Kimura, R. Onose, I. Yamaguchi, and H. Osada. 2002. Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of Clonostachys sp. Biosci. Biotechnol. Biochem. 66:2723-2726. [DOI] [PubMed] [Google Scholar]
- 20.Karlovsky, P. 1994. Inhibition of imidazoleglycerolphosphate dehydratase of Phytophthora parasitica by aminotriazole in situ and after cloning and expression of the respective gene (HIS3) in Escherichia coli. J. Phytopathol. 141:121-126. [Google Scholar]
- 21.Karlovsky, P. 1999. Biological detoxification of fungal toxins and its use in plant breeding, feed and food production. Natural Toxins 7:1-23. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. T., Y. R. Lee, J. Jin, K. H. Han, H. Kim, J. C. Kim, T. Lee, S. H. Yun, and Y. W. Lee. 2005. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol. Microbiol. 58:1102-1113. [DOI] [PubMed] [Google Scholar]
- 23.Lazo, G. R., P. A. Stein, and R. A. Ludwig. 1991. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9:963-967. [DOI] [PubMed] [Google Scholar]
- 24.Long, G. G., and M. A. Diekman. 1986. Characterization of effects of zearalenone in swine during early pregnancy. Am. J. Vet. Res. 47:184-187. [PubMed] [Google Scholar]
- 25.Matsuura, Y., and T. Yoshizawa. 1985. Conversion of zearalenone, an estrogenic mycotoxin, by brewing microorganisms. J. Food Hyg. Soc. Jpn. 26:24-28. [Google Scholar]
- 26.Mattanovich, D., F. Rüker, A. da Camara, A. Machado, M. Laimer, F. Reguer, H. Steinkellner, G. Himmler, and H. Katinger. 1989. Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 17:6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molnar, O., G. Schatzmayr, E. Fuchs, and H. Prillinger. 2004. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 27:661-671. [DOI] [PubMed] [Google Scholar]
- 28.McMullen, J. R. 1977. Microbiological reduction of zearalenone and related compounds. U.S. patent 4,004,978.
- 29.Nelson, R. R. 1971. Hormonal involvement in sexual reproduction in the fungi, with special reference to F-2, a fungal estrogen, p. 181-200. In S. Akai and S. Ouchi (ed.), Morphological and biochemical events in plant-parasite interaction. Phytopathological Society of Japan, Tokyo, Japan.
- 30.Pfohl-Leszkowicz, A., L. Chekir-Ghedira, and H. Bacha. 1995. Genotoxicity of zearalenone, an estrogenic mycotoxin: DNA adduct formation in female mouse tissues. Carcinogenesis 16:2315-2320. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Scott, P. M., S. R. Kanhere, E. F. Daley, and J. M. Farber. 1992. Fermentation of wort containing deoxynivalenol and zearalenone. Mycotoxin Res. 8:58-66. [DOI] [PubMed] [Google Scholar]
- 33.Szuetz, P., A. Mesterhazy, G. Y. Falkay, and T. Bartok. 1997. Early telearche symptoms in children and their reaction to zearalenone contamination in food stuffs. Cereals Res. Commun. 25:429-436. [Google Scholar]
- 34.Takahashi-Ando, N., M. Kimura, H. Kakeya, H. Osada, and I. Yamaguchi. 2002. A novel lactonohydrolase responsible for the detoxification of zearalenone: enzyme purification and gene cloning. Biochem. J. 365:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teperi, E., M. Keskinen, E. Ketoja, and R. Tahvonen. 1998. Screening for fungal antagonists of seed-borne Fusarium culmorum on wheat using in vivo tests. Eur. J. Plant Pathol. 104:243-251. [Google Scholar]
- 36.Windels, C. E., C. J. Mirocha, H. K. Abbas, and W. Xie. 1989. Perithecium production in Fusarium graminearum populations and lack of correlation with zearalenone production. Mycology 81:272-277. [Google Scholar]
- 37.Woerfel, G., and P. Karlovsky. 1998. Hydrolyse von Zearalenon durch Gliocladium roseum, p. 189-192. In J. Wolff and T. Betsche (ed.), 20th Mycotoxin Workshop. Gesellschaft für Mykotoxinforschung, Detmold, Germany.
- 38.Wolf, J. C., and C. J. Mirocha. 1973. Regulation of sexual reproduction in Gibberella zeae (Fusarium roseum “graminearum”) by F-2 (zearalenone). Can. J. Microbiol. 19:725-734. [DOI] [PubMed] [Google Scholar]
- 39.Wolf, J. C., and C. J. Mirocha. 1977. Control of sexual reproduction in Gibberella zeae (Fusarium roseum “Graminearum”). Appl. Environ. Microbiol. 33:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf, J. C., J. R. Liebermann, and C. J. Mirocha. 1972. Inhibition of F-2 (zearalenone) biosynthesis and perithecium production in Fusarium roseum “Graminearum.” Phytopathology 62:937-939. [Google Scholar]
- 41.Wu, L., Q. Wang, S. Wang, and F. Zhou. 1992. Biotransformation of zearalenone. Acta Microbiol. Sin. 32:11-16. [PubMed] [Google Scholar]