Abstract
The ability of a ciliate to inactivate bacteriophage was studied because these viruses are known to influence the size and diversity of bacterial populations, which affect nutrient cycling in natural waters and effluent quality in sewage treatment, and because ciliates are ubiquitous in aquatic environments, including sewage treatment plants. Tetrahymena thermophila was used as a representative ciliate; T4 was used as a model bacteriophage. The T4 titer was monitored on Escherichia coli B in a double-agar overlay assay. T4 and the ciliate were incubated together under different conditions and for various times, after which the mixture was centrifuged through a step gradient, producing a top layer free of ciliates. The T4 titer in this layer decreased as coincubation time increased, but no decrease was seen if phage were incubated with formalin-fixed Tetrahymena. The T4 titer associated with the pellet of living ciliates was very low, suggesting that removal of the phage by Tetrahymena inactivated T4. When Tetrahymena cells were incubated with SYBR gold-labeled phage, fluorescence was localized in structures that had the shape and position of food vacuoles. Incubation of the phage and ciliate with cytochalasin B or at 4°C impaired T4 inactivation. These results suggest the active removal of T4 bacteriophage from fluid by macropinocytosis, followed by digestion in food vacuoles. Such ciliate virophagy may be a mechanism occurring in natural waters and sewage treatment, and the methods described here could be used to study the factors influencing inactivation and possibly water quality.
The impact of viruses on water quality is likely more complex than previously thought as increasingly viruses are being recognized as the most abundant biological entities in water (5). Traditionally, the concern has been for viruses pathogenic to humans and how these can be inactivated, but bacteriophage or phage should also be considered because of their impact on microbial ecology. By killing bacteria, phage act to regulate nutrient cycling and the composition of bacterial communities (30). Killing of host bacteria by lytic phage reduces the amount of bacterial biomass available for transfer to higher trophic levels. The capacity for selective, host-specific killing can influence the types and abundances of bacterial species present in natural waters, the basis of the “kill the winner” hypothesis (29). The hypothesis is that phage with high host specificity influence bacterial community structure by more frequently killing hosts with higher growth rates (those winning the competition for nutrients). This contributes to maintenance of bacterial species richness and thereby prevents competitive exclusion in aquatic systems (18, 29). Phage also could operate in a similar manner in sewage treatment plants, influencing the process and effluent quality (31).
Protozoa could have a role in phage inactivation by “killing the killer of the winner” (18), although such actions currently are poorly understood. Protozoa may affect virus levels by grazing on infected host bacteria and by grazing on free phage particles. In the marine environment, heterotrophic flagellates were found to ingest viruses (12), but the rate was not considered significant (9). The ciliates are another group of protozoa of interest because they can be found in most freshwater habitats, including engineered ones (7). However, their interactions with phage have been studied infrequently (2, 6).
Among the ciliates, Tetrahymena has been convenient for cell biology studies because several species are easily grown axenically (10). They are filter feeders, and much is known about their phagosome-lysosome pathway (13, 22). Food vacuoles form at the base of the buccal cavity during the uptake of particles by phagocytosis and the uptake of fluid by pinocytosis (22). Tetrahymena has been found in sewage treatment plants (7), and as a result, the association between Tetrahymena and several viruses infecting mammals has been studied (3, 15). In one case, Tetrahymena removed poliovirus from a coincubation medium (15). The inactivation of phage by Tetrahymena has been examined only briefly and in a biologically complex, essentially uncontrolled pilot plant (2). Therefore, we devised a system both to examine T4 phage removal by T. thermophila and to explore the cellular mechanism responsible.
MATERIALS AND METHODS
T4 bacteriophage and bacterial host.
T4 bacteriophage (ATCC 11303-B4) stock was maintained at 4°C in 3% (wt/vol) Trypticase soy broth (TSB) (Becton-Dickinson, Sparks, MD). Escherichia coli B (ATCC 11303), the T4 host strain, was grown overnight in TSB medium at 37°C, prior to its use for experimentation. Assessment of phage titer was done using the double-agar overlay, and all samples were plated in duplicate (1). The bottom layer of agar was composed of 3% (wt/vol) TSB, 1.5% (wt/vol) agar (Fisher Scientific, Fair Lawn, NJ), and 0.03% (wt/vol) CaCl2 · 2H2O (Sigma, St. Louis, MO), while the top layer of agar contained the same TSB and CaCl2 · 2H2O concentrations, but only 0.7% (wt/vol) agar.
Tetrahymena thermophila.
The method for culturing Tetrahymena thermophila was adapted from Gilron et al. (11). Batch cultures were maintained at 22°C in 10 ml of proteose peptone, yeast extract-enriched medium (PPYE), composed of 0.125% (wt/vol) dextrose (Sigma), 0.5% (wt/vol) proteose peptone (Becton-Dickinson), and 0.5% (wt/vol) yeast extract (Becton-Dickinson) dissolved in MilliQ water. In preparation for experimentation, 1 ml of the T. thermophila batch was added to 10 ml of fresh PPYE (to maintain the stock) while the remaining 9 ml was added to 50 ml of fresh PPYE in a 75-cm2 non-tissue culture-treated flask and kept at 22°C on an orbital shaker (50 rpm) overnight. Enumeration of ciliate cultures was performed using a Z2 Coulter particle count and size analyzer (Coulter, Luton, United Kingdom), using an upper limit diameter of 50 μm and a lower limit diameter of 20 μm. Preparation of T. thermophila populations for counting involved combining 19 ml of IsoFlow sheath fluid solution (Coulter), 500 μl T. thermophila culture, and 500 μl 10% neutral, buffered Formalin (Sigma). Four counts were done per sample, and these values were added and then multiplied by 20 to convert to cells/ml.
General protocol for bacteriophage-protozoan coincubation.
Following enumeration, the T. thermophila culture was centrifuged for 10 min at 400 × g and supernatant was removed. Pelleted ciliates were washed twice with Osterhout's solution (24, 27) and were resuspended in fresh Osterhout's solution for use in experimentation. T4 bacteriophage stock was added as required. Mixtures were maintained in 15-ml capped, plastic test tubes at 22°C (unless otherwise stated) on an orbital shaker (50 rpm). Such mixtures were then subsampled for analysis as indicated in the following sections.
SGS of the bacteriophage-protozoan mixture.
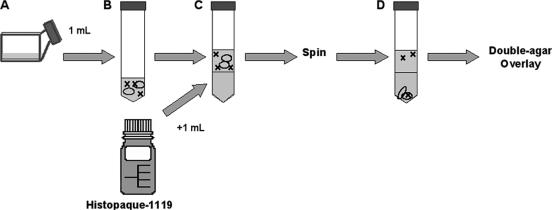
Unassociated phage was separated from protozoans (and any associated phage) using the following technique. Samples were added to sterile, capped test tubes. To this, a sterile Pasteur pipette was used to add 1 ml of Histopaque-1119 (Sigma-Aldrich, St. Louis, MO) for every 1 ml of sample, beneath the protozoan-virus mixture. Subsequent centrifugation at 400 × g for 10 min separated protozoans from any unassociated phage. The supernatant (referring to the Osterhout's phase above the dense Histopaque phase) was diluted with TSB as required for use in the double-agar assay. After overnight incubation at 37°C, plaques were counted to assess titer. The efficiency of this method of separation was tested by inoculating Osterhout's solution with T4 phage (105 PFU/ml), plating the preseparation sample, performing step-gradient separation (SGS), and plating the supernatant along with an undiluted sample of the Histopaque-1119 phase (Fig. 1).
FIG. 1.
Experimental protocol for evaluating Tetrahymena-phage interactions. (A) Experiments began by adding T4 bacteriophage to T. thermophila in medium, usually Osterhout's solution, and incubating this culture in 75-cm2 tissue culture flasks that were maintained on an orbital shaker (50 rpm) at room temperature. (B) At various times afterwards, a 1-ml sample of the culture was transferred to a 15-ml sterile plastic centrifuge tube. (C) A long glass Pasteur pipette was used to layer 1 ml of Histopaque on this sample, after which the tube was centrifuged at 400 × g for 10 min at room temperature. (D) After centrifugation, a pellet was observed and a distinct top layer (the medium) and a distinct bottom layer (the Histopaque) were evident. The distribution of Tetrahymena cells, which are indicated as ovals in the diagram, was monitored by phase-contrast microscopy, and the distribution of phage, which are indicated by X, was monitored by double-agar overlay. After the step gradient centrifugation, all of the Tetrahymena cells were found as a pellet, regardless of the experiment. In contrast, the titer of the phage in panel D, which was usually expressed relative to the titer added in panel A, varied with the experiment.
T4 phage infectivity in various media.
Ten milliliters of Osterhout's solution and PPYE (both sterile and protozoan conditioned) was inoculated with T4 stock to produce a final titer of 105 PFU/ml, in the absence of protozoa. Conditioned PPYE was obtained by centrifuging 2-week-old, PPYE-grown T. thermophila cultures at 400 × g for 10 min. Supernatant was collected and passed through a 0.2-μm syringe filter. All samples were kept in capped, plastic test tubes at 22°C. At 0, 4, 24, 48, and 72 h, samples were serially diluted using TSB and plated using the double-agar overlay. After overnight incubation at 37°C, plaques were counted to assess viral titer. Sterile Histopaque-1119 was also tested for effects on viral infectivity in the same manner, with the time frame being limited to testing at 0, 4, and 24 h.
Assessment of T4 infectivity in the presence of T. thermophila.
Both viable and fixed T. thermophila samples consisted of an overnight culture (35,000 cells/ml final density) transferred to a 15-ml test tube. For the fixed sample, 1 ml of 10% neutral, buffered Formalin was added for every 3 ml of sample and allowed to sit for 10 min; viable samples were treated in the same manner with Osterhout's solution in place of formalin. Samples were inoculated with T4 phage stock to produce a titer of 105 PFU/ml. At 0, 4, 24, 48, 72, and 96 h, the SGS was performed and phage infectivity was assessed by the double-agar overlay. A 1,000-fold dilution of the SGS supernatant was assayed for 0 and 4 h, with decreasing dilutions being plated as phage infectivity decreased.
Saturation of the rate of virophagy.
Using an enumerated, overnight T. thermophila culture, protozoans were transferred to Osterhout's solution to produce a final density of 35,000 cells/ml. T4 phage stock was used to produce samples of 105, 106, 108, 109, and 1010 PFU/ml. At 0, 4, 24, and 48 h, the SGS was performed and change in titer was assayed by the double-agar overlay method.
Pellet assay.
An overnight culture of T. thermophila (35,000 cells/ml) was suspended in Osterhout's buffer and inoculated with T4 phage (105 PFU/ml). At 0, 4, and 24 h, 2 ml of sample was chilled on ice. An SGS was performed, and ciliates were resuspended in 1 ml of 20 mM Tris-HCl and returned to ice to chill. To this was added 2× lysis buffer (1.4 M sucrose, 4 mM EDTA, 2% Triton X-100) in a 1:1 ratio to the Tris-HCl volume, and the sample was vortexed for 1 min. The lysed sample was then plated, and viral titer was assessed through the double-agar overlay. Following the 24-h time point, the remaining phage-ciliate mixture was centrifuged (400 × g for 10 min), washed twice with fresh Osterhout's solution, and resuspended in fresh Osterhout's solution. Twenty-four hours following this transfer to fresh buffer, ciliates were pelleted by the SGS method. The viral titer of the supernatant was assayed, along with that of the ciliates, after pelleting and lysing as described above.
Epifluorescence of T4 phage.
SYBR gold nucleic acid stain (Invitrogen, Eugene, OR) was used as a means of fluorescently labeling T4 phage. Prior to labeling, T4 stock was treated with RNase-free, DNase I (100 U/ml) for 10 min. SYBR gold stock dye solution (provided as a 10,000× concentrate) was diluted and added to the phage stock, resulting in a 2.5× dye solution, and allowed to sit in the dark for 10 min (19). The labeled-phage solution was then passed through a molecular weight 100,000 Centricon concentrator (Fisher) and centrifuged at 2,000 × g for 5 min to remove unbound dye. The labeled-phage solution was rinsed twice by passing 1 ml of Osterhout's solution through the concentrator at 2,000 × g for 5 min. A control solution (consisting of Osterhout's solution, SYBR gold dye solution, and DNase-free RNase I) was also passed through a fresh concentrator and rinsed twice with Osterhout's solution. The concentrate was returned to its original volume using Osterhout's solution. This labeled-phage solution was diluted as required and added to Tetrahymena (35,000 cells/ml), which had previously been suspended in Osterhout's solution, to produce samples of 1010, 109, and 108 PFU/ml. A control sample was also produced, consisting of Tetrahymena suspended with the T4-free dye solution. Samples were kept in the dark for 2 h, after which 60 μl was moved to a microcentrifuge tube and centrifuged for 5 min at 2,000 × g. Thirty microliters of the supernatant was removed, and 3 μl of 10% neutral, buffered formalin were added. After fixation, samples were viewed using a fluorescence microscope equipped with a Nikon B-2A filter (excitation, 450 nm; emission, 520 nm).
Confocal microscopy.
Tetrahymena cells (35,000/ml) were mixed with SYBR gold-labeled T4 phage (final concentration, 108 PFU/ml) and maintained at 22°C for 15 min. At this time, Tetrahymena cells were isolated using centrifugation and viewed using confocal microscopic analysis in a Zeiss LSM 510 META confocal microscope using an argon laser as a light source, at an excitation wavelength of 488 nm line and bandpass filter of 505 nm.
Effect of cytochalasin B on decrease of phage infectivity.
Cytochalasin B (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO) and added to suspensions of T. thermophila cells in Osterhout's solution (final concentration, 35,000 cells/ml) at concentrations of 0.1, 10, and 50 μg/ml; a 0-μg/ml control was made using DMSO only. The final volume of DMSO did not exceed 0.5% (wt/vol) in any sample. T4 phage stock was added to produce a final titer of 105 PFU/ml. At 0, 1, 2, and 24 h, 1 ml was moved to a 15-ml plastic, capped test tube, and, using the SGS, unassociated phage were isolated and their titer was determined. Additionally, SYBR gold-labeled T4 phage (108/ml) were added to Tetrahymena cells (35,000/ml) suspended in Osterhout's solution containing 50 μg/ml cytochalasin B. Two hours later, cells were fixed with 10% neutral, buffered formalin and viewed using a fluorescence microscope.
Effect of temperature on phage inactivation.
Four samples of viable T. thermophila (35,000 cells/ml)-T4 phage (105 PFU/ml) mixtures in Osterhout's solution were prepared for incubation at 4°C, 22°C, 28°C, and 37°C. Protozoan-free controls were also prepared for each temperature. Samples were assayed at 0, 4, and 24 h using the SGS and double-agar overlay. After overnight incubation at 37°C, plaques were counted to assess viral titer. Additionally, SYBR gold-labeled T4 phage (108 PFU/ml) were added to Tetrahymena cells (35,000/ml) suspended in Osterhout's solution that had been previously chilled to 4°C for 10 min. Following inoculation, the mixture was returned to 4°C for 2 h, after which the cells were fixed with 10% neutral, buffered formalin and viewed using the fluorescence microscope.
Statistical analysis.
GraphPad InStat was used for statistical analysis of raw data.
RESULTS
Maintenance of T4 titer in various solutions.
As background for investigating any direct effects of T. thermophila on the phage, the maintenance of T4 titer, or infectivity, in solutions used to grow and manipulate Tetrahymena was tested. T4 continued to be infective after up to 4 days in medium (PPYE) or Osterhout's solution that was used, respectively, to grow or maintain the ciliates (data not shown). T4 infectivity also was undiminished by incubation in PPYE and Osterhout's solution, in which Tetrahymena had been grown or maintained, respectively (data not shown). These solutions are described as being conditioned because potentially they could contain factors released by Tetrahymena. T4 infectivity also was unchanged by incubation in Histopaque-1119, which was to be used in step gradient centrifugation to separate T4 from Tetrahymena (data not shown).
Separation of T4 from Tetrahymena by centrifugation.
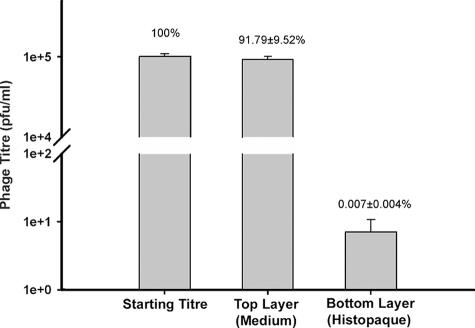
T4 could be separated from Tetrahymena by centrifugation through a step gradient of Histopaque-1119 (Fig. 1). When T4 alone in Osterhout's solution was layered on top of Histopaque-1119 and centrifuged, nearly all the phage as judged by their titer, or infectivity, were found in the top layer and only a miniscule fraction had penetrated into the Histopaque layer (Fig. 2). When Tetrahymena cells alone in Osterhout's solution were layered on top of Histopaque-1119 and centrifuged, all the ciliates as judged by light microscopy were found in the bottom Histopaque layer. In this solution, the Tetrahymena cells were slightly rounded and less motile, but they recovered their shape and motility upon being returned to Osterhout's solution and PPYE.
FIG. 2.
Distribution of T4 titer after mixing the phage with Tetrahymena and immediately subjecting the medium to step gradient centrifugation. Within a minute of 105 PFU of T4 being added to a Tetrahymena culture, Tetrahymena cells were pelleted by centrifugation (see Fig. 1), and the T4 titer was measured in the top (medium) and bottom (Histopaque) layers. The data represent the mean of three separate trials. The means of the starting titer and titer in the top layer were compared using the Tukey-Kramer multiple comparisons test, and variation was not considered to be significantly different from that expected by chance (P > 0.05).
T4 titer after incubation with Tetrahymena.
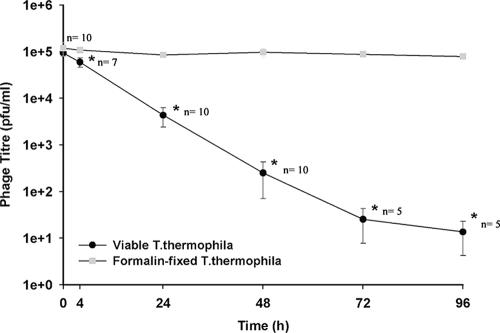
T4 infectivity declined as the time of incubation with Tetrahymena increased (Fig. 3). When the mixture of phage and Tetrahymena was centrifuged and the top layer was assayed for infective T4, the phage titer declined steadily with incubation time (Fig. 3). When the phage was incubated with Tetrahymena that had been killed by formalin, the phage remained in the top layer and showed no decline with time. With living Tetrahymena, the decline in phage titer was seen at the first time point (4 h) examined, and by 24 h, the phage had been decreased by 95.3% ± 2.1% (n = 10). Little or no phage was detected in the bottom layer. When the Tetrahymena pellet was lysed, very few infective phage were released (Table 1). As the coincubation time increased, the titer in the pellet did not increase. When the pellet from a 24-h coincubation was resuspended in buffer without T4 and incubated for a further 24 h, the titer became even lower (Table 1). Overall, the results suggested that Tetrahymena actively removed and inactivated T4 from the coincubation medium.
FIG. 3.
Removal of T4 from medium by live Tetrahymena as the coincubation time with the phage increased. Coincubations of T4 phage (105 PFU/ml) were begun in Osterhout's solution with either living or formalin-fixed Tetrahymena (35,000 cells/ml). At various times, the Tetrahymena cells were removed by centrifugation, and the remaining virus in the medium was measured as PFU The mean numbers of PFU with standard deviation (n = 3 to 10) are plotted. The data were subjected to a Tukey-Kramer multiple comparisons test. The asterisks identify values significantly different from those in the fixed sample (P < 0.05).
TABLE 1.
T4 titer in pellet of Tetrahymena after centrifugation through Histopaquea
| Coincubation time at 22°C before centrifugation (h) | Treatment of Tetrahymena pelletb | Subsequent centrifugationsc | Titer (mean PFU ± SD)d |
|---|---|---|---|
| 0 | Lysede | None | 2,867 ± 702 |
| 4 | Lysed | None | 3,550 ± 900 |
| 24 | Lysed | None | 3,060 ± 198 |
| 24 | Resuspended | Supernatant | 8 ± 6 |
| in buffer | Pellete | 28 ± 24 | |
| for 24 hf |
The T4 titer at the start was 100,000 PFU for each of the coincubation conditions before centrifugation.
Pellet formed after centrifugation through step gradient of Histopaque.
After 24 h, Tetrahymena cells were collected by centrifugation but not through Histopaque and the supernatant and pellet were assayed for PFU.
Titer expressed as mean of two experiments ± standard deviations (SD).
Pellet was lysed with Triton X-100.
Pellet was resuspended in Osterhout's buffer.
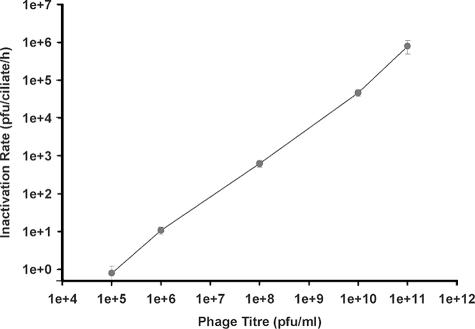
When the titer at the start of the coincubation was increased and the number of Tetrahymena cells was kept constant, the inactivation rate increased, without saturation (Fig. 4).
FIG. 4.
Removal of T4 by Tetrahymena from medium in which the starting phage titer varied. Coincubations of T4 phage and Tetrahymena cells (35,000/ml) were begun in Osterhout's solution at differing T4/Tetrahymena ratios, from a low of 10 to 1 to a high of 5.0 × 106 to 1. After 4 h, the Tetrahymena cells were removed by centrifugation and the remaining phage in buffer was measured as PFU. The removal of PFU was termed the inactivation rate and is expressed as PFU/ciliate/h (n = 4).
Visualizing T4-Tetrahymena interactions.
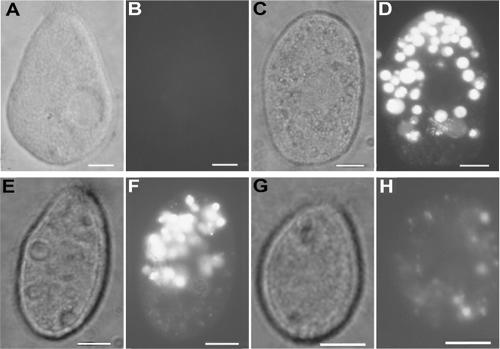
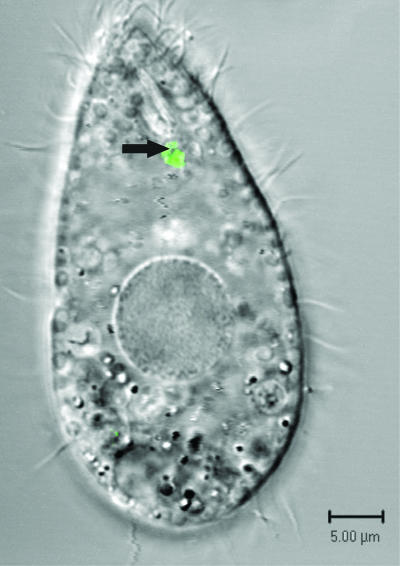
Tetrahymena cells and T4 phage that had been fluorescently labeled with SYBR gold were incubated together and viewed by fluorescence and confocal microscopy. When examined by fluorescence microscopy 2 h after the initiation of the coincubation, Tetrahymena contained pockets of fluorescence that had the approximate size and shape of food vacuoles (Fig. 5D, F, and H). When examined by confocal microscopy 15 min after the initiation of the coincubation, some fluorescent particles were seen within vacuoles forming at the base of the buccal cavity (Fig. 6). The exclusive location of fluorescence staining in food vacuoles was lost with longer incubations. Nuclear staining of Tetrahymena occurred after overnight incubations.
FIG. 5.
Visualizing T4-Tetrahymena interactions by fluorescence microscopy. Tetrahymena cells (35,000/ml) were exposed to SYBR gold nucleic acid stain solution (A and B) or to SYBR gold-labeled T4 phage at 1010 PFU/ml (C and D), 109 PFU/ml (E and F), and 108 PFU/ml (G and H) for 2 h at room temperature. Cultures were fixed in Formalin immediately prior to microscopic analysis. Note that phase-contrast photomicrographs are shown in panels A, C, E, and G; fluorescence photomicrographs are shown in panels B, D, F, and H. Bars are 10 μm in all panels.
FIG. 6.
Visualizing T4-Tetrahymena interactions by confocal microscopy. SYBR gold-labeled T4 phage (final concentration, 108 PFU/ml) were added to Tetrahymena (35,000 cells/ml). After 15 min, ciliates were separated from unassociated phage by centrifugation, isolated, and viewed by confocal microscopy. An arrow indicates phage in a food vacuole. The micrograph is a two-channel image taken in fluorescence and differential interference contrast modes. Bar, 5 μm.
Effect of cytochalasin B and temperature on T4-tetrahymena interactions.
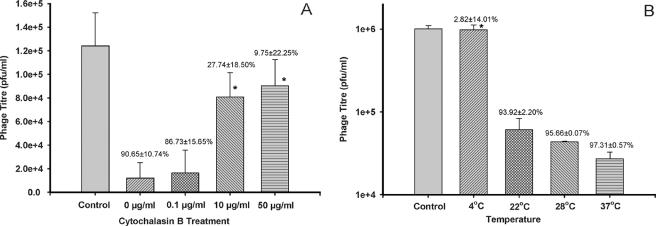
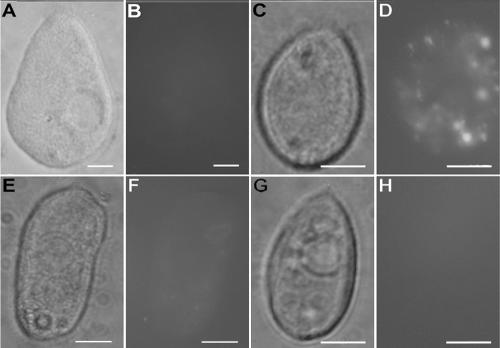
Cytochalasin B inhibited the removal of T4 by Tetrahymena. As the concentration of cytochalasin B increased, the number of phage persisting in the coincubation medium increased (Fig. 7A). The titer in the top layer after 24 h with 10 and 50 μg/ml of cytochalasin B was not significantly different from the starting titer in this layer (Fig. 7A). The amount of fluorescence in Tetrahymena after incubation with SYBR gold-labeled T4 was decreased by cytochalasin B (Fig. 8E and F).
FIG. 7.
Effect of cytochalasin B and temperature on removal of T4 by Tetrahymena. Coincubations of Tetrahymena cells and T4 phage were begun in Osterhout's solution with a known amount of T4 phage and allowed to proceed under different conditions for 24 h, at which time, cultures were centrifuged to remove the Tetrahymena cells and to permit measurement of the remaining virus as PFU. The mean PFU with standard deviation (n = 2 to 4) are plotted, with the percent decrease in PFU from time zero listed above the bars. The data were subjected to a Tukey-Kramer multiple comparisons test. (A) The coincubations were begun at 105 PFU and proceeded with cytochalasin B at 0.1, 10, or 50 μg/ml or without cytochalasin B (0 μg/ml). The control bar represents cell-free, phage-inoculated medium. Significant inhibition of phage inactivation occurred at 10 and 50 μg/ml of cytochalasin B, as determined by Tukey-Kramer multiple comparisons test (P < 0.05). (B) The coincubations were begun at 106 PFU and proceeded at 4°C, 22°C, 28°C, and 37°C. The control bar represents cell-free, phage-inoculated medium maintained at 4°C. Low temperature (4°C) was found to significantly inhibit phage inactivation, as determined by Tukey-Kramer multiple comparisons test (P < 0.05).
FIG. 8.
Effect of cytochalasin B and temperature on T4-Tetrahymena interactions as visualized by phase-contrast and fluorescence microscopy. Tetrahymena cells (35,000/ml) were exposed to SYBR gold nucleic acid stain solution (A and B) or to SYBR gold-labeled T4 phage at 108 PFU/ml (C and D) for 2 h at room temperature in the presence of cytochalasin B (E and F) or at 4°C (G and H). Cultures were fixed in formalin immediately prior to microscopic analysis. Phase-contrast photomicrographs are shown in panels A, C, E, and G, and fluorescence photomicrographs are shown in panels B, D, F, and H. Bars are 10 μm in all panels.
Low temperature (4°C) also inhibited the removal of T4 by Tetrahymena. When Tetrahymena and T4 were coincubated at 4°C for 24 h, the titer in the top layer remained unchanged, whereas when the incubations were done at 22, 28, or 37°C, the titer in this layer was reduced by over 90% (Fig. 6B). In coincubations at 4°C with T4 that had been stained with SYBR gold, no fluorescence was seen associated with Tetrahymena (Fig. 8G and H).
DISCUSSION
Coincubations of T4 phage and Tetrahymena were terminated rapidly by centrifugation through a discontinuous or step gradient of Histopaque to pellet the Tetrahymena with associated T4 and to leave only free or unassociated T4 in the coincubation medium. Gradient centrifugation has been used to separate ciliates from bacteria (4), but appears not to have been used for viruses, although viruses have been separated from spermatozoa by discontinuous gradient centrifugation (14). In the case of T4 and Tetrahymena, simple differential centrifugation, which does not involve a gradient, would likely have accomplished the same separation, but because Tetrahymena cells rapidly swim up, the precise separation of pellet and supernatant is harder to achieve, especially if multiple cultures are being handled at the same time. In contrast, with the step gradient, the Tetrahymena cells remained in the Histopaque for many minutes and the supernatant is clearly defined by the interface with Histopaque. The supernatant can be easily drawn off and the viral titer assayed to monitor the disappearance or removal of T4 from the coincubation medium.
T4 phage were removed from medium only if living Tetrahymena cells were present. When Tetrahymena was Formalin fixed, unreduced numbers of T4 remained in the supernatant. As well, when coincubations were done at 4°C, unreduced numbers of T4 phage were found in the supernatant. This is supported by epifluorescence microscopy, which clearly showed a lack of fluorescence either on or in Tetrahymena cells when phagocytosis was inhibited by temperature. These observations suggest that adsorption to the Tetrahymena was not the mechanism behind T4 removal. The rate of removal of T4 by living Tetrahymena cells suspended in Osterhout's solution was exponential for at least 48 h.
T4 phage were not inactivated by the medium in which Tetrahymena had been grown. This suggests that T4 removal was not due to the release of antiviral components. A similar conclusion was reached in the case of poliovirus being removed by Tetrahymena (15). Thus, the disappearance of T4 from the coincubation medium by Tetrahymena was most likely due to the active engulfment of T4 by the ciliates.
The cellular process behind T4 removal by Tetrahymena appeared to involve food vacuole formation. First, T4 removal was inhibited by cytochalasin B, which has been shown to inhibit food vacuole formation in Tetrahymena by disrupting actin organization (21, 32). Second, T4 removal did not occur at 4°C, a temperature shown to inhibit formation of food vacuoles in Tetrahymena (20). Finally, the time course of T4 removal was consistent with the development of food vacuoles. The earliest coincubation time point to be examined was at 4 h, and the decline in PFU at this point was significant. This time period falls well within the Tetrahymena digestive cycle, which takes 2 h from food vacuole formation to egestion (22). As well as forming food vacuoles, Tetrahymena cells also form receptor dependent, smaller vesicles with an apparent clathrin coat at the parasomal sacs of Tetrahymena (8, 23). These appear not to be involved in T4 removal by Tetrahymena because removal could not be saturated through modification of the phage/protozoan ratio and fluorescence was never observed on the cell surface onto which the parasomal sacs open.
In the natural habitat, Tetrahymena would typically feed on bacterium-sized food particles, probably with an optimum clearance rate on particles of about 0.5 to 1.0 μm in diameter (10). These food particles are ingested in vacuoles that are considered to be phagosomes, and their formation is phagocytosis (22). In fluid axenic medium, Tetrahymena can form food vacuoles with culture fluid. These fluid-filled food vacuoles are called pinosomes and their formation is pinocytosis, which is the process of internalizing fluid (22). It has long been known that rapid growth of Tetrahymena in fluid culture medium is maximized if there is particulate material present, which presumably increases food vacuole formation (16, 25). Thus, the particulate nature of T4 may stimulate the ingestatory functions of Tetrahymena. Phagocytosis is generally defined as involving particles greater than 500 nm in diameter (25). T4 viruses are approximately 250 nm long with a diameter of 85 nm (17). Therefore, T4 removal by Tetrahymena would not fit the definition of phagocytosis, but could be a consequence of at least one form of pinocytosis, which is the process of internalizing fluid. Micropinocytosis involves vesicles less than 200 nm in diameter (26). These dimensions make their involvement in T4 removal seem unlikely. The other form of pinocytosis is macropinocytosis, which creates vesicles larger than 1 μm in diameter by an actin-dependent process (26, 28). Tetrahymena does create vesicles with these characteristics in the cytopharynx of the oral apparatus. These are fluid-filled food vacuoles (20), which by current terminology can be considered macropinosomes. Therefore, macropinocytosis in the cytopharynx of the oral apparatus is the likely mechanism behind the removal of T4 phage by Tetrahymena.
The removal of T4 by Tetrahymena appears to result in T4 inactivation. First, PFU declined continuously with time, suggesting that Tetrahymena offered no transitory refuge. Previously, a mammalian rotavirus was found to adsorb to Tetrahymena, but with time, the virus was released again (3). Second, although PFU were detected from the pellet of Tetrahymena, there was a significant decrease in pellet PFU at 24 h following the transfer of Tetrahymena from phage-inoculated solution to phage-free solution. Finally, transmission electron microscopy failed to provide unequivocal evidence of T4 being associated with Tetrahymena (data not shown). This is likely due to the rapid loss of their characteristic morphology by digestion in secondary lysosomes (22). At least one ciliate, Euplotes, has been reported to digest viral particles or phage under natural conditions. When Euplotes from a lake were examined over a year by transmission electron microscopy, some had food vacuoles with bacteria that were lysing and releasing phage particles, which were then digested, in the food vacuoles (6). Overall the removal of T4 by Tetrahymena likely involves T4 engulfment by macropinocytosis and subsequent T4 digestion in secondary lysosomes.
Conclusions.
Tetrahymena cells were able to remove T4 phage from solutions in which the ciliate and virus had been coincubated. Metabolically active ciliates were required for removal, and the cellular process responsible had the characteristics of macropinocytosis. Removal appeared to coincide with inactivation. Ciliates are known to continuously undergo macropinocytosis, which could mean that they are continuously inactivating phage. Phages, in turn, kill their bacterial hosts. By influencing both participants in phage-host interactions—by inactivating phage as well as grazing on bacteria—the ciliates might have a more important and intricate role in determining the microbial ecology of water, and in turn water quality, than suggested from their relatively low numbers in the environment.
Acknowledgments
This research was funded by grants from Earth and Environmental Technologies (ETech) of Ontario Centres for Excellence (OCE) and the Canadian Water Network (CWN).
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophage. Interscience Publishers, New York, NY.
- 2.Benyahya, M., J. Bohatier, H. Laveran, J. Senaud, and M. Ettayebi. 1998. Les virus des eaux usées et leur élimination au cours des traitments des effluents pollués. Année. Biol. 78:95-105. [Google Scholar]
- 3.Benyahya, M., H. Laveran, J. Bohatier, J. Senaud, and M. Ettayebi. 1997. Interactions between the ciliated protozoan Tetrahymena pyriformis and the simian rotavirus SA11. Eur. J. Protohistol. 33:211-213. [Google Scholar]
- 4.Berk, S. G., P. Guerry, and R. R. Colwell. 1976. Separation of small ciliate protozoa from bacteria by sucrose gradient centrifugation. Appl. Environ. Microbiol. 31:450-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 113:278-284. [DOI] [PubMed] [Google Scholar]
- 6.Clark, K. J. 1998. Virus particle production in lysogenic bacteria exposed to protozoan grazing. FEMS Microbiol. Lett. 166:177-180. [Google Scholar]
- 7.Curds, C. R. 1973. A theoretical study of factors influencing the microbial population dynamics of the activated sludge process. I. The effects of diurnal variations of sewage and carnivorous ciliated protozoa. Water Res. 7:1269-1284. [Google Scholar]
- 8.Elde, N. C., G. Morgan, M. Winey, L. Sperling, and A. P. Turkewitz. 2005. Elucidation of clathrin-mediated endocytosis in Tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLOS Genet. 1:514-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenchel, T. 1980. Suspension feeding of ciliated protozoa: structure and function of feeding organelles. Arch. Protistenkd. 123:239-260. [Google Scholar]
- 10.Frankel, J. 2000. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62:27-125. [DOI] [PubMed] [Google Scholar]
- 11.Gilron, G., S. G. Gransden, D. H. Lynn, J. Broadfoot, and R. Scroggins. 1999. A behavioral toxicity test using the ciliated protozoan Tetrahymena thermophila. I. Method description. Environ. Toxicol. Chem. 18:1813-1816. [Google Scholar]
- 12.Gonzalez, J. G., and C. A. Suttle. 1993. Grazing by marine nanoflagellates on viruses and viral-sized particles: ingestion and digestion. Mar. Ecol. Prog. Ser. 94:1-10. [Google Scholar]
- 13.Gortz, H. D., H. W. Kuhlmann, M. Mollenbeck, A. Tiedtke, J. Kusch, H. J. Schmidt, and A. Miyake. 1999. Intra- and intercellular communication systems in ciliates. Naturwissenschaften 86:422-434. [DOI] [PubMed] [Google Scholar]
- 14.Hanabusa, H., N. Kuji, S. Kato, H. Tagami, S. Kaneko, H. Tanaka, and Y. Yoshimura. 2000. An evaluation of semen processing methods for eliminating HIV-1. AIDS 14:1611-1616. [DOI] [PubMed] [Google Scholar]
- 15.Kim, T. D., and H. Unno. 1996. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 33:243-250. [Google Scholar]
- 16.Kiy, T., and A. Tiedke. 1992. Mass cultivation of Tetrahymena thermophila yielding high densities and short generation times. Appl. Microbiol. Biotechnol. 37:576-579. [Google Scholar]
- 17.Mesyanzhinov, V. V. 2004. Bacteriophage T4: structure, assembly, and initiation of infection studied in three dimensions. Adv. Virus Res. 63:287-352. [DOI] [PubMed] [Google Scholar]
- 18.Miki, T., and N. Yamamura. 2005. Intraguild predation reduces bacterial species richness and loosens the viral loop in aquatic systems: ‘kill the killer of the winner’ hypothesis. Aquat. Microb. Ecol. 40:1-12. [Google Scholar]
- 19.Mosier-Boss, P. A., S. H. Lieberman, J. M. Andrews, F. L. Rohwer, L. E. Wegley, and M. Breitbart. 2003. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl. Spectrosc. 57:1138-1144. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson, J. R. 1972. Further studies on vacuole formation in Tetrahymena pyriformis GL. Comp. Rend. Trav. Lab. Carlsberg 39:83-110. [PubMed] [Google Scholar]
- 21.Nilsson, J. R. 1977. Fine structure and RNA synthesis of Tetrahymena during cytochalasin B inhibition of phagocytosis. J. Cell Sci. 27:115-126. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson, J. R. 1979. Phagotrophy in Tetrahymena, p. 339-379. In M. Levandowsky and S. H. Hutner (ed.), Biochemistry and physiology of protozoa, 2nd ed. Academic Press, New York, NY.
- 23.Nilsson, J. R., and B. van Deurs. 1983. Coated pits and pinocytosis in Tetrahymena. J. Cell Sci. 63:209-222. [DOI] [PubMed] [Google Scholar]
- 24.Orias, E., and L. Rasmussen. 1979. Dual capacity for nutrient uptake in Tetrahymena. V. Utilization of amino acids and proteins. J. Cell Sci. 36:343-353. [DOI] [PubMed] [Google Scholar]
- 25.Osterhout, W. J. V. 1906. On the importance of physiologically balanced solutions for plants. I. Marine plants. Bot. Gaz. 43:127-134. [Google Scholar]
- 26.Rupper, A., and J. Cardeli. 2001. Regulation of phagocytosis and endo-phagosomal trafficking pathways in Dictyostelium discoideum. Biochim. Biophys. Acta 1525:205-216. [DOI] [PubMed] [Google Scholar]
- 27.Society of Protozoologists. 1958. A catalogue of laboratory strains of free-living and parasitic protozoa. J. Protozool. 5:1-8. [Google Scholar]
- 28.Swanson, J. A., and C. Watts. 1995. Macropinocytosis. Trends Cell Biol. 5:424-428. [DOI] [PubMed] [Google Scholar]
- 29.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 30.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 31.Withey, S., E. Cartmell, L. M. Avery, and T. Stephenson. 2005. Bacteriophage—potential for application in wastewater treatment processes. Sci. Total Environ. 339:1-18. [DOI] [PubMed] [Google Scholar]
- 32.Zackroff, R. V., and L. A. Hufnagel. 2002. Induction of anti-actin drug resistance in Tetrahymena. J. Eukaryot. Microbiol. 49:475-477. [DOI] [PubMed] [Google Scholar]