Abstract
Methanogenesis in cold marine sediments is a globally important process leading to methane hydrate deposits, cold seeps, physical instability of sediment, and atmospheric methane emissions. We employed a multidisciplinary approach that combined culture-dependent and -independent analyses with geochemical measurements in the sediments of Skan Bay, Alaska (53°N, 167°W), to investigate methanogenesis there. Cultivation-independent analyses of the archaeal community revealed that uncultivated microbes of the kingdoms Euryarchaeota and Crenarchaeota are present at Skan Bay and that methanogens constituted a small proportion of the archaeal community. Methanogens were cultivated from depths of 0 to 60 cm in the sediments, and several strains related to the orders Methanomicrobiales and Methanosarcinales were isolated. Isolates were psychrotolerant marine-adapted strains and included an aceticlastic methanogen, strain AK-6, as well as three strains of CO2-reducing methanogens: AK-3, AK7, and AK-8. The phylogenetic positions and physiological characteristics of these strains are described. We propose a new species, Methanogenium boonei, with strain AK-7 as the type strain.
More than 85% of the ocean's organic carbon is deposited in shallow, anoxic marine sediments (8). Methanogens play an important role in the degradation of this organic matter by converting low-molecular-weight compounds into methane. In marine sediments, methane is present in dissolved form in the pore water, as free gas, or trapped within methane hydrates. Molecular and isotopic data suggest that most of the methane produced in marine sediments is of biogenic origin (31); however, methanogens that inhabit permanently cold marine sediments have been poorly characterized.
Several studies have investigated the microbial diversity in cold marine sediments (12, 43, 46); however, the role that these microbes play in mediating geochemical processes is still not completely understood. In addition to methanogens, uncultivated lineages of other Archaea have been identified in marine sediments (22, 29, 32, 37, 41, 67). These include phylotypes belonging to the marine benthic groups B (MBG-B), C, and D and the anaerobic methane-oxidizing (ANME) archaeal groups ANME-1 and ANME-2.
Skan Bay has previously served as a model system for studying low-temperature microbial processes in marine sediments (2, 3, 47, 51, 66). The sediments contain clear environmental gradients (66), and the complete redox sequence from oxygen to methane occurs within 0.5 m of the surface, allowing for good depth resolution of microbial activities (20). Previous analyses of Skan Bay sediments have shown that microbes mediate important geochemical processes, including sulfate reduction, methane production, and the anaerobic oxidation of methane (AOM) (66). The specific aim of this study was to identify and cultivate Archaea involved in methane cycling at Skan Bay. Cultivation-independent and -dependent techniques were employed to characterize the archaeal populations at various depths and therefore along geochemical gradients and also to characterize the methanogens present in such sediments.
MATERIALS AND METHODS
Sample collection and source of inocula.
All enrichments and strains described here were obtained from sediment samples collected from Skan Bay, Alaska (57°N, 167°W). Skan Bay is located on the northwest side of Unalaska Island in the Aleutian Island chain. Skan Bay is 65 m deep at the sampling site, and the temperature of the sediment remains between 1°C and 6°C year round. Large amounts of organic matter (2 to 3% of sediment by dry weight), sedimented from diatoms and kelp, are present.
Sediment was collected during three cruises to Skan Bay that occurred during September 1997; September to October 2001; and August to September 2003. Sediment was collected using a box corer or gravity corer at the deepest part of the Bay (65 m). Box cores were subcored as described by Alperin and Reeburgh (4). Sediment samples were collected from discrete depths from the sediment-water interface to the bottom of each core by inserting the barrel of a syringe into predrilled holes in polycarbonate core tubes. The sediment samples were immediately sealed in cans (while excluding air and gas) and stored at 4°C during transport to the laboratory. Sediment samples used to prepare clone libraries were preserved in a phosphate-buffered saline solution or 50% ethanol and stored at −20°C.
Geochemical analyses.
High-precision carbon and nitrogen weight percent determinations were made using a Control Equipment Corporation automated organic elemental analyzer, model 440XL. Instrument calibration was performed using acetanilide (organic analytical standard grade); quality control procedures included initial confirmation of calibration accuracy with a control standard and periodic checking of calibration drift with check standards. Frozen samples were dehydrated, crushed to a fine powder, and treated with phosphoric acid to remove CaCO3, prior to analysis. The weight percent of carbon and nitrogen was converted to a molar value in order to calculate the C-to-N ratio. Standard errors were ±3% for total organic carbon and ±4% for total nitrogen, based on replicate injections of standards.
The depth distributions of C1 to C5 volatile organic acids were quantified in pore fluids extracted from freshly collected sediment by using high-performance liquid chromatography. The method is based on that of Albert and Martens (1), as modified previously (65), and involves chemical derivatization and preconcentration of the organic acids prior to chromatographic separation and detection. Depth distributions of dissolved sulfate were quantified in pore fluids from freshly collected cores using the nephelometric method of Gieskes et al. (18) and modified as previously described (69). This method involves the addition of BaCl2 to an aliquot of the sample and the quantification of turbidity caused by the precipitation of BaSO4. Depth distributions of dissolved methane were quantified in pore fluids from freshly collected cores by using a headspace equilibration technique similar to that described previously (64). Sediment samples were collected laterally from preperforated core liners using 3-ml-volume syringes with their tips removed. The contents of the syringes were immediately injected into a serum bottle containing 10 ml of a degassed solution of saturated NaCl, then capped and crimped. The samples were shaken and allowed to equilibrate for 24 h prior to analysis. Quantification was performed using a gas chromatograph equipped with a flame ionization detector (model 8A; Shimadzu). Depth distributions of dissolved sulfides were approximated in pore fluids from freshly collected cores by using a visual color assay involving lead acetate paper. Digital images were collected for later comparison. Standard errors were estimated conservatively at ±10% for organic acids, ±5% for sulfate, and ±5% for methane, based on repeated injections of standards.
Culture techniques and media.
The anaerobic culture techniques of Hungate (23), as modified by Sowers and Noll (56), were used in this study. MSH enrichment medium (39) was used as the basal medium for characterization of the strains. The medium was prepared at pH 7.0, with a gas phase mixture of N2 and CO2 (7:3). The salinity of the medium was 0.3 M Na+. To prevent O2 from diffusing through the stoppers during the long incubation times necessary for growth, cultures were incubated in metal paint cans, as described previously (26), at 15°C.
Measurement of growth and determination of growth characteristics.
Growth was estimated from the accumulation of methane in the headspace gas as measured by gas chromatography (34) and taking into account the methane produced during the growth of the inoculum (45). The specific growth rate was calculated by fitting the Gompertz equation (73) to the rate of methane production. The optimal temperature for growth was determined from the specific growth rates of cultures incubated at various temperatures. All growth rate experiments were performed in triplicate.
Enrichment and isolation of methanogens.
Samples collected during the 1997 cruise to Skan Bay were used for all isolations described in the present paper. A 1-g sample from each sediment depth interval was inoculated into MSH enrichment medium (pH 7) containing H2, acetate, or trimethylamine (TMA) as catabolic substrate; these three substrates were intended to enrich for CO2-reducing, aceticlastic, and methylotrophic methanogens, respectively. The suspension was mixed and serially diluted to a final dilution of 10−3. The cultures were serially diluted, incubated for 6 to 12 months at 15°C, and inoculated onto roll tube media (29). Colonies were picked, serially diluted, and subjected to a second roll tube isolation in order to ensure purity. The purity of the cultures was checked microscopically and by ensuring that only a single colony type was seen in the roll tube medium.
Enrichment cultures for determining the distribution of methanogens were prepared for 10 distinct sediment depth intervals, ranging from a surface interval (0 to 5 cm) to a deep interval (63 to 69 cm), using the substrates described above (see Table S1 in the supplemental material). In total, 270 tubes were inoculated (10 depths × three substrates × three dilutions × three replicates). Methane production was monitored over the course of 1 year.
Electron microscopy.
Cells in the late-exponential growth phase were directly fixed in culture medium by adding an equal volume of 5% glutaraldehyde. The cells were fixed for 30 min at 4°C and then collected by centrifugation. The cells were fixed for 30 min in cacodylate buffer (pH 7.2) containing 1% osmium tetroxide, dehydrated in an ethanol series, and embedded in low-viscosity epoxy resin for sectioning. Ultrathin sections were poststained with uranyl acetate and lead citrate and viewed with a Zeiss EM-10CA transmission electron microscope.
DNA extraction.
DNA was extracted from pure cultures of AK-3, AK-6, AK-7, and AK-8 by following QIAamp DNA mini kit (QIAGEN, Inc.) procedures. DNA from enrichment cultures and sediment samples was extracted by using a modified cetyltrimethylammonium bromide method (7) as described previously (26). Samples were resuspended in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) with 7.5% Chelex (Sigma), 0.05 EDTA (pH 7.0), 1% sodium dodecyl sulfate (SDS), and 200 μg of proteinase K for a final volume of 750 μl. Samples were incubated while being slowly rotated for 1 h at 37°C. Chelex was removed by centrifugation. Then 100 μl of 5 M NaCl and 80 μl of 10% (wt/vol) cetyltrimethylammonium bromide in 0.7 M NaCl were added, and the samples were incubated for 30 min at 65°C. The DNA solution was extracted once with chloroform:isoamyl alcohol (24:1) and then twice with phenol:chloroform:isoamyl alcohol (25:24:1). DNA was precipitated with an equal volume of 2-propanol, washed with 70% ethanol, and resuspended in 10 mM Tris buffer (pH 8.0).
DNA for determination of base composition and DNA hybridization experiments was extracted and purified by a modified Marmur method (35) as described previously (53). Briefly, cells were hypotonically lysed, and the lysate was mixed with chloroform-isoamyl alcohol. DNA was extracted into the aqueous phase and then precipitated by mixing 1:1 (vol/vol) with cold ethanol. The DNA was spooled onto a glass rod and dissolved into saline citrate solution (0.15 M NaCl, 0.015 M trisodium citrate, pH 7.0).
Amplification and sequencing of 16S rRNA genes.
A portion of the 16S rRNA gene was amplified by PCR from genomic DNA by using the archaeum-specific primer 4F (5′-TCCGGTTGATCCTGCCRG-3′) and the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′). PCR was performed by standard methods (50). PCR products were purified with an UltraClean PCR clean-up kit (Mo Bio Laboratories, Carlsbad, CA).
Purified PCR products from the plasmid clones were used as the templates for sequence analysis. Sequencing reactions were performed by using an ABI PRISM Big Dye Terminator cycle sequencing kit and an ABI 3100 Avant genetic analyzer. The complete sequences of both strands were obtained by using the primers 4F (described above), 515F (5′-GTGCCAGCMGCCGCGGTAA-3′), 1100F (5′-GGCAACGAGCGMGACCC-3′), 1492R (described above), 907R (5′-CCGTCAATTCCTTTRAGTTT-3′), and 340RA (5′-CCCCGTAGGGCCYGG-3′) to produce an overlapping set of sequences that were assembled into one contiguous sequence by using the AutoAssembler program (ABI).
Amplification and sequencing of the mcrA gene.
DNA was extracted from a pure culture of AK-7 and from pure cultures of the type strains of the following closely related methanogens: Methanogenium marinum, Methanogenium organophilum, Methanogenium cariaci, Methanogenium frigidum, and Methanoculleus submarinus, which were obtained from the Oregon Collection of Methanogens (OCM). The mcrA gene was amplified by PCR from genomic DNA. Reaction mixtures and procedures were performed as described for the 16S rRNA, substituting the primers mcrA S (5′-TAYGAYCARATHTGGYT-3′, where Y means C or T, R means A or G, and H means A, C, or T) and mcrA AS (5′-ACRTTCATNGCRTARTT-3′).
DNA base composition.
The G+C content of the genomic DNA was calculated by using the thermal denaturation method developed by Marmur and Doty (36). The melting temperature (Tm) of each isolate's DNA was determined by thermal denaturation in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) buffer with a Varian Cary UV spectrophotometer. Escherichia coli (ATCC 1174), with a Tm of 51.7°C (57), was used as the standard during each determination. The Tm value was then substituted into the Sly equation (54) to calculate the moles percent of guanine plus cytosine.
DNA-DNA hybridization.
DNA-DNA hybridization values of AK-3, AK-7, and AK-8 with Methanogenium cariaci, M. marinum, and M. organophilum were determined quantitatively by following the method described by De Ley et al. (16). The DNA was diluted to a final concentration of 50 μg/ml in 1× SSC buffer (24). DNA was sheared into 700-base-pair fragments by sonication (Microson TM SL 2000; Misonix) for 2 min. The fragment size was verified by electrophoresis and comparison to a 1-kb ladder (Invitrogen).
Cloning and RFLP.
The PCR products were cloned into the pCR2.1 vector by using the TOPO-TA cloning kit (Invitrogen, Carlsbad, Calif.), amplified, and separated as described previously (26). A total of 448 clones were analyzed, and at least one phylotype from each group (as determined by restriction fragment length polymorphism [RFLP] banding pattern and/or ≥97% sequence similarity) was subject to a complete nucleotide analysis.
Richness and coverage estimators.
To estimate richness and coverage, sequences from each depth were collected into operational taxonomic units (OTUs) based on ≥97% sequence similarity. OTU richness and coverage estimators were calculated by using the software program EstimateS (15) with default settings and 100 random sample repetitions. Species richness was estimated by using the nonparametric model of Chao (13).
Phylogenetic analyses.
Nearly complete rRNA gene sequences were manually aligned to related sequences obtained from GenBank. First the sequences were automatically aligned with the program Clustal_X (61), and then the alignments were checked manually in MacClade version 4.0 (33). Ambiguous nucleotides were omitted in the final analyses. We used the method described by Ashelford et al. (6) to identify chimeras. Six chimeras were detected in the clone libraries and excluded from further analyses. The phylogenetic relationships of the environmental clones were determined by using neighbor-joining analysis in PAUP* (59). The sequences used in the final analysis were between 1,232 and 1,427 bases long. The phylogenetic relationships of the cultivated methanogens were determined for the 16S rRNA and the mcrA genes by maximum-likelihood analysis using the HKY-85 model of evolution in PAUP* (59). Bootstrap analyses were performed using 1,000 trial replications to provide confidence estimates for branch support.
Nucleotide sequence accession numbers.
Sequences for the following were deposited in GenBank (accession numbers are in parentheses): the AK-7 16S rRNA gene (DQ177343), the AK-3 16S rRNA gene (DQ177345), the AK-8 16S rRNA gene (DQ250386), the AK-6 16S rRNA gene (DQ250385), the CO2-reducing and methylotrophic enrichment cultures (DQ280483 to DQ280487), the clone libraries (DQ522901 to DQ522945 and DQ640134 to DQ640236), and the mcrA gene (DQ229156 to DQ229161).
RESULTS
Sediment geochemistry.
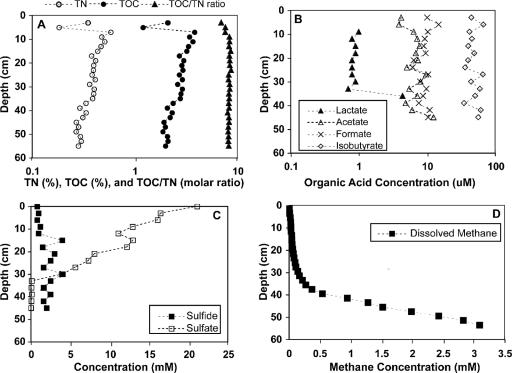
In order to provide an environmental context for the microbiological studies, geochemical depth distributions for a variety of chemical parameters were determined. These parameters included total organic carbon and nitrogen, short-chain volatile organic acids, sulfate, sulfide, and methane (Fig. 1). Because Skan Bay is a silled basin, the bottom water is generally low in oxygen, and the sediment is permanently anoxic (47). Sulfate serves as the primary electron acceptor in the surficial sediments, and alternative electron acceptors such as Fe(III), NO3−, and Mn(IV) are depleted within 1 to 2 cm of the sediment-water interface (2). The high levels of organic material and the presence of sulfide in the surface sediment support this interpretation (Fig. 1A and C). Depth distributions of sulfate and methane (Fig. 1C and D) indicate zonation within the sediments, including an upper sulfate-reducing zone (∼0 to 30 cm), a methane-oxidizing/sulfate-reducing zone (∼30 to 45 cm), and a methanogenic zone (>45 cm). The deeper sediments are characterized by abundant methane (Fig. 1D) and are presumed to be actively methanogenic. A biogenic origin for the methane is supported by the δ13C of methane (∼−80‰) (66) and the trend of δ13C enrichment in cooccurring CO2 (2). Methane is oxidized anaerobically at the interface between the zones of methanogenesis and sulfate reduction, at depths of between 30 and 45 cm (63). The geochemical gradients reported here are consistent with results from previous cruises to Skan Bay (4, 47, 51); therefore, Skan Bay is a stable environment to study depth-related microbial processes.
FIG. 1.
Depth distributions of chemical parameters from the sediments of Skan Bay, AK. (A) Weight percent of total organic carbon (TOC) and total nitrogen (TN) as well as the molar ratio of carbon to nitrogen in the bulk sediment. Note the log scale. (B) Concentrations of lactate, acetate, formate, and isobutyrate in sediment pore fluids. Identities of the organic acids are based on retention time. Note the log scale. (C) Concentrations of sulfate and sulfide dissolved in sediment pore fluids. (D) Concentration of methane dissolved in the sediment pore fluid.
Clone libraries.
Archaeal 16S rRNA gene libraries were constructed from the shallow, mid, and deep zones in the sediment. Zones were determined based on geochemical gradients in the sediment and the available electron acceptors (Fig. 1C and D). The shallow zone included sediment sampled from 3 to 28 cm below the sediment water interface (the zone of sulfate reduction), the mid zone included sediment collected from depths of 33 to 45 cm (the zone where AOM occurs), and the deep zone included sediment collected from depths of 56 to 87.5 cm (the zone of methanogenesis).
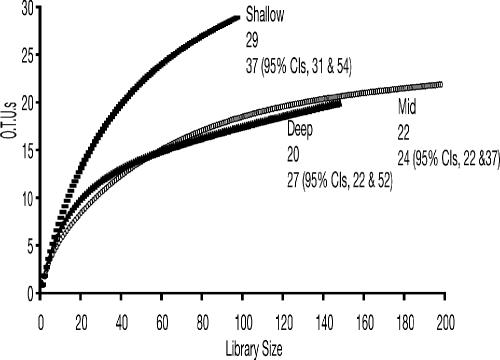
A total of 442 clones (Table 1) were included in the final analyses. RFLP analysis was used to distinguish clones. At all depths, all clones that produced unique restriction fragments were partially sequenced (≥600 bp), and a 16S rRNA gene sequence similarity of ≥97% was chosen to group phylotypes (25). All clones that were included in the final phylogenetic analyses were almost completely sequenced (>1,200 bp). Rarefaction analysis was used to determine sampling effort. A strongly curvilinear rarefaction curve provides evidence that diversity was exhaustively sampled (25). The analysis suggested that diversity was well sampled in all zones (Fig. 2). A nonparametric analysis of estimated species richness did not predict significant differences in the number of phylotypes present at each zone in the sediment (Fig. 2).
TABLE 1.
Comparison of clone abundance by depth and phylogenetic divisiona
| Phylogenetic group | No. of clones
|
|||
|---|---|---|---|---|
| Total | Shallow | Mid | Deep | |
| Euryarchaeota | 328 | 73 | 167 | 88 |
| ANME-1 | 3 | 1 | 2 | 0 |
| ANME-2 | 69 | 0 | 54 | 15 |
| MBG-D | 226 | 65 | 97 | 64 |
| Methanomicrobiales | 4 | 1 | 3 | 0 |
| Methanosarcinales | 1 | 0 | 1 | 0 |
| Other Euryarchaeota | 25 | 6 | 10 | 9 |
| Crenarchaeota | 114 | 24 | 29 | 61 |
| MBG-B1 | 50 | 9 | 9 | 32 |
| MBG-B2 | 58 | 15 | 14 | 29 |
| MBG-C | 6 | 0 | 6 | 0 |
| Total | 442 | 97 | 196 | 149 |
Shallow-, mid-, and deep-zone sediments were collected from 3 to 28, 33 to 45, and 56 to 87.5 cm below the sediment-water interface, respectively.
FIG. 2.
Rarefaction curves for the clone libraries prepared from shallow, mid, and deep zones in the sediments. The top numbers at each depth are observed OTUs. Numbers on the bottom line represent predicted OTUs with 95% upper and lower confidence intervals, respectively, in parentheses.
The majority of the clones were most closely related to uncultivated lineages (Table 1; see Fig. S1 and S2 in the supplemental material). The phylotypes that grouped within the Euryarchaeota were the most abundant overall and the most abundant detected at each depth (Table 1). The most common phylotype clustered within MBG-D (Table 1). Although no members of this group have been cultivated, MBG-D Archaea are ubiquitous in marine environments: they have been detected at hydrothermal vents (60), in methane seep sediments (22, 29, 42), and in deep marine sediments (67).
Clones that were related to the ANME-1 and ANME-2 groups of methanotrophic Archaea were also detected (Table 1). The ANME-1 clones were a small proportion of the clone library, and this group was detected only in the shallow- and mid-zone sediments. ANME-2 clones were more abundant, and these sequences were only detected in sediments from the mid and deep zones. ANME-1 is distantly related to cultivated methanogens, and the ANME-2 group is related to characterized methanogens of the order Methanosarcinales. Both groups are suspected to mediate AOM (9, 19). These groups of Archaea have been detected in marine environments characterized by high-methane partial pressures (i.e., seep and hydrate sediments) (22, 29, 32) and in shallow marine sediments with methane levels of ca. 1 atm (62). The presence of methane-oxidizing Archaea in the sulfate-depleted deep sediments was surprising, as the oxidation of methane is coupled to sulfate reduction (9).
Only five clones in all the libraries were associated with cultivated methanogens (Table 1). Four clones that were related to the Methanomicrobiales were detected in the shallow- and mid-zone sediments. Cultivated methanogens belonging to the Methanomicrobiales gain energy by CO2 reduction with hydrogen or sometimes formate, but they require supplemental organic material for growth (11). One clone that was isolated from the mid zone grouped with environmental sequences that were most closely related to the methanogenic genus Methanosaeta. Cultivated methanogens belonging to this genus catabolize only acetate for energy (11).
The remaining euryarchaeotal clones were not affiliated with a specific clade (see Fig. S1 in the supplemental material). The most closely related sequences were obtained from cold seep sediments (5).
The Crenarchaeota sequences clustered into three distinct clades (Table 1). The majority of the clones clustered into two related clades that were labeled MBG-B1 and MBG-B2. Sequences grouping within the clades MBG-B1 and MBG-B2 were detected at every depth in the sediment. These Archaea have been detected in numerous marine environments, including hydrothermal vents (60), methane seeps (42), deep sediments (67), hydrate-bearing sediments (37), and freshwater sediments (21).
The remaining sequences were related to clones previously identified as MBG-C (67) (Table 1). These clones (n = 6) were unique to the mid-zone sediments and constituted a minor proportion of the library. MBG-C sequences have been recovered from deep-sea sediments (67) and methane seep sites (32).
Cultivation, isolation, and characterization of methanogens.
Growth of methanogens cultivated from Skan Bay was monitored for 1 year, at which time DNA was extracted and amplified from the highest dilutions that produced methane. At least one enrichment culture from each phylogenetic group (≥97% sequence similarity) was subjected to 16S rRNA gene sequencing (>1,300 bp) and phylogenetic analysis. The sequencing chromatographs suggested that the Archaea in each of these enrichment cultures each had a single 16S rRNA gene, such that each of these enrichments contained a single kind of methanogen as the only detectable phylotype.
Methylotrophic methanogens.
Based on phylogenetic analyses of the enrichment cultures, methylotrophic methanogens were present at all depths in the sediment. All cultures that produced methane were phylogenetically closely related to described species of the genus Methanococcoides (see Table S1 in the supplemental material). Methylated compounds are the only substrates known to support growth of all characterized species within this genus (11).
Aceticlastic methanogen.
An aceticlastic methanogen, strain AK-6, was isolated from the interval 32 to 39 cm deep in the sediment. This strain has been deposited in the OCM as OCM 805 and in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) as DSM 17370. Efforts to cultivate aceticlastic methanogens from other depths in the sediment were not successful (see Table S1 in the supplemental material).
Colonies of strain AK-6 formed after 8 months of incubation at 15°C. The colonies were brown in color, irregularly shaped, and 0.5 to 1.0 mm in diameter. Strain AK-6 was a nonmotile sarcina and was seen as aggregates when viewed with a light microscope. Cells autofluoresced and measured 0.8 to 2.0 μm in diameter.
Strain AK-6 grew at temperatures between 10° and 35°C, at the pH range of 5.2 to 7.2, and from 0.2 to 0.9 M Na+. Strain AK-6 grew when acetate, methanol, or trimethylamine was added as a catabolic substrate. Strain AK-6 did not catabolize H2 plus CO2 or formate. Strain AK-6 did not require additional nutrients; however, growth was stimulated by the addition of yeast extract or Trypticase peptone to the medium.
Phylogenetic analysis of the 16S rRNA gene indicated that strain AK-6 was most closely related to strain FRX-1, which was isolated from freshwater sediments in Lake Fryxell, Anarctica (53), and to Methanosarcina lacustris, which was isolated from freshwater sediments in Switzerland (52) (see Fig. S3 in the supplemental material). The gene sequence similarities were 99% and 98%, respectively. Several phenotypic differences between strain AK-6 and Methanosarcina lacustris exist. Methanosarcina lacustris grows over broader pH and temperature ranges; salinity growth ranges were not reported for Methanosarcina lacustris (33). Additionally, Methanosarcina lacustris grew on H2 plus CO2 but did not catabolize acetate and required organic growth factors (33). Although strain AK-6 can be distinguished phenotypically from M. lacustris, DNA-DNA hybridization experiments were not conducted; thus, it cannot be concluded that strain AK-6 represents a new species (70).
CO2-reducing methanogens.
CO2-reducing methanogens were cultivated from samples collected at all depths in the sediment (see Table S1 in the supplemental material), and these methanogens were phylogenetically related to previously described species of the genus Methanogenium. Three strains of CO2-reducing methanogens were isolated: strain AK-7 was isolated from the interval 39 to 41 cm deep in the sediment, and strains AK-3 and AK-8 were isolated from the interval 32 to 39 cm deep in the sediment. These strains have been deposited in the OCM as OCM 787 (AK-7), OCM 773 (AK-3), and OCM 792 (AK-8). Strain AK-7 has also been deposited in the DSMZ as DSM 17338.
Morphology.
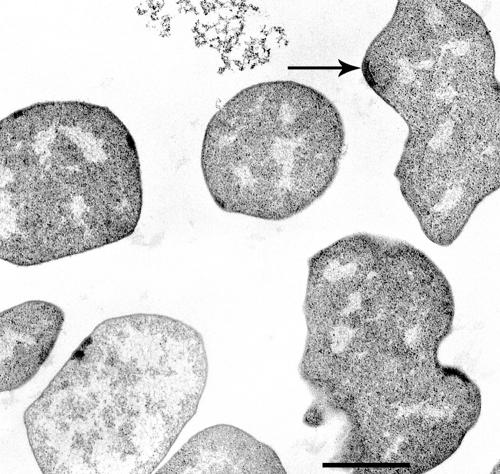
Strain AK-7 formed colonies after 3 months of incubation at 15°C, at which time the surface colonies were colorless, transparent, and round, measuring 0.5 mm in diameter. Typical cells were 1.0 to 2.5 μm in diameter, irregularly coccoid, and singly occurring (Fig. 3). Transmission electron microscopy showed the absence of a cell well other than an S-layer (data not shown). In several cells, a plaque indicating a putative flagellum basal area was observed near the periphery of the cell. However, no flagella were detected, and these cells were nonmotile when viewed under a light microscope. The cells were autofluorescent and stained gram negative. The cells lysed immediately when 0.1 g of SDS per liter of culture was added, indicating a proteinaceous cell wall (10).
FIG. 3.
Electron micrograph of strain AK-7. Transmission electron micrograph shows the irregular coccoid morphology. The arrow indicates a flat plaque that appears to be a flagellum basal area. Bar, 1 μm.
Strain AK-3 formed colonies after 4 months of incubation at 15°C. The colonies measured 0.5 to 1.0 mm in diameter and were light yellow and transparent with an entire edge. The cells were irregular cocci that measured 1.5 to 3.0 μm in diameter and autofluoresced. The cells stained gram negative and lysed when SDS was added to the medium. Strain AK-8 formed colonies after 6 months of incubation at 15°C. The colonies were light yellow, round, and transparent and measured 1.5 mm in diameter. The cells lysed when SDS was added to the medium.
Growth characteristics.
Minimal, optimal, and maximum values for temperature, salinity, and pH that supported the growth of strain AK-7 were determined (see Fig. S4 in the supplemental material). The growth of AK-7 was measured from 5°C to 30°C. Strain AK-7 grew optimally at 19.4°C; the minimum growth temperature measured was 5°C, and the maximum growth temperature was 25.6°C. Strain AK-7 grew most rapidly at a Na+ concentration of 0.3 M, and cells grew in the range 0.1 M to 0.8 M. Growth of strain AK-7 occurred in the pH range from 6.4 to 7.8 but not at pHs of 6.2 or 8.0.
Strains AK-3 and AK-8 grew from 15°C to 25°C; no growth occurred at 30°C, and temperatures lower than 15°C were not tested. Strain AK-3 grew in the pH range from 5.1 to 7.25. Strain AK-3 grew fastest at 0.3 to 0.5 M Na+, and cells grew in the range from 0.2 to 0.9 M. Salinity and pH growth experiments were not conducted for strain AK-8.
The catabolic range was determined by amending cultures with common methanogenic substrates, including CO2 plus H2 (70 kPa), 10 mM acetate, 20 mM TMA, and 40 mM formate. Strains AK-7, AK-3, and AK-8 grew by reducing CO2 with H2 and formate, and no growth occurred when either acetate or TMA was added as a catabolic substrate. Additional substrate tests were undertaken for strain AK-7 by amending the cultures with 10 mM 1-propanol, 10 mM 2-propanol, 20 mM ethanol, 40 mM methanol, 10 mM 1-butanol, or 10 mM 2-butanol. Strain AK-7 did not catabolize any of these substrates.
Each of the three strains required organic growth factors. Strain AK-7 required acetate in addition to complex organic matter for growth. This strain was able to grow when yeast extract (0.2%) was added to the medium along with acetate, but neither acetate alone nor a combination of acetate and Trypticase peptone supported growth.
mol% G+C.
The Tm values for strains AK-7, AK-3, and AK-8 were 85.7, 83.2, and 84.2, respectively, which correspond to DNA G+C contents of 49.7, 43.6, and 46.2 mol%, respectively.
Phylogenetic analysis and DNA hybridization.
The 16S rRNA gene sequences for strains AK-7, AK-3, and AK-8 indicated that these strains belong to the genus Methanogenium (see Fig. S3 in the supplemental material). Strain AK-7 was most closely related to CO2-reducing methanogens cultivated from the Hydrate Ridge (26) and to Methanogenium cariaci, a methanogen isolated from marine sediments in the Cariaco Trench (98% sequence similarity) (49). Strain AK-3 and strain AK-8 were both most closely related to Methanogenium marinum, a methanogen isolated previously from Skan Bay (14). Phylogenetic analysis of the mcrA gene also revealed that strain AK-7 clustered within the genus Methanogenium (see Fig. S5 in the supplemental material).
DNA hybridization indicated that the genomic sequence similarity between strain AK-7 and Methanogenium cariaci, M. marinum, or M. organophilum was less than 70%, and these results suggest that strain AK-7 should be classified as a new species if it can also be distinguished by phenotypic characteristics (70). DNA hybridization results suggest that strains AK-3 and strain AK-8 should be classified as the same species, and that strain AK-8 may represent a new species; however, further physiological tests were not pursued during the present study.
DISCUSSION
The sediments at Skan Bay are permanently cold and strictly anoxic (47). Geochemical investigations of this location presented in the present study and previously (66) indicate the presence of a sulfate-reducing zone extending to a depth of ∼30 cm, a methane-oxidizing zone extending from 30 to 45 cm, and an underlying methanogenic zone. The maximum rates of sulfate reduction are located near the surface at a depth of ∼5 cm, with a secondary maximum around 31 cm in the methane-oxidizing zone. Although distinct geochemical gradients exist in the sediment, no significant differences among archaeal populations were evident using molecular methods. These results are in agreement with another study that compared microbial diversity in marine sediments at various depths (48).
Geochemical analyses of Skan Bay sediments have indicated that AOM occurs in the mid-zone depths of the sediments (66), and this study confirms the presence of methanotrophic Archaea. Although the ANME-1 phylotypes were present in very low numbers, the ANME-2 group constituted about 16% of all the clones analyzed. ANME-1 and ANME-2 groups may coexist, or one group may be more prevalent than the other group at particular sites (29). The low-methane partial pressure at Skan Bay may favor the ANME-2 group. Phylogenetic and isotopic data have shown a specific association between ANME-2 Archaea and sulfate-reducing bacteria (SRB) (42); thus, the detection of members of the ANME-2 group in deep sediments where sulfate was exhausted was surprising. These microbes may gain energy via an alternative mode of metabolism or may be simply residual from sediment burial. Given the lack of bioturbation at Skan Bay, and a sedimentation rate of 0.9 cm per year (58), the cells present in the deep interval are 10 to 50 years removed from the methane-oxidizing zone. Whether rRNA genes would remain intact for this period of time in inactive cells is not clear.
Methanogens seem to be a minor component of the archaeal community, as only five phylotypes related to cultivated methanogens were detected in the sediments. We isolated one strain of aceticlastic methanogen, strain AK-6, from the sediments at Skan Bay. Although several strains of aceticlastic methanogens have been isolated from marine sediments (17, 55, 68), isotopic evidence suggests that acetate is a minor precursor to methanogenesis in most marine sediments (71). The low concentrations of acetate in Skan Bay (Fig. 1B) are below the kinetic threshold values of cultivated aceticlastic methanogens (72). The carbon isotopic composition of methane at Skan Bay is ∼−80‰, which is inconsistent with acetate being a primary source. Strain AK-6 also catabolized methyl compounds; thus, its role in methane production in marine sediments warrants further study.
Methylotrophic methanogens that catabolize noncompetitive substrates (28) not used by SRB may be responsible for the limited amounts of methane production that occurs concurrently with sulfate reduction (40). Indeed, methylotrophic methanogens that grouped within the genus Methanococcoides were cultivated from all depths in the sediments. These methanogens are only able to catabolize methylated compounds (11).
Stable isotope data for carbon and hydrogen suggest that CO2 reduction is the most important pathway for biological methane production in marine sediments (71), and studies of marine sediments at the Nankai Trough and Hydrate Ridge have shown that CO2 reduction was the predominant mode of methanogenesis (26, 38). Our cultivation data are consistent with these results. We enriched and isolated methanogens belonging to the order Methanomicrobiales. These methanogens gain energy strictly by reducing CO2 and oxidizing H2 or formate (11).
Interestingly, we cultivated CO2-reducing methanogens not only from the deeper, sulfate-depleted sediments, but also from the shallow sulfate-containing sediments and from the zone of AOM. Typically, in marine sediments, sulfate reduction is the dominant microbial process in the upper sediments. SRB out-compete methanogens for most low-molecular-weight electron donors, such as hydrogen (and acetate), because of their higher affinity for these substrates (30). As a result, methanogens are active mainly in deeper sediments, where sulfate is depleted. However, methane production from CO2 plus H2 in sediments with high sulfate concentrations has been detected in other marine sediments (26, 44), and the results of this study are congruous with these data. Possibly, these methanogens are able to gain energy by growing syntrophically with propionate- and butyrate-oxidizing bacteria (27).
By combining cultivation-independent and -dependent techniques, we were able to assess the archaeal diversity in marine sediments at Skan Bay. The enrichment cultures allowed for the detection of methanogens that are present in low numbers in the sediment and also allowed for the gleaning of physiological information that cannot necessarily be derived from sequence data. Finally, the distribution of uncultivated Archaea highlights significant gaps in our understanding of the biogeochemistry and microbial ecology of such environments.
Description of Methanogenium boonei sp. nov.
Methanogenium boonei (boo′ne.i. N.L. gen. n. boonei of Boone; named in honor of David R. Boone, who has made many contributions to the ecology, physiology, and taxonomy of methanogens). The organism takes the form of irregular cocci 1.0 to 2.5 μm in diameter, occurring singly, and is nonmotile. CO2 plus H2 or formate serves as the sole catabolic substrate, with methane as the end product. The fastest growth occurred at 19.4°C, with a salinity of 0.3 to 0.5 M Na+ and a pH of 6.4 to 7.8. It was isolated from permanently cold, anoxic marine sediments at Skan Bay, Alaska. The type strain is AK-7 (OCM 787/DSMZ 17338).
Supplementary Material
Acknowledgments
We thank Bill Reeburgh, John Kessler, Cameron Adams, and the crew of the R/V Alpha Helix for help in sample collection. We are grateful to Henry Aldrich (University of Florida) for preparing the electron micrographs. We also thank Susan Masta for help with the phylogenetic analysis and Ken Stedman for critical reading of the manuscript.
This work was supported by a grant from the National Science Foundation's LExEn program (0085607) and by an NSF IGERT Earth Subsurface Biosphere fellowship awarded to M.M.K.
This paper is dedicated to the late David R. Boone.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Albert, D. B., and C. S. Martens. 1997. Determination of low-molecular-weight organic acid concentrations in seawater and pore-water samples via HPLC. Mar. Chem. 56:27-37. [Google Scholar]
- 2.Alperin, M. J. 1989. The carbon cycle in an anoxic marine sediment: concentrations, rates, isotope ratios, and diagenetic models. Ph.D. thesis. University of Alaska, Fairbanks.
- 3.Alperin, M. J., and W. S. Reeburgh. 1984. Geochemical observations supporting anaerobic methane oxidation, p. 282-289. In R. Crawford and R. Hanson (ed.), Microbial growth on C-1 compounds. American Society for Microbiology, Washington, DC.
- 4.Alperin, M. J., and W. S. Reeburgh. 1985. Inhibition experiments on anaerobic methane oxidation. Appl. Environ. Microbiol. 50:940-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arakawa, S., and C. Kato. 27 July 2005, posting date. Microbial diversity of cold-seep sediments in Japan Trench. [Online.] http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=60417930.
- 6.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least one in twenty 16S rRNA sequence records currently held in public repositories estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY.
- 8.Berner, R. A. 1982. Burial of organic carbon and pyrite in the modern ocean: its geochemical and environmental significance. Am. J. Sci. 28:193-206. [Google Scholar]
- 9.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 10.Boone, D. R., and W. B. Whitman. 1988. Proposal of minimal standards for describing new taxa of methanogenic bacteria. Int. J. Syst. Bacteriol. 38:212-219. [Google Scholar]
- 11.Boone, D. R., W. B. Whitman, and Y. Koga. 2001. Order III: methanosarcinales, p. 268-293. In D. R. Boone and R. W. Castenholz, G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 12.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 13.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 14.Chong, S. C., Y. Liu, M. Cummins, D. L. Valentine, and D. R. Boone. 2002. Methanogenium marinum sp. nov., a H2-using methanogen from Skan Bay, Alaska, and kinetics of H2 utilization. Antonie Leeuwenhoek 81:263-270. [DOI] [PubMed] [Google Scholar]
- 15.Colwell, R. 1997. EstimateS: statistical estimation of species richness and shared species from samples. User's guide, version 5, and application. [Online.] http://viceroy.eeb.uconn.edu/EstimateS.
- 16.De Ley, J., H. Cattoir, and A. Reynaerts. 1970. The quantitative measurement of DNA hybridization from renaturation rates. Euro. J. Biochem. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 17.Elberson, M. A., and K. R. Sowers. 1997. Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae. Int. J. Syst. Bacteriol. 47:1258-1261. [DOI] [PubMed] [Google Scholar]
- 18.Gieskes, J., T. Gamo, and H. Brumsack. 1991. Chemical methods for interstitial water analysis: Joides resolution. Texas A&M University, College Station.
- 19.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 20.Henrichs, S. M., and W. S. Reeburgh. 1987. Anaerobic mineralization of marine sediment organic matter: rates and the role of anaerobic processes in the oceanic carbon economy. Geomicrobiol. J. 5:191-237. [Google Scholar]
- 21.Hershberger, K. L., S. M. Barns, A.-L. Reysenbach, S. C. Dowson, and N. R. Pace. 1996. Wide diversity of Creanarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 22.Hinrichs, K.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 23.Hungate, R. E. 1966. The rumen and its microbes. Academic Press, Inc., New York, NY.
- 24.Huss, V. A. R., H. Festl, and K. H. Schleifer. 1983. Studies on the spectrometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4:184-192. [DOI] [PubMed] [Google Scholar]
- 25.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 26.Kendall, M. M., and D. R. Boone. 2006. Cultivation of methanogens from shallow marine sediments at Hydrate Ridge, Oregon. Archaea 2:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kendall, M. M., Y. Liu, and D. R. Boone. 2006. Butyrate- and propionate-degrading syntrophs from permanently cold marine sediments in Skan Bay, Alaska, and description of Algorimarina butyrica gen. nov., sp. nov. FEMS Microbiol. Lett. 262:107-114. [DOI] [PubMed] [Google Scholar]
- 28.King, G. M. 1984. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl. Environ. Microbiol. 48:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knittel, K., T. Losekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kristjansson, J. K., P. Schoenheit, and R. K. Thauer. 1982. Different Ks values for hydrogen of methanogenic bacteria and sulfate-reducing bacteria: an explanation for the apparent inhibition of methanogenesis by sulfate. Arch. Microbiol. 131:278-282. [Google Scholar]
- 31.Kvenvolden, K. A., and T. D. Lorenson. 2001. The global occurrence of natural gas hydrate, p. 3-18. In C. K. Paull and W. P. Dillon (ed.), Natural gas hydrates: occurrence, distribution, and detection, vol. 124. American Geophysical Union, Washington, DC. [Google Scholar]
- 32.Lanoil, B. D., M. T. La Duc, M. Wright, M. Kastner, K. H. Nealson, and D. H. Bartlett. 2005. Archaeal diversity in ODP legacy borehole 892b and associated seawater and sediments of the Cascadia Margin. FEMS Microbiol. Ecol. 54:167-177. [DOI] [PubMed] [Google Scholar]
- 33.Maddison, D. L., and W. P. Maddison. 2003. MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates, Inc., Sunderland, MD.
- 34.Maestrojáun, G. M., and D. R. Boone. 1991. Characterization of Methanosarcina barkeri MST and 227, Methanosarcina mazei S-6T, and Methanosarcina vacuolata Z-761T. Int. J. Syst. Bacteriol. 41:267-274. [Google Scholar]
- 35.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 36.Marmur, J., and P. Doty. 1962. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5:109-118. [DOI] [PubMed] [Google Scholar]
- 37.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 38.Newberry, C. J., G. Webster, B. A. Cragg, R. J. Parkes, A. J. Weightman, and J. C. Fry. 2004. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 6:274-287. [DOI] [PubMed] [Google Scholar]
- 39.Ni, S., and D. R. Boone. 1991. Isolation and characterization of a dimethyl sulfide-degrading methanogen, Methanolobus siciliae HI350, from a oil well, characterization of M. siciliae T4/MT, and emendation of M. siciliae. Int. J. Syst. Bacteriol. 41:410-416. [DOI] [PubMed] [Google Scholar]
- 40.Oremland, R. S., and S. Polcin. 1982. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl. Environ. Microbiol. 44:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 43.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkes, R. J., B. A. Cragg, J. C. Fry, R. A. Herbert, and J. W. T. Wimpenny. 1990. Bacterial biomass and activity in deep sediment layers from the Peru margin. Philos. Trans. R. Soc. Lond. A 331:139-153. [Google Scholar]
- 45.Powell, G. E. 1983. Interpreting gas kinetics of batch culture. Biotechnol. Lett. 5:437-440. [Google Scholar]
- 46.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeburgh, W. S. 1980. Anaerobic methane oxidation: rate depth distributions in Skan Bay sediments. Earth Planet. Sci. Lett. 47:345-352. [Google Scholar]
- 48.Reed, D. W., Y. Fujita, M. E. Delwiche, D. B. Blackwelder, P. P. Sheridan, T. Uchida, and F. S. Colwell. 2002. Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68:3759-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romessar, J. A., R. S. Wolfe, F. Mayer, E. Speiss, and A. Walther-Mauruschat. 1979. Methanogenium, a new genus of marine methanogenic bacteria, and characterization of Methanogenium cariaci sp. nov., and Methanogenium marisnigri sp. nov. Arch. Microbiol. 121:147-153. [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatinds. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Shaw, D. G., M. J. Alperin, W. S. Reeburgh, and D. J. McIntosh. 1984. Biogeochemistry of acetate in anoxic sediments of Skan Bay, Alaska. Geochim. Cosmochim. Acta 48:1819-1825. [Google Scholar]
- 52.Simankova, M. V., S. N. Parshina, T. P. Tourova, T. V. Kolganova, A. J. B. Zehnder, and A. N. Nozhevnikova. 2001. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst. Appl. Microbiol. 24:362-367. [DOI] [PubMed] [Google Scholar]
- 53.Singh, N., M. M. Kendall, Y. Liu, and D. R. Boone. 2005. Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska, description of Methanococcoides alaskense sp. nov., and emendation of Methanosarcina baltica. Int. J. Syst. Evol. Bacteriol. 55:2531-2538. [DOI] [PubMed] [Google Scholar]
- 54.Sly, L. I., L. L. Blackall, P. C. Kraat, T. Tian-Shen, and V. Sangkhobol. 1986. The use of second derivative plots for the determination of mol% guanine plus cytosine of DNA by the thermal denaturation method. J. Microbiol. Methods 5:139-156. [Google Scholar]
- 55.Sowers, K. R., S. F. Baron, and J. G. Ferry. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sowers, K. R., and K. M. Noll. 1995. Techniques for anaerobic growth, p. 15-47. In F. T. Robb, K. R. Sowers, H. J. Schreier, S. DasSarma, and E. M. Fleischmann (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 57.Starr, M. P., and M. Mandel. 1968. DNA base composition and taxonomy of phyotophathgenic and other enterobacteria. J. Gen. Microbiol. 56:113-123. [DOI] [PubMed] [Google Scholar]
- 58.Sugai, S. F., M. J. Alperin, and W. S. Reeburgh. 1994. Episodic deposition and Cs-137 immobility in Skan Bay sediments: a 10-year Pb-210 and Cs-137 time-series. Mar. Geol. 116:351-372. [Google Scholar]
- 59.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony and other methods, version 4. Sinauer Associates, Inc., Sunderland, MA.
- 60.Takai, K., and K. Horikoshi. 1999. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valentine, D. L. 2002. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Leeuwenhoek 81:271-282. [DOI] [PubMed] [Google Scholar]
- 64.Valentine, D. L., D. C. Blanton, W. S. Reeburgh, and M. Kastner. 2001. Water column methane oxidation adjacent to an area of active hydrate dissociation, Eel River Basin. Geochim. Cosmochim. Acta 65:2633-2640. [Google Scholar]
- 65.Valentine, D. L., M. Kastner, G. D. Wardlaw, X. Wang, A. Purdy, and D. H. Bartlett. 10 November 2005, posting date. Biogeochemical investigations of marine methane seeps, Hydrate Ridge, Oregon. J. Geophys. Res. 110:GO2005. doi: 10.1029/2005JG000025. [DOI] [Google Scholar]
- 66.Valentine, D. L., and W. S. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 67.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Klein, D., H. Arab, H. Volker, and M. Thomm. 2002. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6:103-110. [DOI] [PubMed] [Google Scholar]
- 69.Wardlaw, G. D., and D. L. Valentine. 2005. Evidence for salt diffusion from sediments contributing to increasing salinity in the Salton Sea, California. Hydrobiologia 533:77-85. [Google Scholar]
- 70.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 71.Whiticar, M. J., E. Faber, and M. Schoell. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 50:693-709. [Google Scholar]
- 72.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry, and genetics. Chapman & Hall, New York, NY.
- 73.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. von'tRiet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.