Abstract
In nutrition research the number of human in vivo experiments is limited because of the many restrictions and the high costs of testing in humans. Up to now predictive computer models aiming to enhance research have been rare or too complex, with many nonmeasurable adjustable parameters. This study aimed to develop a basic physicochemical computer model for a first quantitative interpretation of results obtained from in vivo intestinal experiments with bacteria. This new modeling approach is validated with results obtained from gut infection studies in vivo. The design of the model is described, and its ability to reproduce experimental data is evaluated. The model predictions are compared with new experimental data. The phenomena that take place in the gastrointestinal tract are summarized by model constants for growth, adherence, and release of bacteria. Although the model is far from describing all details and many processes in the intestine are combined, the model calculation results lead to reasonable conclusions and interesting hypotheses. One of these hypotheses concluded from the model outcomes is that Escherichia coli bacteria have a much lower intestinal growth rate in humans than in rats. Extra laboratory validation experiments proved the reliability of this hypothesis predicted by the model. In addition, the known protective effect of dietary calcium and detrimental effect of clindamycin on the growth and adherence of Salmonella bacteria could be quantified. From these results it is clear that the model enhances the interpretation of in vivo gastrointestinal experiments and will facilitate research trajectories towards new functional foods that improve resistance to pathogenic bacteria in humans.
In science more and more mathematical models are being described for use in experimental design, interpretation of experimental results, prediction of new experimental results, or scaling up from small-scale experiments to industrial-scale production plants. The first application of these mathematical models in general could be found in the chemical industry in the 1960s. The food industry started to use models in the early 1990s. Nowadays predictive computer (in silico) models play a major role in the design of food products, aiming at reducing the number of expensive experiments on the pilot or industrial scale (10).
In human nutrition research the use of mathematical models is very limited. The first reason is the extreme complexity of the system to be modeled: the human being. Especially in the case of infection studies, most of the research is performed with model studies with animals or with in vitro experiments. The number of human in vivo experiments is limited because of the many restrictions and the high costs of testing in humans. There are relatively few reports in the literature of attempts to model the gastrointestinal tract. Most of the articles are focused on the residence time distribution (RTD) in the stomach and the gut (11, 18, 26, 29). Wilkinson (29, 30) performed an extensive modeling study of the gastrointestinal tract. In this study a large number of phenomena were taken into account, such as oxygen tolerance, metabolism, and biofilm production by the gut microflora, and described by differential equations. However, this model had too many parameters which could not be validated by measurements. Modeling studies of parts of the gastrointestinal tract have led to interesting results such as the effect of pH and residence time on the inactivation of Escherichia coli in the stomach (26). Another approach is described by Minekus (18, 19), presenting a dynamic physical in vitro model simulating the successive dynamic conditions in the stomach and intestine of humans and monogastric animals. The disadvantage of this physical model is that new uncertainties are introduced by the hardware (e.g., piping, hollow fibers, pumps, and valves). Moreover, the model is constructed with material (i.e., glass or metal) that has a completely different behavior than that of the gut mucosa. As a consequence the effect of adhesion of microorganisms to the mucosa cannot be taken into account. Therefore, it is not suitable for determination of the relevant physiological constants such as adhesion constants of bacteria in the gut.
In food production much progress has been achieved in modeling complex phenomena in processing equipment. Some of these models incorporate the interaction between phenomena in the flowing liquid and the equipment walls. For example, recently a validated model that predicts contamination in a processing system as a function of bacterial characteristics (growth, adhesion, and death kinetics), wall behavior (release kinetics), and local conditions (temperature) was described (8). It is clear that a number of analogies can be recognized from intestinal studies where the effect of food components on the course of an intestinal bacterial infection is determined (5, 6, 7). Both in processing equipment and in the gastrointestinal tract, inactivation, growth, adhesion, and release of bacteria take place. Moreover, it is known that growth and adhesion phenomena are very relevant for in vivo infection studies (5). From a modeling point of view the most important differences with processing equipment are the large residence time distribution in the gastrointestinal tract and the time-dependent properties of the system.
This study aimed to examine the possibility of developing a basic in silico model for a first-order approximation of the colonization of bacterial pathogens in the gut by using the experimental results of in vivo animal and human infection studies. The hypothesis driving the study is that a computer model with a limited number of parameters gives information in addition to the measured course of the target microorganism in the feces. For reasons of the biological complexity of the in vivo system and the difficulty of validation of biological phenomena taking place, the model is simplified as much as possible. On the other hand and in contrast with many other computer models, the model is built with mechanism-based equations only. As a result, each parameter has a physiological meaning and no model constant or correction factor is added to shape the predictions. The “fitted” model parameters for the growth, adherence, and release rate of the target microorganisms are the combined quantitative factors describing the way in which, for example, bacteria pass, adsorb to, and are released from the gut mucosa after oral intake. In fact the model should give information in addition to the measured course of fecal pathogen excretion in humans and animals. The presented model is developed for (i) interpretation of rat infection experiments and its extrapolation to the human situation; (ii) determination of the role of the several parts of the gastrointestinal tract with respect to the adhesion, growth, and release of microorganisms; (iii) testing of hypotheses followed by setting up experiments to verify these hypotheses (improvement of experimental design); and (iv) further development of the in silico model describing the phenomena in more detail.
In this article the in silico model is described and its ability to reproduce experimental data is evaluated. Finally, model predictions are compared with new experimental data.
MATERIALS AND METHODS
Design of the in silico model.
The model for bacterial growth, mucosal adhesion, and release of bacteria and spores within the gastrointestinal tract is largely based on a validated model for processing equipment (8). It is known that adherence of microorganisms and bacterial spores on heat exchanger surfaces in cheese and liquid milk factories is an important source of bacterial contamination of dairy products. The increase in the levels of bacteria in the product during process operation is partly the result of growth in the product, but release of bacteria that grow on equipment walls also plays a significant role (4, 16, 20, 24, 27, 31). In general terms this computer model is now used to simulate the behavior of pathogenic bacteria in the gastrointestinal tract. However, there is one important difference: the main outcome of the model is not the concentration of bacteria based on a known value for growth, adherence rate of the target microorganism, but vice versa. In addition to the oral intake of bacteria and the dimensions of the stomach and the small and large intestine, the measured concentrations of the target bacteria in time are used as an input. The adherence, release, and growth parameters are fitted to the measured concentration of the target bacteria in the feces. Model outputs are the value of the adherence (ka), growth (μT), and release (kr) parameters in the small and large intestine by minimization of the differences between the measured and calculated fecal excretion. Since no meaningless adjustable constants are used, the determined parameters quantify the adherence, release, and growth characteristics of the bacteria in the digestive tract.
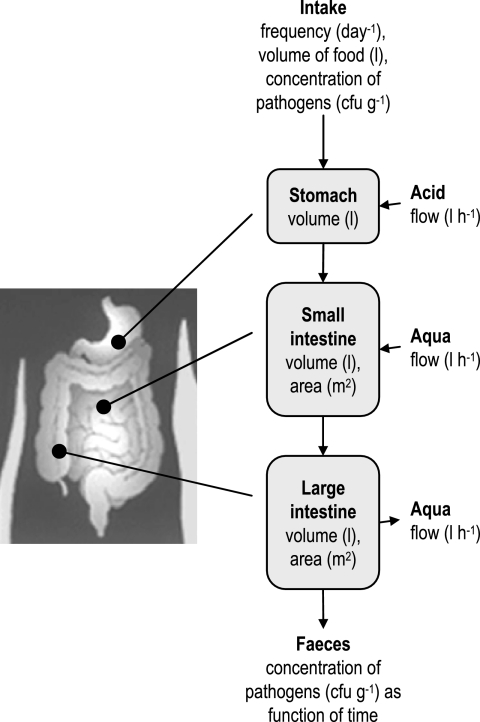
The computer model consists of four major parts: (i) the oral intake of bacteria; (ii) the residence time distribution and inactivation of bacteria in the stomach; (iii) the residence time distribution and growth, adhesion, and release of bacteria in the small and large intestine; and (iv) the concentration of bacteria in feces.
The oral intake of the target bacteria is described by the frequency of eating, the volume per time of eating, and the concentration of target bacteria in the food. With these data and accounting for gastric secretion, the volume flow in the stomach is known. In Fig. 1 a simplified flow diagram of the gastrointestinal tract as used by the model is shown.
FIG. 1.
Flow diagram of the gastrointestinal tract used for the model.
Modeling the residence time distribution.
The local concentration of the target bacteria in the gut is highly dependent on the residence time distribution. Due to mixing of phenomena some bacteria stay longer in the gut than others. In general, there are limited data available about the residence time distribution in humans and rats. The model described below is used as a first best estimate. In the literature the RTD in the gastrointestinal tract has been described in different ways (11, 17, 29). Most of these are based on exponential decay equations the summarized distributed intervals of which result in S-shaped, cumulative curves. These equations have a black box nature, and the equation constants have no direct physiological meaning. From chemical engineering the reactor model is an appropriate way to describe the residence time distribution in arbitrary systems, accounting for the volume and the flow behavior of a system. In this model the flow in the system is defined by a fictive number of N ideally mixed tank reactors in series. For example, when a system is described by N = 1, this means that the flow in this system can be described by one ideally mixed tank reactor. If N = ∞, there is no mixing at all and the flow is characterized by plug flow (28). The residence time distribution curve of bacteria or spores in a system, as, for example, the small intestine, is then estimated by
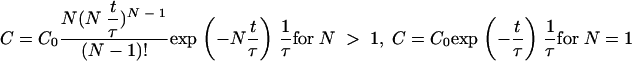 |
(1) |
where C is the concentration in CFU · ml−1 (e.g., in the case of bacteria), t is time in h, and τ is the average residence time in the system in h (e.g., in the small intestine). The parameter τ is defined by the volume of the system. As a consequence the residence time model accounts for the different volumes between, for example, the intestines of rats and humans. In the case that two or more systems are connected with each other (e.g., small and large intestine), the residence time distribution curve of the last system is obtained by convolution of the separated curves
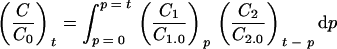 |
(2) |
where p is a time parameter in h (28). In this case C1.0 and C2.0 are the inlet concentrations of bacteria in the small and large intestine, respectively.
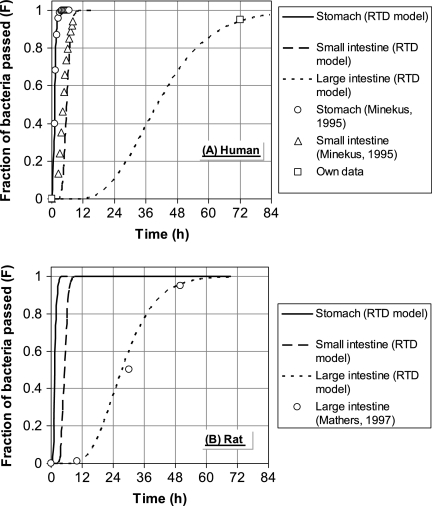
In order to estimate the fictive number of stirred tank reactors necessary to describe the RTD in the gastrointestinal parts according to the work of Westerterp et al. (28), in vivo experimental data from the work of Minekus (18) and Mathers et al. (17) were used. It was assumed that the mixing behavior is roughly the same as the behavior in humans. By evaluating equations 1 and 2 the number N was fitted with the experimental data for the stomach and the small and large intestine, reproducing the S-shaped curves from the literature. The results are shown in Fig. 2. For example, in the case of humans the RTD model predicts that 72 h after oral intake 95% of the target bacteria has passed the large intestine. It turned out that both for humans and for rats the RTD in the stomach, small intestine, and large intestine could be described by 7, 14, and 5 reactors in series, respectively. For the stomach and the small intestine of rats no data were available.
FIG. 2.
Estimated RTD in the stomach, small intestine, and large intestine of humans (A) and rats (B) compared with data from literature.
Modeling the growth, adherence, and release of bacteria.
From the literature it is known that a number of bacteria are inactivated in the stomach. Depending on, for example, the pH, 0.1% to several percent of bacteria such as E. coli survive (26). Although during the experiments the exact survival rate could not be measured, it is assumable that this value was constant. During the model simulations the inactivation rate (ki) was kept constant at 3.2 h−1 which is in line with values reported in the literature (26). The survival rate was calculated by
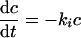 |
(3) |
As spores are much more resistant, their inactivation rate in the stomach was taken as 0.
The surviving bacteria or spores enter the small intestine. The number of bacteria adsorbed to the wall of the small and large intestine is determined by both the transport of bacteria to the surface and the adhesion reaction with the surface. The rate of adhesion is given by
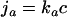 |
(4) |
where ka is the adhesion rate constant and c the concentration of bacteria near the surface (i.e., the gut lumen). Since each compartment of the gastrointestinal tract has its own characteristics (dimensions and RTD) in the model, the tract is divided into three compartments (model reactors): stomach, small intestine, and large intestine. As a result of the RTD in the gastrointestinal tract, it is not valid to assume that all bacteria have the same residence time in the three compartments. In order to account for the existence of RTD, the total volume of each compartment is subdivided into smaller-volume fractions with a certain number of bacteria. Each fraction has a defined residence time, surface area, volume, and concentration of local bacteria. In each part, growth and/or inactivation, adhesion, and release of bacteria can occur. The adsorbed microorganisms grow on the mucosa and may be released to the intestinal contents, resulting in an increasing bacterial concentration in intestinal contents. The growth rate at the mucosal surface is assumed to be equal to that of “free” bacteria in the intestinal contents. Two mass balances form the basis of the model: one bacterial balance for the mucosa to which bacteria adhere and one for the gastrointestinal contents. Bacterial growth as a function of the time t at position x on the gut mucosa in a volume fraction is defined by the transfer equation as the change in bacterial colonization of the mucosa with time = bacteria produced at the surface − released + adhered:
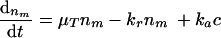 |
(5) |
where nm is the mucosa colonization in CFU m−2, μT is the bacterial growth rate at temperature T in h−1, kr is the release constant of bacteria at the mucosa in h−1, and ka is the adhesion constant in m h−1. The local concentration of the bacteria c in CFU m−3 at operating time t follows from the component (bacterial) balance: change in concentration of bacteria in the intestine with position = released − adsorbed + produced in the intestine:
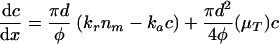 |
(6) |
where φ is the feed or food flow in m3 h−1 and d the hydraulic diameter of the volume part in m. More information about the background of this equation is given in earlier publications (8).
In order to evaluate the model outcomes with the experimental results, all equations within the model described above were built into the user-friendly software NIZO Premia (10, 25).
Model calculation procedures.
In general, five model parameters were determined: the bacterial growth in the small and large intestine, the adherence rate in the small and large intestine, and the release rate. In order to determine the five values of the model parameters per experiment, an advanced fit procedure based on dynamic optimization tools was used in analogy with other modeling work (8, 9). The optimization method is based on an advanced simplex method of the work of Nelder and Mead (21) and works as follows. First, the basic model data from Table 1 and Table 2 are loaded into the software. Then the experimental data set obtained from in vivo experiments and used for comparison of the measured and calculated values is loaded. After initiation (first guess) of the constants for growth, adhesion, and release, the optimization starts. This means that the error function that represents the sum of squared errors (SSE) regarding the measured data is minimized by changing the value of the model constants via a smart mathematical algorithm (simplex method). The error function was defined as the SSE:
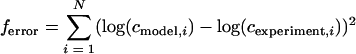 |
(7) |
where N is the number of data points, i is the index, and c is the concentration of bacteria in feces. In the ideal situation the value of ferror → 0, meaning that the difference between all the measured values and the model simulations is nil.
TABLE 1.
Estimated dimensions of the gastrointestinal tract in humans and rats used as parameters in the modela
| Compartment | Human
|
Rat
|
||||||
|---|---|---|---|---|---|---|---|---|
| L (mm) | D (mm) | V (ml)b | A (cm2)c | L (mm) | D (mm) | V (ml)b | A (cm2)c | |
| Duodenum | 250 | 35d | 491 | 393 | 98 | 2.8 | 0.58 | 8.42 |
| Jejunum | 3,000 | 35d | 5,890 | 4,710 | 1,125 | 4.5 | 17.9 | 159 |
| Ileum | 3,000 | 35d | 5,890 | 4,710 | 30 | 4.0 | 0.38 | 3.77 |
| Total small intestine | 12,271 | 9,813 | 18.8 | 171 | ||||
| Cecum | 200 | 70 | 770 | 440 | 60 | 10 | 4.7 | 18.8 |
| Colon | 1,500 | 50 | 2,945 | 2,355 | 100 | 6.5 | 3.3 | 20.4 |
| Total large intestine | 3,715 | 2,795 | 8.0 | 39.3 | ||||
Data are derived from the work of Kararli (14) unless otherwise noted. Abbreviations: L, length; D, diameter; V, volume; A, area.
Calculated values (0.25 · πD2L).
Hydrodynamic surface; including the crypts of the surface area might result in a value that is higher by a factor of 100 (14).
Data are derived from the work of Avraham (3).
TABLE 2.
Miscellaneous data for the gastrointestinal tract in humans and rats
Statistical analyses and robustness of the model.
To use the calculated values of the physical model parameters (growth, adherence, and release) for interpretation of experimental data, it is useful to know the confidence intervals of the parameter values. However, since the in silico model is nonlinear and multidimensional, the confidence intervals cannot be calculated according to conventional statistics. In principle the confidence intervals and the sensitivity of the model output to the parameter values can be estimated by performing a large number (e.g., >1,000) of repeated model simulations (Monte Carlo) with slightly varying parameter values. However, the computing of one simulation cycle for calculation of the growth, adherence, and release constants already takes 30 to 60 min. Thus, this approach is not feasible. In order to overcome this problem a limited number of computer simulations have been performed giving an indication of the significance of the estimated model parameters (growth, adherence, and release factors). The following statistical indicators were calculated: (i) correlation coefficient (R2) of the parameter fit; (ii) standard error of the logarithm of the estimated parameter values (SElog); (iii) sensitivity of the model parameters, defined as the second derivative of SSE with respect to a change in the value of the model parameter (δ2SSE/δP2); and (iv) correlation between the different model parameters.
Experimental setup of in vivo studies.
For determination of the model constants for growth, adhesion, and release of bacteria in the gastrointestinal tract six experimental data sets have been used: (i) gastrointestinal transit of Bacillus stearothermophilus spores in rats, (ii) Salmonella infection experiments in rats, (iii) Salmonella infection experiments in rats fed a high-calcium diet, (iv) Salmonella infection experiments in rats pretreated with the antibiotic clindamycin, (v) E. coli infection experiments in rats, and (vi) E. coli infection experiments in humans.
Rat experiments.
In most of the rat infection experiments mentioned below, the animals were fed a purified low-calcium control diet. In one experiment rats were fed this diet supplemented with calcium (5, 7). After a period of adaptation to the experimental diets, the rats were orally infected with 108 to 109 CFU of Salmonella enterica serovar Enteritidis or enterotoxigenic Escherichia coli (ETEC) to mimic a food-borne infection. In an additional experiment, “inert” thermophilic Bacillus stearothermophilus spores were administered orally to the rats as gastrointestinal passage markers (15). Fresh fecal samples were collected at several time points after infection to quantify fecal pathogen (spore) excretion by standard culturing on specific agar plates. In an additional study, rats were adapted to the low-calcium control diet and orally pretreated with the antibiotic clindamycin (15 mg/kg of body weight dissolved in 1 ml of saline and administered by gastric gavage) for four consecutive days. This antibiotic generally suppresses the anaerobic flora, leading to overgrowth of aerobic microorganisms. Two days after termination of the clindamycin treatment, the rats were orally infected with S. enterica serovar Enteritidis to study the effect of this intervention on colonization resistance of the rats.
Human experiment.
Healthy human subjects consumed their habitual diet with either regular-milk-supplemented products with a naturally high calcium content or placebo low-calcium milk products of identical appearance. After an adaptation period of 10 days, the volunteers were orally infected with a live but attenuated ETEC strain. This strain induces mild short-lived infection symptoms such as diarrhea. The subjects regularly collected 24-h fecal samples for quantification of fecal ETEC output on days after oral infection (5).
Additional model validation experiments.
ETEC or Salmonella enterica serovar Enteritidis was added to pooled rat or human fecal water obtained from the in vivo experiments described above. Fecal water was prepared by centrifuging feces of rats and humans collected before oral infection as described elsewhere (6). These fecal waters were sterile. Stock suspensions of the above-mentioned bacterial pathogens in saline were added to pooled fecal waters or brain heart infusion broth (positive controls) to obtain a final concentration of nearly 107 CFU · ml−1. After incubation for 0, 2, 4, and 6 h at 37°C in an anaerobic cabinet, a small sample was taken from the incubates, diluted in saline, and plated on brilliant green agar modified agar for Salmonella detection and MacConkey agar supplemented with streptomycin for ETEC quantification as described earlier (5, 6). Plates were incubated overnight at 37°C. The whole experiment was performed in duplicate. In an additional in vitro experiment, it was tested whether growth inhibition of ETEC and salmonellas in fecal waters was due to substrate limitation or to the presence of growth-inhibiting factors in fecal water. For that purpose, samples of the same human and rat fecal waters were supplemented with glucose (final concentration, 0.5%) and incubated with Salmonella and ETEC as described above. Growth of these pathogens was monitored analogously.
Anatomic data and assumptions.
The anatomic data for the human and rat gastrointestinal tracts were derived from the literature (14, 18). The volume of the rat stomach is assumed to be 8 ml; the food intake was 2 × 9 g per day. Further, it was assumed that no bacterial growth occurs in the stomach and the adhesion and release of bacteria were ignored there.
It should be noted that the model takes the release of bacteria as a continuous process. This means that after passage of the infected food there is still release of bacteria or spores from the gut mucosa proportional to the amount of adherent bacteria or spores at the intestinal mucosa.
To calculate the surface of the small and large intestine, the hydrodynamic surface area (πDL) is used. The exact surface area is not known. For example, by including the crypts of mucosa the surface area might be higher by a factor of 100 (14). Using the hydrodynamic surface area implies that the adherence constant estimated by the computer model accounts indirectly for the surface characteristics of the mucosa. A high value of the adherence factor means that there are relatively many positions in the mucosa (e.g., within the crypts) at which bacteria can be adsorbed.
It should be noted that the model is considered a tool for comparison and interpretation of in vivo data. For that reason the model has been simplified as much as possible. A number of model constants, such as the inactivation in the stomach and the mucosa characteristics, are not equal for different host-pathogen combinations. Even during one in vivo experiment some model constants (e.g., growth rate and flow characteristics) might change. Hence, the comparison between model parameter values of different experiments is based on the average conditions during the experiment.
RESULTS
In Table 3 the determined values of bacterial growth, adhesion, and the release constants are given for the different evaluated data sets. Table 4 shows the results of related statistical analyses. From the related correlation coefficients and the plots in Fig. 3 it is clear that the model is able to reproduce the experimental data quite well. It appears that the release constant in both the small and the large intestine can be taken as equal to fit the experimental results. This implies that only five physical constants are used without adjustable correction factors to fit the model. As a consequence, the model can be used to derive five physical constants for a given set of experimental data. Based on the values of the determined constants as shown in Fig. 3, the following conclusions and hypotheses can be drawn.
TABLE 3.
Estimated model parameters
| Expt | Model parameter
|
||||
|---|---|---|---|---|---|
| Growth rate (h−1)
|
Adherence rate (m · h−1)
|
Release rate (h−1) | |||
| Small intestine | Large intestine | Small intestine | Large intestine | ||
| Rats with | |||||
| Spores | 1.1 × 10−3 | 1.2 × 10−5 | 4.2 × 10−2 | ||
| Salmonella | 1.1 × 100 | 5.9 × 10−3 | 1.9 × 10−6 | 1.2 × 10−6 | 1.1 × 100 |
| Plus calcium | 2.9 × 10−1 | 4.7 × 10−4 | 2.9 × 10−7 | 4.6 × 10−7 | 3.0 × 10−1 |
| After clindamycin | 5.3 × 10−1 | 2.0 × 10−3 | 2.2 × 10−4 | 1.6 × 10−4 | 5.6 × 10−1 |
| ETEC | 5.4 × 10−1 | 4.1 × 10−2 | 6.1 × 10−6 | 2.5 × 10−6 | 6.0 × 10−1 |
| Humans with ETEC | 2.3 × 10−1 | 3.6 × 10−5 | 3.1 × 10−3 | 3.5 × 10−2 | 3.0 × 10−1 |
TABLE 4.
Statistical analyses of the estimated model parameters
| Expt | Model parametera
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth rate (h−1)
|
Adherence rate (m · h−1)
|
Release rate (h−1)
|
Measured variation in log concns (σ) (n) | dfb | R2 | ||||||||
| Small intestine
|
Large intestine
|
Small intestine
|
Large intestine
|
||||||||||
| SElog | δ2SSE/δP2 | SElog | δ2SSE/δP2 | SElog | δ2SSE/δP2 | SElog | δ2SSE/δP2 | SElog | δ2SSE/δP2 | ||||
| Rats with | |||||||||||||
| Spores | 0.07 | 2.2 | 0.25 | <0.1 | 0.03 | 10.7 | 0.15 (8) | 6 | 0.88 | ||||
| Salmonella | <0.01 | 1,186.2 | >2c | 0.1 | 0.61 | 1.0 | >2c | 0.0 | <0.01 | 1,065.9 | 0.24 (8) | 1 | 0.98 |
| Plus calcium | 0.03 | 120.0 | >2c | 0.0 | 1.17 | 0.7 | >2c | 0.0 | <0.01 | 486.5 | 0.19 (8) | 1 | 0.96 |
| After clindamycin | 59.0 | 0.0 | 4.0 | 0.0 | 214.0 | 0.18 (8) | 0.98 | ||||||
| ETEC | 0.02 | 281.9 | 0.27 | 5.5 | 1.30 | 1.1 | 1.99 | 0.0 | 0.01 | 816.5 | 0.26 (8) | 1 | 1.00 |
| Humans with ETEC | 0.07 | 73.2 | 0.75 | 0.0 | 0.60 | 6.0 | 0.91 | 9.5 | 0.03 | 279.2 | 0.27 (16) | 2 | 0.98 |
SElog is the standard error of the log value of the estimated parameter. δ2SSE/δP2 is a measure for the sensitivity of the value of the model parameter value (P) to the estimated concentrations expressed as SSE; a high value means a large impact of the parameter value on the model output.
Every data point is the average of 8 (rats) or 16 (human) measurements.
Parameters were highly correlated (r > 0.9) in this experiment.
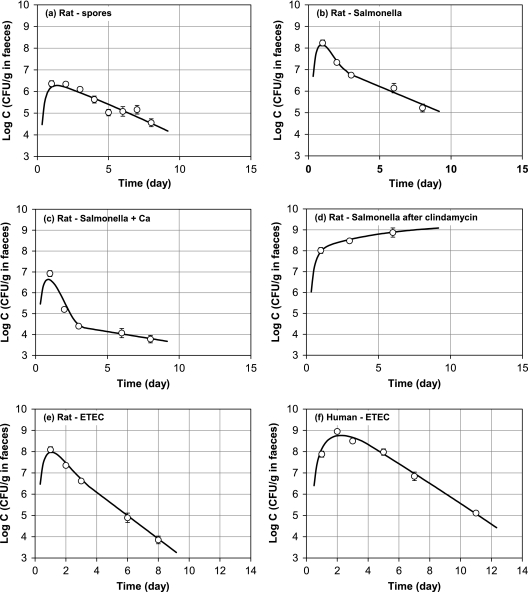
FIG. 3.
Comparison between the model calculation results and the experimental data points. ○, experimental data; —, model calculations. (a) B. stearothermophilus spores in rats (intake, 2.1 × 107 CFU); (b) Salmonella in rats (intake, 5.1 × 108 CFU); (c) Salmonella in rats on a high-calcium diet (intake, 5.1 × 108 CFU); (d) Salmonella plus clindamycin in rats (intake, 8.0 × 108 CFU); (e) ETEC in rats (intake, 2.1 × 109 CFU); (f) ETEC in humans (intake, 1.1 × 1010 CFU).
In Fig. 3a the model calculation results are compared with the measured concentration of spores in rat feces. As expected based on the experimental data the computer model estimates that no growth occurs in the gastrointestinal tract. It is remarkable that the model also indicates that a substantial number of the spores adhere to the gut mucosa (Table 3). For example, in the small intestine the adsorption rate is calculated as 1.10 × 10−3 m · h−1, meaning that according to equation 6 at a local concentration of 106 CFU · g−1 every second 30 spores are transported to 1 cm2 mucosa.
By comparison of Fig. 3b with Fig. 3c and the related values in Table 3 it is clear that the supplementation of the rat diet with calcium has a significant effect on the fecal Salmonella excretion. The fecal concentration of Salmonella is reduced by more than 1 log by calcium supplementation. In terms of growth and adherence the computer model calculated decreased values for the related constants. Especially in the large intestine pathogen adhesion is lower by a factor of 23 (in the small intestine lower by a factor of 10) on a calcium-supplemented diet. Also the Salmonella growth is approximately 10 times lower on a high-calcium diet.
In Fig. 3d it is shown that pretreatment with clindamycin results in high adherence rates of Salmonella: compared to the control (Fig. 3b) the adherence rate is increased by a factor of 70 to 80. The impact on the Salmonella growth rate is much less; this rate is decreased by a factor of only 2 to 3. Figure 3d shows that with clindamycin the fecal concentration of Salmonella does not decrease during the first 6 to 8 days. From model calculations it is reasonable to assume that this is due to the relatively high adhesion rates. This results in a high bacterial load of the intestinal mucosa and apparently sufficient release to hold the fecal excretion at a high level for at least a week.
In Fig. 3e and Fig. 3f the results are given for infection with E. coli (ETEC) in rats and humans, respectively. This enables comparison of the growth, adherence, and release rates in humans and rats. It is remarkable that the growth rate of E. coli in the human large intestine is calculated to be very low in comparison to that of rats (1,000-fold difference). In contrast ETEC adherence to the intestinal mucosa is much higher in humans than in rats.
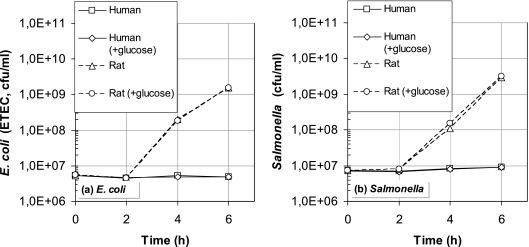
Figure 4 gives the results of the extra validation experiments to test the counterintuitive outcome of the computer model that the growth rate of ETEC in the human gut is much lower than in the rat intestine. From this figure it is clear that the “wet” experimental data support the validity of this prediction: both the growth rate of E. coli and that of Salmonella in human fecal water, mimicking colonic contents, are close to zero. The absence of pathogen growth observed in human fecal water was not abolished by glucose supplementation. These results indicate that substrate limitation is not a major factor and that the bacteriostatic effect of human fecal water is likely due to the presence of growth inhibitors in the intestinal tract of humans. In addition, glucose supplementation did not further increase growth of ETEC or Salmonella in rat fecal water, as similar exponential growth curves were observed in rat fecal waters with and without additional glucose.
FIG. 4.
Growth experiments with E. coli (a) and Salmonella (b) in rats and human fecal water.
DISCUSSION
Adhesion of Bacillus stearothermophilus spores.
It is assumed that B. stearothermophilus spores are not destroyed in the stomach and do not germinate and grow in the gastrointestinal tract and large intestine. Initially it was also assumed from the literature (17) that no adhesion would take place in the intestine. However, a closer look at the experimental data and the model outcomes (Fig. 3a) indicates that there must be some delay in the spore flow through the gastrointestinal tract to explain the course of the spore concentration in feces. In general, the majority of the food intake is removed from the gastrointestinal tract within 3 days. Assuming that thermophilic spores do not grow and adhere, it is obvious that also the majority of the spores should have been excreted in feces. This is not the case. As the computer model indicated, it is most likely that this is due to adhesion of spores at the gut mucosa. It is remarkable that from the literature it is known that bacterial spores have the capability to adhere to, for example, steel surfaces in equipment (1, 2, 22, 23). Apparently this is also the case in the intestine. In conclusion, Bacillus stearothermophilus spores cannot be considered as inert and thus cannot be used as a transit marker, for example, to determine the residence time of species in the gastrointestinal tract.
Effect of dietary calcium.
Figure 3b and especially Fig. 3c show a discontinuity in the slope of the fecal excretion of Salmonella around 3 days after infection. A closer look at the model calculations indicates that before day 3 the concentration in feces is mainly determined by the growth in the small and large intestine. After day 3 the concentration in feces is determined by the amount of bacteria released from the intestinal mucosa. It is remarkable that the release constant is lower with calcium than without calcium. However, this effect is overruled by the lower adherence and growth rates with calcium resulting in lower quantities of released Salmonella (released = release constant × bacterial colonization of the mucosa). This finding is in fact a quantification of results earlier reported in the literature (7). From the statistical data in Table 4 it can be concluded that the model could not discriminate between the growth and adherence rates in the large intestine. The two model parameters turned out to be correlated. However, this is largely due to the fact that both the growth and adherence rates in the large intestine are relatively low and close to nil.
Effect of clindamycin.
From Fig. 3d it can be concluded that the fecal Salmonella concentration does not decrease during the first 6 to 8 days. This is the result of the high bacterial load of the intestinal mucosa and apparently sufficient release to keep the fecal excretion at a high level for at least a week. Although the significance of the model parameter values could not be determined because of a lack of experimental data (Table 4), the computer model indicates that the high bacterial load is mainly due to high adhesion rates. From a physiological point of view higher adhesion rates are obvious after antibiotic treatment. Clindamycin suppresses the normal gut flora, especially the anaerobes. After treatment with clindamycin, more mucosal surface and growth substrate are available in the gut for food-borne Salmonella (12, 13).
Comparison of ETEC infection in humans and rats.
A first look at Fig. 3e and Fig. 3f suggests that in the case of E. coli there is no big difference between the two curve shapes for rats and humans. In humans the ETEC excretion peak is much broader, which is explicable from a dimensional point of view: the flow/volume ratio in the human intestine is lower than that in rats. Comparing ETEC infection in humans and rats, it should be noted that some assumptions might not be valid anymore. For example, it is probable that the inactivation in the stomach will differ for such different host-pathogen combinations. However, it is still remarkable that the estimated growth rate of ETEC in the human colon is relatively low. The results from the validation experiments with fecal water presented in Fig. 4 indeed show that the growth rate of ETEC in humans is nil compared to the growth rate in rats. This implies that the rat is not an appropriate model to quantify the behavior of ETEC in the colonic lumen of humans.
Comparison of Salmonella and ETEC.
From the values for the growth, adherence, and release rates it can also be concluded that the intestinal behavior of the pathogens Salmonella (Fig. 3b) and ETEC (Fig. 3e) does not differ greatly. An exception seems to be the growth rate in the large intestine. The growth rate of ETEC in that intestinal compartment is higher than that of Salmonella by approximately a factor of 7. However, looking at the poor significance of especially the estimated growth and adherence rates of Salmonella in the large intestine (Table 4), no further conclusions can be drawn.
Release factor.
Comparing all the computer estimations, it can be concluded that the bacterial release rates in rats and humans are estimated as more or less equal. The release rate is on the order of 0.1 to 1 h−1 (0.1 h−1 means that in the case of a mucosal colonization of 106 bacteria cm−2 after 1 h 105 bacteria are released to the contents of the intestine), and this value is not much influenced by the type of bacteria adsorbed. It is noteworthy that these values of the release factor are comparable with those found in biofilms in milk pasteurizers (8). Apparently the release is not mainly determined by the system in which the bacteria are adherent but probably by shear forces.
Conclusions.
In summary, modeling the behavior of bacteria in the gastrointestinal tract based on a sound chemical engineering approach turns out to be able to reproduce the results of in vivo experiments by application of only five physicochemical parameters. No adjustable constants are used to manipulate the fitting results. Although the model is far from describing all details and many processes in the intestine are combined, the model calculation results lead to reasonable conclusions or interesting hypotheses that need to be tested further. One of these hypotheses is that E. coli bacteria have a much lower growth rate in the human colon than in the rat colon. Extra validation experiments proved the reliability of this hypothesis predicted by the model. In addition, the effect of calcium and treatment with clindamycin on the growth and adherence of Salmonella could be quantified.
From the results it is clear that the developed model is a useful tool for the interpretation of in vivo gastrointestinal experiments. To obtain a certain amount of knowledge concerning the behavior of bacteria in the intestine, it is expected that fewer in vivo experiments are needed when using the computer model. The approach presented might facilitate research trajectories, for example, towards development of new functional foods that improve resistance to pathogenic bacteria in humans. Because of the generic design of the computer model, it is also expected to be applicable for quantification of the behavior of probiotic ingredients in the gastrointestinal tract. The model can also give more insight into the release rate and position of encapsulated bioactive ingredients in foods and pharmaceuticals.
Acknowledgments
This study was supported by the Wageningen Centre for Food Sciences, The Netherlands.
We thank René van der Heijden from NIZO food research for his contribution to the discussion on model statistics.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Andersson, A., P. E. Granum, and U. Ronner. 1998. The adhesion of Bacillus. Int. J. Microbiol. 39:93-99. [DOI] [PubMed] [Google Scholar]
- 2.Austin, J. W., and G. Bergeron. 1995. Development of bacterial biofilms in dairy processing lines. J. Dairy Res. 62:509-519. [DOI] [PubMed] [Google Scholar]
- 3.Avraham, R. 2000. The digestive system, p. 52. Chelsea House Publications, New York, NY.
- 4.Bouman, S., D. B. Lund, F. M. Driessen, and D. G. Schmidt. 1982. Growth of thermoresistant streptococci and deposition of milk constituents on plates of heat exchangers during long operating times. J. Food Prot. 45:806-812. [DOI] [PubMed] [Google Scholar]
- 5.Bovee-Oudenhoven, I. M. J., M. L. G. Lettink-Wissink, W. Van Doesburg, B. J. M. Witteman, and R. Van der Meer. 2003. Diarrhea caused by enterotoxigenic Escherichia coli infection of humans is inhibited by dietary calcium. Gastroenterology 125:469-476. [DOI] [PubMed] [Google Scholar]
- 6.Bovee-Oudenhoven, I. M. J., S. J. M. Ten Bruggencate, M. L. G. Lettink-Wissink, and R. Van der Meer. 2003. Dietary fructo-oligosaccharides and lactulose inhibit intestinal colonisation but stimulate translocation of salmonella in rats. Gut 11:1572-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bovee-Oudenhoven, I. M. J., M. L. G. Wissink, J. T. M. Wouters, and R. Van der Meer. 1999. Dietary calcium phosphate stimulates intestinal lactobacilli and decreases the severity of a salmonella infection in rats. J. Nutr. 129:607-612. [DOI] [PubMed] [Google Scholar]
- 8.De Jong, P., M. C. Te Giffel, and E. H. Kiezebrink. 2002. Prediction of the adherence, growth and release of thermo-resistant streptococci in production chains. Int. J. Microbiol. 74:13-25. [DOI] [PubMed] [Google Scholar]
- 9.De Jong, P., and H. J. L. J. Van der Linden. 1998. Polymerization model for prediction of heat-induced protein denaturation and viscosity changes in milk. J. Agric. Food Chem. 46:2136-2142. [Google Scholar]
- 10.De Jong, P., M. Verschueren, M. M. M. Vissers, J. Straatsma, and F. Smit. 2002. Hybrid modelling for development and optimisation of food production chains including costs and food quality, p. 13-17. In B. O'Connor and D. Thiel (ed.), Proceedings of the 2nd International Conference on Simulation in Food and Bio Industries. SCS Europe, Ghent, Belgium.
- 11.Haruta, S., K. Kawai, R. Nishii, S. Jinnouchi, K. Ogawara, K. Higaki, S. Tamura, K. Arimori, and T. Kimura. 2002. Prediction of plasma concentration-time curve of orally administered theophylline based scintigraphic monitoring of gastrointestinal transit in human volunteers. Int. J. Pharm. 233:179-190. [DOI] [PubMed] [Google Scholar]
- 12.Hentges, D. J., A. J. Stein, S. W. Casey, and J. U. Que. 1985. Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect. Immun. 47:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooker, K. D., and J. T. Di Piro. 1988. Effect of antimicrobial therapy on bowel flora. Clin. Pharm. 7:878-888. [PubMed] [Google Scholar]
- 14.Kararli, T. T. 1995. Comparison of the gastrointestinal anatomy, physiology and biochemistry of humans and commonly used laboratory animals. Biopharm. Drug Dispos. 16:351-380. [DOI] [PubMed] [Google Scholar]
- 15.Klijn, N., A. H. Weerkamp, and W. M. De Vos. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61:2771-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langeveld, L. P. M., R. M. G. E. Van Montfort-Quasig, A. H. Weerkamp, R. Waalewijn, and J. S. Wever. 1995. Adherence, growth and release of bacteria in a tube heat exchanger for milk. Neth. Milk Dairy J. 49:207-220. [Google Scholar]
- 17.Mathers, J. C., H. Smith, and S. Carter. 1997. Dose-response effects of raw potato starch on small-intestinal escape, large-bowel fermentation and gut transit time in the rate. Br. J. Nutr. 78:1015-1029. [PubMed] [Google Scholar]
- 18.Minekus, M. 1995. A multi-compartmental dynamic computer controlled model simulating the stomach and small intestine. Altern. Lab. Anim. 23:197-209. [Google Scholar]
- 19.Minekus, M. 1998. Development and validation of a dynamic model of the gastrointestinal tract. Ph.D. dissertation. University of Utrecht, Utrecht, The Netherlands.
- 20.Mozes, N., F. Marchal, M. Hermesse, J. L. Van Haecht, L. Reuliaux, A. J. Léonard, and P. G. Rouxchet. 1987. Immobilization of micro-organisms by adhesion; interplay of electrostatic and non-electrostatic interactions. Biotechnol. Bioeng. 30:439-450. [DOI] [PubMed] [Google Scholar]
- 21.Nelder, J. A., and R. Mead. 1965. A simplex method for function minimization. Comp. J. 7:308-313. [Google Scholar]
- 22.Parkar, S. G., S. H. Flint, J. S. Palmar, and J. D. Brooks. 2002. Factors influencing attachment of thermophilic bacilli to stainless steel. J. Appl. Microbiol. 90:901-908. [DOI] [PubMed] [Google Scholar]
- 23.Ronner, U., U. Husmark, and A. Hendriksson. 1990. Adhesion of Bacillus spores in relation to hydrophobicity. J. Appl. Bacteriol. 69:550-556. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg, E. 1986. Microbial surfactants. Crit. Rev. Biochem. 3:109-132. [Google Scholar]
- 25.Smit, F., P. De Jong, J. Straatsma, and M. Verschueren. 2001. NIZO-Premia enables knowledgement in the food industry. Part 1: background and application of the system. Voedingsmiddelentechnologie 34:(22)23-26. (In Dutch.) [Google Scholar]
- 26.Takumi, K., R. De Jonge, and A. Havelaar. 2000. Modelling inactivation of Escherichia coli by low pH: application to passage through the stomach of young and elderly people. J. Appl. Microbiol. 89:935-943. [DOI] [PubMed] [Google Scholar]
- 27.Te Giffel, M. C., R. R. Beumer, L. P. M. Langeveld, and F. M. Rombouts. 1997. The role of heat exchangers in the contamination of milk with Bacillus cereus in dairy processing plants. Int. J. Dairy Technol. 50:43-47. [Google Scholar]
- 28.Westerterp, K. R., W. P. M. Van Swaaij, and A. A. C. M. Beenackers. 1984. Chemical reactor design and operation. John Wiley & Sons, New York, N.Y.
- 29.Wilkinson, M. H. F. 2002. Model intestinal micro flora in computer simulation: a simulation and modelling package for host-micro flora interactions. IEEE Trans. Biomed. Eng. 49:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson, M. H. F. 1998. Ordinary differential equations for modelling bacterial interaction in the gut. MIMICS Technical Report. University of Groningen, Groningen, The Netherlands.
- 31.Zottola, E. A., and K. C. Sasahara. 1994. Microbial biofilms in the food processing industry—should they be a concern? Int. J. Food Microbiol. 23:125-148. [DOI] [PubMed] [Google Scholar]