Abstract
This investigation provides molecular analyses of the periodontal microbiota in health and disease. Subgingival samples from 47 volunteers with healthy gingivae or clinically diagnosed chronic periodontitis were characterized by PCR-denaturing gradient gel electrophoresis (DGGE) with primers specific for the V2-V3 region of the eubacterial 16S rRNA gene. A hierarchical dendrogram was constructed from band patterns. All unique PCR amplicons (DGGE bands) were sequenced for identity. Samples were also analyzed for the presence of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis by multiplex PCR. Associations of patient age, gender, and smoking status together with the presence of each unique band and putative periodontal pathogens with disease were assessed by logistic regression. Periodontal pockets were colonized by complex eubacterial communities (10 to 40 distinct DGGE bands) with substantial individual variation in the community profile. Species diversity in health and disease was determined by the Shannon-Weaver index of diversity and compared by the Mann-Whitney U test. Sequence analyses of DGGE amplicons indicated the occurrence of many nontypical oral species and eubacteria previously associated with this environment. With the exception of T. forsythensis, the putative pathogens were not detected by DGGE. Multiplex PCR, however, detected T. forsythensis, A. actinomycetemcomitans, and P. gingivalis in 9% 16%, and 29% of the patients with disease, respectively. The presence of A. actinomycetemcomitans was significantly associated with disease (P < 0.01). Statistical analyses indicated that the presence of Treponema socranskii and Pseudomonas sp. was a significant predictor of disease (P < 0.05) and that there was no significant difference (P > 0.05) in terms of eubacterial species diversity between health and disease.
Periodontitis is a generic term relating to inflammation of the tissues supporting the teeth but is widely attributed to succession by polymicrobial communities (36, 58, 74). The etiology of the condition is further complicated by the presence of a complex resident subgingival microbiota that underlies both periodontal health and disease (22, 45). Periodontitis is often self-limiting; invasion of bacteria beyond the gingival tissue is rare (32). No single etiologic agent has been identified; rather, specific groups and combinations of bacteria including Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythensis have been strongly associated with pathology (11, 32, 58). Emerging research now implicates both host genetic and immunological factors as being important in disease susceptibility (9, 11, 23, 24), further demonstrating the complex nature of this condition.
Plaque accumulates in the mouth at sites such as the gingival margin, where shear forces are low (36). Chronic bacterial colonization of this site, often in the absence of effective oral hygiene, leads to inflammation of the adjacent gingival tissue, termed gingivitis. Chronic gingivitis has been associated with both qualitative and quantitative changes in the subgingival microbiota, but the mechanisms of initiation and progression are poorly understood (32, 45). According to the ecological-plaque hypothesis (36), increased gingival crevicular fluid flow in moderate gingival inflammation is responsible for early changes in the population dynamics of microorganisms at this site. This causes a shift from a largely gram-positive community (e.g., streptococci and actinomycetes) to one characterized by higher numbers of putative periodontal pathogens such as fusobacteria (4), Porphyromonas spp., Prevotella spp., T. forsythensis (50), and spirochetes such as Treponema spp. (7, 10).
Culture-independent methods have proved to be useful tools to aid the understanding of polymicrobial processes and have highlighted the limitations of isolation. Quantitative DNA-DNA hybridization (checkerboard), for example, while enabling high-throughput analysis of the abundance for ca. 40 defined, previously isolated target species (60), does not necessarily detect hitherto uncultured genotypes. With such techniques, the relative abundance of ca. 40 subgingival taxa was determined in more than 13,000 plaque samples in a single study (58). Cloning and sequencing, on the other hand, while nonquantitative, enable high-quality phylogenetic information to be obtained about both culturable and nonculturable species. Paster et al. (45) used this technique to identify 2,522 bacterial clones obtained from the subgingival plaque of 31 human volunteers, 62% of which were novel species. Similarly, Hutter et al. (26) analyzed 578 sequences obtained from 26 subjects, 30% of which were from novel species. Both real-time PCR and DNA hybridization methods have also been used to investigate complex bacterial communities and have detected the uncultivated bacterial division TM7 in subgingival plaque (6).
In a number of studies, denaturing gradient gel electrophoresis (DGGE) has produced highly reproducible fingerprints of consortia associated with the human mouth (15, 39, 74) and the general environment (40). A further enhancement of this technique is the application of image analysis to construct dendrograms by the unweighted-pair group method using average linkages (UPGMA) based on lane-matching profiles (5, 27, 73). This allows the identification of band pattern motifs that are characteristic of particular states or conditions.
The aim of this investigation was cross-sectional analysis of bacterial consortia in health and periodontal disease. PCR-DGGE was combined with image analysis to give insights into the microbial diversity of the sites, while UPGMA dendrogram construction (14, 68) and sequencing (37-41) were done to test for disease-associated DGGE motifs and taxa. Associations of Actinobacillus actinomycetemcomitans, P. gingivalis, and T. forsythensis with disease were tested by multiplex PCR (16, 66).
MATERIALS AND METHODS
Subjects.
Patients (n = 47) attending the periodontal clinic at the University Dental Hospital of Manchester participated in this study. The use of human subjects in this investigation was approved by the Central Manchester Ethics Committee (reference 02/CM/166). All subjects were required to read and sign a consent form approved by the Central Manchester Ethics Committee. Individuals known to be human immunodeficiency virus positive or those who had received antibiotics in the previous 3 months were excluded from this study because these factors have been associated with shifts in the periodontal microbiota (6). None of the subjects taking part in this study had been recently treated for chronic periodontal disease. Twenty-nine of the patients (9 males and 20 females; age range, 20 to 55 years; average age, 40.1 years) had chronic periodontitis (diagnosed by the presence of periodontal pockets with depths of >3 mm and bleeding upon probing). The remaining 18 volunteers (10 males and 8 females; age range, 18 to 77 years; average age, 40.6 years) showed no signs of chronic periodontitis (no periodontal pockets >3 mm deep and no evidence of attachment loss at any sites in the mouth).
Specimen collection.
Samples from periodontal pockets were taken with a sterile curette. One sample was taken from each patient from the selected periodontal crevice. The age, gender, and smoking status of each patient were recorded along with pocket depth, sample site, and bleeding on probing. Samples were placed in a 2-ml plastic vial containing sterile phosphate-buffered saline (1 ml) and transported to the laboratory within 30 min. Samples were vortexed vigorously for 1 min and then archived at −60°C for subsequent PCR-DGGE analysis.
DNA extraction.
DNA was extracted from the archived periodontal samples with a DNeasy Tissue Kit (QIAGEN Ltd., West Sussex, United Kingdom) in accordance with the manufacturer's instructions. The amounts and quality of DNA extracted were estimated by electrophoresis of 5-μl aliquots on a 0.8% agarose gel and comparison to a molecular weight standard (stained with ethidium bromide). DNA extracts were stored at −60°C prior to analysis in nuclease-free containers.
PCR amplification for DGGE analysis.
The V2-V3 region of the 16S rRNA gene (corresponding to positions 339 to 539 of Escherichia coli) was amplified with eubacterium-specific primers HDA1-GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTATTA CCG CGG CTG CTG GCA C-3′) as previously described (70). The reactions were performed in 0.2-ml tubes with a DNA thermal cycler (model 480; Perkin-Elmer, Cambridge, United Kingdom). In all cases, reactions were carried out with Red Taq DNA polymerase ready mix (25 μl; Sigma, Poole, Dorset, United Kingdom), HDA primers (2 μl of each, 5 μM), nanopure water (16 μl), and extracted community DNA (5 μl, corresponding to ca.10 ng). Optimization studies done as described by Muyzer and Smalla (43) showed that extracted community DNA required a maximum of a 1:10 dilution to ensure reliable PCRs. The thermal program was 94°C (4 min) followed by 30 thermal cycles of 94°C (30 s), 56°C (30 s), and 68°C (60 s). The final cycle incorporated a 7-min chain elongation step (68°C).
Multiplex PCR analysis.
Multiplex PCR analysis for A. actinomycetemcomitans, P. gingivalis, and T. forsythensis was carried out as previously described (16, 66). The master mixture contained 10.3 mM Tris-HCl; 51.3 mM KCl (10× PCR buffer II); 2.9 mM MgCl2; 0.15 μM primer AaF; 0.74 μM primer BfF; 0.49 μM primer PgF; 0.47 μM primer C11R; 10 U of AmpliTaq Gold (Applied Biosystems); 0.2 mM dATP, dCTP, and dGTP; and 600 mM dUTP (Promega). The final volume of each PCR mixture was 53.6 μl (containing 33.6 μl of the master mixture and 20 μl of template). The DNA template material used for the multiplex PCR was the extracted DNA from the periodontal samples. The PCR was carried out in a Whatman Biometra T Gradient thermal cycler (Biometra, Göttingen, Germany) and consisted of 40 cycles of 95°C for 1 min (10 min for the first cycle), 61°C for 1 min, and 72°C for 5 min (10 min for the last cycle). Each PCR was run with a negative (sterile deionized water) and a positive (genomic DNA extracted from pure cultures of A. actinomycetemcomitans NCTC 9710, P. gingivalis NCTC 11834, and T. forsythensis ATCC 4303) control.
Statistical analyses.
The chi-square test was used to compare the frequencies of detection of A. actinomycetemcomitans, P. gingivalis, and T. forsythensis in healthy patients and those with evidence of periodontitis and to determine any significant association of the presence of periodontitis with age (greater or less than 40 years), smoking status, or gender. The data were arranged in two-by-two contingency tables before being subjected to a Microsoft Excel macro. A forward stepwise logistic regression was built with SPSS version 11.5 to examine the relationship of, age, gender, smoking status, the presence of each unique band, or the presence of the widely acknowledged periodontal pathogens A. actinomycetemcomitans, P. gingivalis, and T. forsythensis with periodontitis (58, 66). The fit of the model was determined by analyzing the Hosmer-Lemeshow test statistic.
The Shannon-Weaver index of diversity (H′) (12, 17) was used to determine the diversity of eubacteria present in the subgingival pockets of patients with and without disease by the following equation:
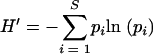 |
where s is the number of species (species richness) and pi is the proportion of species in sample i. H′ was compared for subjects in both health and disease by the Mann-Whitney U test performed with SPSS version 11.5 (SPSS, Chicago, IL).
DGGE analysis.
PCR products derived from community samples were resolved with a D-Code universal mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom) with 10% polyacrylamide gels (16 by 16 cm, 1 mm deep) run with 1× TAE buffer diluted from 50× TAE buffer (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA). Separation parameters were optimized by running PCR products from selected pure cultures of bacteria and PCR amplicons from extracted community DNA on gels with a 0 to 100% denaturation gradient perpendicular to the direction of electrophoresis (a 100% denaturing solution contained 40% [vol/vol] formamide and 7.0 M urea). Denaturing gradients were formed with two 10% acrylamide (acrylamide-bis ratio, 37.5:1) stock solutions (Sigma, Dorset, United Kingdom). On this basis, a denaturation gradient for parallel DGGE analysis ranging from 30 to 60% was selected for community sample analyses. A DGGE standard mixture was prepared from a number of commonly isolated oral bacteria that showed good separation at the above parameters. The standard was prepared from bacterial colonies (one to three) of F. nucleatum ATCC 10953, Streptococcus oralis SK139, and P. gingivalis W50 grown anaerobically on Wilkins Chalgren agar and aseptically transferred from the surface of the plate and homogenized in a reaction tube containing nanopure water (100 μl). The bacterial suspension was heated to 100°C in a boiling-water bath for 10 min and then centrifuged for 10 min (10,000 × g). The supernatant was used as a template for PCR. The resulting mixed-template PCR amplicon was used as an internal standard. Electrophoresis was carried out at 150 V and 60°C for approximately 4.5 h. All gels were stained with SYBR Gold stain [diluted to 10−4 in 1× TAE; Molecular Probes (Europe), Leiden, The Netherlands] for 30 min. Gels were viewed and images documented with a BioDocit system (UVP, Inc., Upland, CA).
Dendrogram construction for cluster analysis.
Gel images were analyzed with the Phoretix 1D software package (Nonlinear Dynamics, Newcastle, United Kingdom). Initially, lane boundaries were created and correction factors were applied to eliminate error due to smiling of gel lanes. The bands present in each lane were detected visually. Manually detected bands were subsequently used to create a synthetic reference lane. Each lane on the gel was then compared to the reference lane, allowing a matching profile for each lane to be generated. The matching profiles for each lane were used to produce a UPGMA dendrogram so that potential clustering patterns could be observed. For each band represented in the synthetic reference lane, corresponding bands that had been excised were selected for sequencing.
Sequencing of bacterial isolates and excised gel bands.
Resolved bands representative of every unique band position (Fig. 1) were cut out from the polyacrylamide gels with a sterile scalpel under UV illumination and incubated at 4°C for 20 h together with 20 μl of nanopure water in nuclease-free universal bottles. Bands were well resolved on gels, enabling effective excision. Portions (5 μl) were removed and used as a template for a PCR identical to that outlined for DGGE analysis. PCR products were purified with QIAquick PCR purification kits (QIAGEN Ltd., West Sussex, United Kingdom) and sequenced with the reverse (non-GC clamp) primer (HDA2). The sequencing protocol was 94°C (4 min) followed by 25 cycles of 96°C (30 s), 50°C (15 s), and 60°C (4 min). Once chain termination was complete, sequencing was done in a Perkin-Elmer ABI 377 sequencer. DNA sequences were compiled with CHROMAS-LITE (Technelysium Pty. Ltd., Helensville, Queensland, Australia) to obtain consensus sequences or to check and edit unidirectional sequences. For excised DGGE band PCRs, the fidelity of derived sequences was used as an indictor of the purity of the target sequence and the presence of a GC clamp upon sequence analyses confirmed that the correct target, rather than a contaminant, had been reamplified.
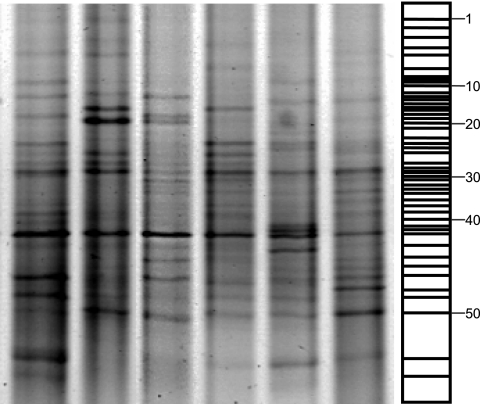
FIG. 1.
Representative negative DGGE image of six selected lanes showing the positions of all excised gel bands corresponding to the synthetic reference lane. Bands numbered 1 to 52 correspond to the numbered bands in Table 1.
RESULTS
DGGE analysis of periodontal samples.
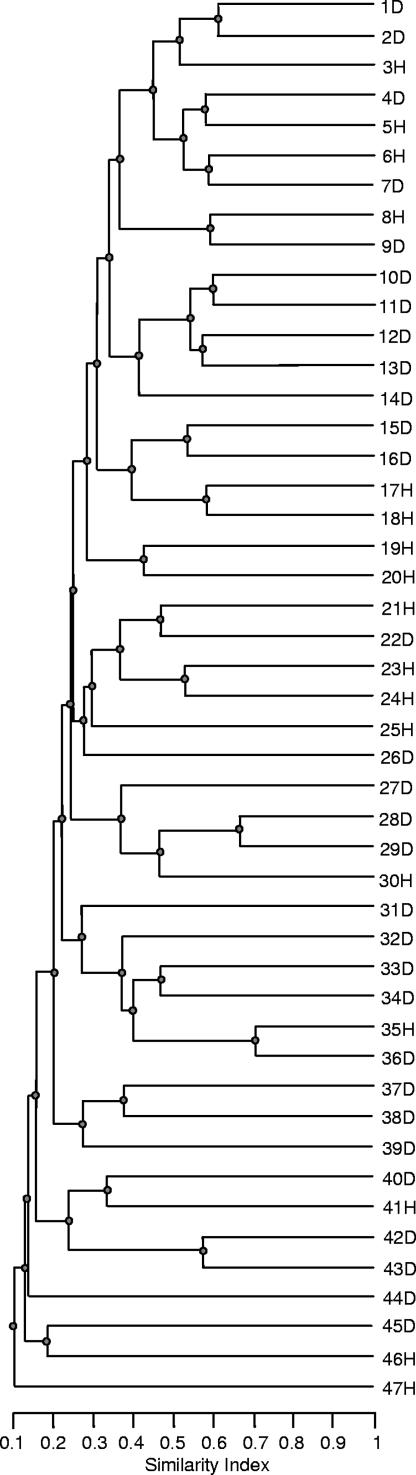
Complex DGGE fingerprints were observed for all patients, regardless of disease status. Some lanes exhibited up to 40 discrete bands of various densities. In general, most DGGE fingerprints were characterized by the presence of approximately five dominant bands with backgrounds of up to 40 distinct but less intense bands (Fig. 1). Analysis of DGGE fingerprints, where the number of gel bands is broadly proportional to eubacterial diversity, did not reveal any association between community diversity and severity of periodontal disease. Figure 2 shows a dendrogram of the periodontal community DGGE profiles created by UPGMA. This comparison of DGGE profiles showed that the highest level of similarity between two individuals was 70%, with matching between the majority of lanes occurring at a 20 to 50% similarity level. Image analysis did not reveal overt motifs associated with either periodontal health or disease, as evidenced by the absence of significant similarity between samples from uniquely periodontally healthy or diseased subjects.
FIG. 2.
UPGMA dendrogram showing the percent DGGE fingerprint matching of 47 samples of periodontally healthy (H) and diseased (D) samples derived from the subgingival crevice.
Sequence analysis of selected DGGE amplicons.
The synthetic reference lane generated by image analysis was used to determine which excised bands to sequence. For each band in the synthetic reference lane, corresponding excised bands were identified for subsequent sequence analysis. A diagrammatic representation of the synthetic reference lane is indicated in Fig. 1. Data in Table 1 show the closest relatives based on results of BLAST searches with DNA sequences obtained from DGGE gel bands identified by cluster analysis, excised from the gels, and sequenced. Bands in the same position but in different lanes were excised and sequenced to confirm that they had the same identity (data not shown). The data in Table 1 include the incidence of each bacterium in healthy and diseased subjects. Corynebacterium matruchotii (accession no. X82065) was uniquely associated with healthy patients (frequency, 16.9%), while Streptococcus mutans (AE014854) occurred in 17% of healthy patients and in none of those with disease. Overall, of 11 different streptococcal strains identified, 9 were more prevalent in health than in disease. Staphylococcus sp. (AB167056) and Alcaligenes latus (D88007) were identified in 17% of diseased subjects but not in healthy persons. Several DNA amplicons that had no significant homology to known bacteria were detected, as were a number of bacteria not normally considered to be autochthonous oral species, e.g., Pseudomonas sp. (AY3650825), Chryseobacterium indologenes (AY050493), “Citromicrobium” sp. (AY456209), Enterococcus sp. (AB0756928), Sphingomonas sp. (AY834344), and Sphingobium yanoikuyae (AF541931 and AJ627009). The sequences obtained from one excised band did not give significant database matches, indicating the presence of a novel phylotype.
TABLE 1.
Sequences of PCR amplicons derived from DGGE gels and identities based on the BLAST database
| Sequence no. and closest relative (% sequence similarity)a | Sequence length (bp)b | Incidencec
|
|
|---|---|---|---|
| Healthy | Diseased | ||
| 1. Fusobacterium nucleatumAF287812 (89) | 158 (10) | 33 | 31 |
| 2. Streptococcus mitisAY281077 (88) | 185 (3) | 50 | 45 |
| 3. Streptococcus oralisAY281080 (96) | 186 (6) | 39 | 52 |
| 4. Corynebacterium sp. AB242687 (98) | 180 (12) | 33 | 21 |
| 5. Prevotella sp. AY350613 (89) | 164 (7) | 28 | 28 |
| 6. Streptococcus sanguinisAY281086 (96) | 180 (3) | 44 | 38 |
| 7. Enterococcus sp. AB0756928 (90) | 158 (11) | 28 | 21 |
| 8. Fusobacterium nucleatum subsp. vincentiiAJ006964 (95) | 161 (3) | 22 | 48 |
| 9. Bacteroides sp. AF529225 (96) | 176 (2) | 33 | 24 |
| 10. Chryseobacterium indologenesAY050493 (88) | 174 (3) | 28 | 45 |
| 11. Fusobacterium nucleatumAF543300 (97) | 153 (7) | 44 | 38 |
| 12. Pseudomonas sp. AY365082 (99) | 177 (4) | 56 | 38 |
| 13. Sphingomonas sp. AY834344 (97) | 145 (2) | 39 | 24 |
| 14. Streptococcus gordoniiAF481950 (83) | 183 (11) | 17 | 17 |
| 15. Flexibacter roseolusM58787 (97) | 180 (3) | 17 | 10 |
| 16. Porphyromonas endodontalisAY253728 (97) | 180 (4) | 39 | 31 |
| 17. Streptococcus parasanguinisAY281087 (87) | 180 (6) | 33 | 31 |
| 18. Bacillus subtilisDQ301917 (98) | 180 (11) | 11 | 14 |
| 19. Treponema amylovorumY09959 (99) | 180 (2) | 6 | 17 |
| 20. Streptococcus cristatusAY2810904 (97) | 181 (2) | 39 | 24 |
| 21. Prevotella nigrescensAF414844 (98) | 168 (4) | 33 | 3 |
| 22. Prevotella sp. DQ188617 (83) | 185 (6) | 11 | 7 |
| 23. Putative new phylotyped | 177 (8) | 0 | 3 |
| 24. Staphylococcus sp. AB167056 (93) | 188 (8) | 17 | 0 |
| 25. Veillonella sp. DQ188766 (90) | 180 (10) | 0 | 24 |
| 26. Alcaligenes latusD88007 (98) | 180 (3) | 0 | 17 |
| 27. Streptococcus oligofermentansAY099095 (95) | 185 (3) | 28 | 24 |
| 28. Shewanella sp. AB059264 (95) | 130 (2) | 22 | 7 |
| 29. Treponema sp. AF023044 (85) | 180 (9) | 17 | 14 |
| 30. Holdemania filiformisY11466 (91) | 178 (4) | 28 | 17 |
| 31. Scardovia genomospecies AY278626 (100) | 180 (6) | 22 | 24 |
| 32. Bulleidia extructaAF22064 (87) | 177 (3) | 33 | 14 |
| 33. Prevotella sp. DQ188583 (94) | 176 (4) | 6 | 0 |
| 34. Streptococcus sp. DQ188664 (94) | 174 (6) | 0 | 37 |
| 35. Streptococcus pneumoniaeAY281084 (98) | 178 (5) | 17 | 7 |
| 36. Sphingomonas sp. AY167827 (92) | 164 (4) | 33 | 21 |
| 37. Veillonella disparAF4396394 (91) | 180 (3) | 33 | 21 |
| 38. Staphylococcus epidermidisAJ717377 (89) | 174 (4) | 0 | 17 |
| 39. Sphingobium yanoikuyaeAJ627009 (98) | 154 (4) | 17 | 0 |
| 40. Leptotrichia sp. AY267541 (87) | 180 (4) | 17 | 28 |
| 41. Corynebacterium matruchotiiX82065 (94) | 157 (8) | 17 | 0 |
| 42. Tannerella forsythensisAB053947 (95) | 180 (10) | 6 | 24 |
| 43. Streptococcus sp. AY494661 (87) | 187 (5) | 28 | 28 |
| 44. Streptococcus mutansAE014854 (84) | 180 (5) | 17 | 0 |
| 45. Sphingobium yanoikuyaeAF541931 (100) | 151 (3) | 28 | 21 |
| 46. Neisseria sp. AJ586614 (87) | 184 (7) | 44 | 24 |
| 47. Capnocytophaga granulosaX97248 (88) | 190 (3) | 11 | 17 |
| 48. Streptococcus oligofermentansAY099095 (91) | 187 (8) | 11 | 3 |
| 49. Treponema socranskii subsp. buccaleAY369255 (99) | 180 (5) | 17 | 10 |
| 50. Citromicrobium sp. AY456209 (100) | 157 (4) | 22 | 7 |
| 51. Anaerococcus sp. AY738694 (92) | 184 (11) | 0 | 3 |
| 52. Veillonella caviaeAY355140 (91) | 197 (3) | 0 | 3 |
Similarities are based on pairwise alignments with published sequences according to BLAST searches and indicate similarity, not guaranteed identity. Sequence numbers 1 to 52 refer to consecutive DGGE band positions from the top to the bottom of the synthetic reference ladder.
The number of ambiguous bases for an individual band is given in parentheses.
Refers to the percent incidence of each unique band in healthy or diseased individuals.
Refers to a sequence that did not generate a database match. Each sequence was derived from a unique, discrete gel band.
Statistical analyses.
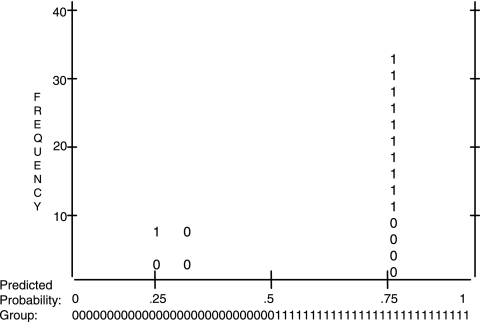
Chi-square analyses revealed that there was a significant relationship between periodontitis and smoking (P < 0.05). No significant associations were revealed between age (greater or less than 40 years) or gender and the presence of periodontal disease. Logistic regression analysis (Fig. 3) indicated that the presence of Treponema socranskii and Pseudomonas sp. was a significant predictor of disease (P < 0.05). The overall percentage of the Hosmer-Lemeshow test for goodness of fit of the model was 74.5%. The Mann-Whitney U test was used to compare the Shannon-Weaver indexes of diversity (H′) of periodontally healthy and diseased patients. This revealed that there was no significant (P > 0.05) difference in terms of biodiversity between the two sample groups.
FIG. 3.
Classification plot derived from logistic regression analysis. The x axis is the predicted probability (Prob), from 0.0 to 1.0, of the dependent being classified “1” (disease). The y axis is frequency, i.e., the number of cases classified. Inside the plot are columns of observed ones and zeros, which stand for disease and health, respectively, with 2.5 cases per symbol.
Multiplex PCR.
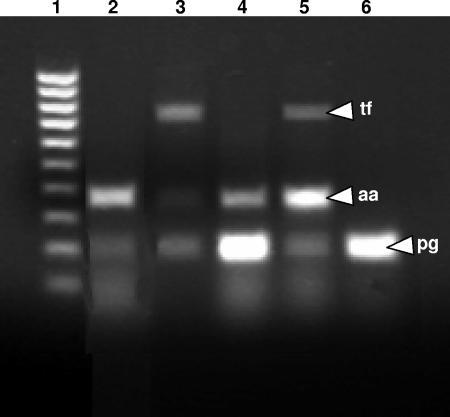
The presence of the periodontal pathogens A. actinomycetemcomitans, P. gingivalis, and T. forsythensis in each of the samples as ascertained by multiplex PCR analysis was indicated by the presence of a clear band of the expected size (Fig. 4). The frequencies of detection of all three pathogens in healthy and diseased subjects are indicated in Table 2. Statistical analyses (chi square) revealed that there was a significant association between A. actinomycetemcomitans (P < 0.05) and periodontal disease but not P. gingivalis (P = 0.67) or T. forsythensis (P = 0.09).
FIG. 4.
Multiplex PCR of the periodontal pathogens A. actinomycetemcomitans (aa; 360 bp), P. gingivalis (pg; 197 bp), and T. forsythensis (tf; 745 bp). Lane 1, molecular size marker (100 to 1,000 bp). Lanes 2, 3, 4, and 6, amplified DNA from periodontal samples. Lane 5, positive control derived from the type strains A. actinomycetemcomitans NCTC 9710, P. gingivalis NCTC 11834, and T. forsythensis ATCC 43037.
TABLE 2.
Incidence of periodontal pathogens in clinically healthy and diseased subjects as determined by multiplex PCR
| Subject group | No. of subjects | No. of positive subjects
|
||
|---|---|---|---|---|
| A. actinomycetemcomitans | P. gingivalis | T. forsythensis | ||
| Healthy | 29 | 4 | 7 | 2 |
| Diseased | 18 | 6a | 14 | 8 |
Significant difference (P < 0.05).
DISCUSSION
Current developments in molecular analysis have overcome many of the restrictions associated with culture-based techniques. The requirement for samples to be processed quickly, the need for specialized anaerobic techniques, and low cell recoveries are particularly problematical when studying microbial ecosystems associated with the periodontal pocket.
In the present study, the bacterial diversity of the human subgingival crevice in health and disease was investigated by combining DGGE of the 16S rRNA gene with image and cluster analysis and sequencing of key PCR amplicons, together with statistical analyses and multiplex PCR of three periodontal pathogens. The strength of this approach is that it will theoretically detect the presence and identity of any amplifiable target sequence above the detection threshold (43) whereas hybridization techniques are arguably more suited to measuring the abundance of a finite number of previously cultured, relatively abundant species (25, 59, 60).
Although DGGE analysis has previously been used in a limited number of studies to produce fingerprints of periodontal bacterial diversity (15, 74) to date, few studies have combined DGGE, cluster analysis, and sequencing in a relatively large subject group. DGGE data indicate that the periodontal pockets were colonized by diverse eubacterial communities. The amount of diversity, however, did not correlate with either periodontal health or disease, which supports the hypothesis that periodontal disease is associated with a shift in the balance of the subgingival microbiota rather than the action of a single pathogen (36) or a simple increase in diversity. Equally, cluster analysis of DGGE fingerprints did not reveal overt band pattern motifs associated with either health or disease and statistical analyses indicated that there was no significant (P > 0.05) difference in species diversity between health and disease. Furthermore, despite the acknowledged presence of resident species within the periodontal pocket in health and disease (23, 63, 72), only two fingerprints shared ca. 70% homology while the majority of fingerprints were less strongly related.
Figure 2 shows that most of the clustering between individuals occurred at the 20 to 50% level. Since no correlation between these clusters and the presence of extant disease was observed, it is possible that these clusters relate to variables beyond the scope of this investigation, such as the genetic subgroup of the subject (e.g., tissue type), immune types, ethnicity, geographical location, and successional events during oral colonization. It is also possible that distinct stability classes exist within periodontal microbial ecosystems.
Race (47), age (53), diabetes (57), and smoking status (3, 19, 42, 62) have all been shown to be important in relation to various levels of antibody production associated with periodontitis. In this respect, it is generally accepted that host factors including genetic susceptibility and the gingival immune responses (18, 31, 32, 52) play a significant role in the etiology of periodontitis. Hart et al. (23) measured the expression of immune genes to demonstrate the role of the host immune response in periodontitis, while early colonization events (28) could also account for the high levels of interindividual variation observed in the complex bacterial community of the periodontal pocket.
Sequence analysis of all unique DGGE bands enabled the association of specific bacterial genotypes with health or disease to be investigated (Table 1), although this depends on the validity of community analysis and the accuracy of the sequence data derived from the technique. Accordingly, it has previously been demonstrated that comigrating bands generally correspond to identical sequences (29). This was verified in this study by excising bands that corresponded to the migration positions of known reference bands and carrying out sequence analysis to ensure that the same identification was obtained.
The apparent association of C. matruchotii with healthy gingivae is noteworthy (Table 1). This observation is in agreement with Kumar et al. (30). Our data also demonstrate the unique but putative association of S. mutans, Staphylococcus sp., and an unidentified bacterium with health. To our knowledge, this has not been previously reported, although it is notoriously difficult to differentiate between indirect positive or negative associations and direct involvement in the disease process. The association of gram-positive bacteria with health is, however, in agreement with the ecological-plaque hypothesis (36). Conversely, the association of Staphylococcus epidermidis, A. latus, and Streptococcus sp. with disease and the lack of such an association for the putative periodontal pathogens P. gingivalis, T. forsythensis, and T. denticola do not prove that these species are unimportant in disease progression, since their relative abundance might be low. They do, however, indicate the complexity of the disease process and a probable multifactorial etiology (23). An important issue relating to progression to periodontitis and the bacteria that have previously been associated with the process is the population density of the potential causative organisms. It is possible that differences in the sensitivity of the PCR technique in comparison to hybridization methods mean that putative pathogens strongly associated with disease only at high abundances could have been detected even when below a critical threshold.
One sequence did not generate a database match and has been designated a putative new phylotype. Since up to half of the bacteria present in the mouth remain uncultured (45), they have not been characterized with respect to metabolism and pathogenesis, other than by molecular informatics. It is therefore possible that a hitherto uncharacterized bacterium plays a role in the pathogenesis of oral diseases and may also be a useful marker of disease.
A number of nontypical oral genotypes were identified; these included, Pseudomonas sp., C. indologenes, S. epidermidis, Sphingomonas sp., Enterococcus sp., A. latus, S. yanoikuyae, “Citromicrobium” sp., and Shewanella sp. The detection of nontypical oral genotypes in the samples reinforces the utility of DGGE over hybridization methods and real-time PCR since there is no experimental bias toward typical resident oral species. DGGE will theoretically identify any amplifiable target sequence above the detection threshold (43), whereas hybridization techniques measure the abundance of a finite number of species (25, 59, 60). It is important to note that DGGE is not a truly quantitative technique and that band density does not necessarily relate to target abundance. It is therefore possible that subtle associations between species abundance and diseases would not necessarily be identified. Care should be taken when making phylogenetic inferences from sequenced DGGE bands, since derived sequences are short and may be of variable quality (Table 1). Such ambiguities probably arise from amplification of different phylotypes with similar or identical electrophoretic mobilities. The relatively short sequences derived from DGGE also reduce the refinement of phylogenetic determination. Despite these concerns, DGGE is currently one of the few techniques that allow reproducible visual comparisons of profiles from microbial communities to be derived and has been successfully applied to a wide variety of microbial ecosystems (51, 67, 70).
Statistical analyses confirmed significant associations between smoking and the presence of disease. This finding substantiates previous studies that have linked smoking to an increase in periodontitis (20, 21, 42, 69). The identification of the presence of T. socranskii as a significant predictor of disease supports previous analyses that have demonstrated a potentially pathogenic role for this spirochete (46, 71). It has previously been detected at higher frequencies in periodontitis patients than in healthy patients (61). The association of Pseudomonas spp. with periodontitis is supported by previous studies that have indicated that periodontal patients harbor pseudomonads (2, 54). Since pseudomonads are markedly less susceptible to chlorhexidine than are other members of the periodontal microflora (55), this could account for their increased prevalence in periodontal disease during chemotherapeutic intervention.
The specific detection of periodontal pathogens by a multiplex PCR assay revealed the presence of A. actinomycetemcomitans, P. gingivalis, and T. forsythensis. Interestingly, both A. actinomycetemcomitans and P. gingivalis were undetected by PCR-DGGE, suggesting that their relative abundance was low in the periodontal pocket and that they remained below the detection threshold, in contrast to T. forsythensis, which was detected by PCR-DGGE and was more prevalent among diseased patients. This is in agreement with previous studies that have detected T. forsythensis at higher frequencies in diseased subjects compared to healthy controls (22, 72). A. actinomycetemcomitans has been widely linked to periodontitis (13, 56, 64) and was significantly associated with periodontal disease in this study. All three pathogens were present in both health and disease, which indicates that the amount of these bacteria is more significant with regard to the disease process. Furthermore, significant associations between certain bacteria and disease are arguably more likely to be ascertained in larger-cohort studies.
The etiology of periodontal disease has remained poorly understood despite considerable research attention. Pioneering work (1, 8, 33-35, 58, 59, 65) using DNA-DNA hybridization “checkerboards” to measure the presence and increased abundance of key species has enabled associations to be made between certain species and periodontal disease. For example, P. gingivalis, T. denticola, and T. forsythensis have been most strongly associated with periodontal disease (48, 61). Paster et al., for example, identified a number of putative periodontal pathogens from eight different phyla by a clone library technique (45). Importantly, this approach identified the unculturable division TM7 as strongly disease associated (6, 44). Significantly, these researchers documented that human subgingival plaque harbors several hundred bacterial species or phylotypes (26, 45, 49), the majority of which are nonculturable.
Despite a number of elegant studies of the bacterial diversity present in periodontal pockets, attempts to differentiate between the association of certain phyla with disease and causality have met with only limited success, although studies which integrate microbiology and immunity are providing new insights into this complex process (23).
Conclusions.
This study demonstrates the efficacy of DGGE combined with cluster analysis to interrogate resident communities in oral health and disease by providing reproducible community patterns. High levels of interindividual variation of bacterial subgingival communities were observed with no association between microbial complexity and disease. No clear association between putatively pathogenic species and disease emerged during DGGE analysis. For the detection of specific pathogens that may be present at low abundance, specific PCR protocols have greater sensitivity, as evidenced by data derived from multiplex PCR analysis for A. actinomycetemcomitans, P. gingivalis, and T. forsythensis.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Ali, R. W., A. C. Johannessen, G. Dahlen, S. S. Socransky, and N. Skaug. 1997. Comparison of the subgingival microbiota of periodontally healthy and diseased adults in northern Cameroon. J. Clin. Periodontol. 24:830-835. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, F. C., M. P. Mayer, E. Saba-Chujfi, and S. Cai. 2001. Subgingival occurrence and antimicrobial susceptibility of enteric rods and pseudomonads from Brazilian periodontitis patients. Oral Microbiol. Immunol. 16:306-310. [DOI] [PubMed] [Google Scholar]
- 3.Barbour, S. E., K. Nakashima, J. B. Zhang, S. Tangada, C. L. Hahn, H. A. Schenkein, and J. G. Tew. 1997. Tobacco and smoking: environmental factors that modify the host response (immune system) and have an impact on periodontal health. Crit. Rev. Oral Biol. Med. 8:437-460. [DOI] [PubMed] [Google Scholar]
- 4.Bolstad, A. I., H. B. Jensen, and V. Bakken. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 9:55-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boon, N., W. De-Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 6.Brinig, M. M., P. W. Lepp, C. C. Ouverney, G. C. Armitage, and D. A. Relman. 2003. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 69:1687-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo, A. P., A. D. Haffajee, F. E. Dewhirst, B. J. Paster, C. M. Smith, M. A. Cugini, and S. S. Socransky. 1998. Clinical and microbiological features of refractory periodontitis subjects. J. Clin. Periodontol. 25:169-180. [DOI] [PubMed] [Google Scholar]
- 9.Craig, R. G., R. Boylan, J. Yip, D. Mijares, M. Imam, S. S. Socransky, M. A. Taubman, and A. D. Haffajee. 2002. Serum IgG antibody response to periodontal pathogens in minority populations: relationship to periodontal disease status and progression. J. Periodontal Res. 37:132-146. [DOI] [PubMed] [Google Scholar]
- 10.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 11.Donley, C. L., R. Badovinac, S. Sapir, L. Shapira, Y. Houri, A. Kantarci, M. L. Warbington, S. Dibart, T. E. Van Dyke, H. L. Needleman, N. Karimbux, and E. Bimstein. 2004. IgG antibody levels to Porphyromonas gingivalis and clinical measures in children. J. Periodontol. 75:221-228. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, M. L., A. K. Lilley, T. H. Timms-Wilson, I. P. Thompson, and I. Cooper. 2001. Characterisation of the culturable heterotrophic bacterial community in a small eutrophic lake (Priest Pot). FEMS Microbiol. Ecol. 35:295-304. [DOI] [PubMed] [Google Scholar]
- 13.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 14.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto, C., H. Maeda, S. Kokeguchi, S. Takashiba, F. Nishimura, H. Arai, K. Fukui, and Y. Murayama. 2003. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 38:440-445. [DOI] [PubMed] [Google Scholar]
- 16.Gafan, G. P., V. S. Lucas, G. J. Roberts, A. Petrie, M. Wilson, and D. A. Spratt. 2004. Prevalence of periodontal pathogens in dental plaque of children. J. Clin. Microbiol. 42:4141-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gafan, G. P., V. S. Lucas, G. J. Roberts, A. Petrie, M. Wilson, and D. A. Spratt. 2005. Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J. Clin. Microbiol. 43:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genco, R. J. 1992. Host responses in periodontal diseases: current concepts. J. Periodontol. 63:338-355. [DOI] [PubMed] [Google Scholar]
- 19.Gulsvik, A., and M. K. Fagerhoi. 1979. Smoking and immunoglobulin levels. Lancet i:449. [DOI] [PubMed] [Google Scholar]
- 20.Haber, J. 1994. Smoking is a major risk factor for periodontitis. Curr. Opin. Periodontol. 1:12-18. [PubMed] [Google Scholar]
- 21.Haber, J., J. Wattles, M. Crowley, R. Mandell, K. Joshipura, and R. L. Kent. 1993. Evidence for cigarette smoking as a major risk factor for periodontitis. J. Periodontol. 64:16-23. [DOI] [PubMed] [Google Scholar]
- 22.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 23.Hart, G. T., D. J. Shaffer, S. Akilesh, A. C. Brown, L. Moran, D. C. Roopenian, and P. J. Baker. 2004. Quantitative gene expression profiling implicates genes for susceptibility and resistance to alveolar bone loss. Infect. Immun. 72:4471-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 25.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67-75. [DOI] [PubMed] [Google Scholar]
- 27.Ibekwe, A. M., S. K. Papiernik, J. Gan, S. R. Yates, C. H. Yang, and D. E. Crowley. 2001. Impact of fumigants on soil microbial communities. Appl. Environ. Microbiol. 67:3245-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 29.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 31.Landi, L., S. Amar, A. S. Polins, and T. E. Van Dyke. 1997. Host mechanisms in the pathogenesis of periodontal disease. Curr. Opin. Periodontol. 4:3-10. [PubMed] [Google Scholar]
- 32.Loesche, W. J., and N. S. Grossman. 2001. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 14:727-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López, N. J., S. S. Socransky, I. Da Silva, M. R. Japlit, and A. D. Haffajee. 2004. Subgingival microbiota of Chilean patients with chronic periodontitis. J. Periodontol. 75:717-725. [DOI] [PubMed] [Google Scholar]
- 34.Mager, D. L., A. D. Haffajee, and S. S. Socransky. 2003. Effects of periodontitis and smoking on the microbiota of oral mucous membranes and saliva in systemically healthy subjects. J. Clin. Periodontol. 30:1031-1037. [DOI] [PubMed] [Google Scholar]
- 35.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 36.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 37.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Effects of a chlorhexidine gluconate-containing mouthwash on the vitality and antimicrobial susceptibility of in vitro oral bacterial ecosystems. Appl. Environ. Microbiol. 69:4770-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Effects of triclosan-containing rinse on the dynamics and antimicrobial susceptibility of in vitro plaque ecosystems. Antimicrob. Agents Chemother. 47:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Growth and molecular characterization of dental plaque microcosms. J. Appl. Microbiol. 94:655-664. [DOI] [PubMed] [Google Scholar]
- 40.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, B. B. Price, and P. Gilbert. 2003. Exposure of sink drain microcosms to triclosan: population dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 69:5433-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, A. H. Rickard, S. A. Symmons, and P. Gilbert. 2003. Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl. Environ. Microbiol. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mooney, J., P. J. Hodge, and D. F. Kinane. 2001. Humoral immune response in early-onset periodontitis: influence of smoking. J. Periodontal Res. 36:227-232. [DOI] [PubMed] [Google Scholar]
- 43.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 44.Ouverney, C. C., G. C. Armitage, and D. A. Relman. 2003. Single-cell enumeration of an uncultivated TM7 subgroup in the human subgingival crevice. Appl. Environ. Microbiol. 69:6294-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paster, B. J., F. E. Dewhirst, B. C. Coleman, C. N. Lau, and R. L. Ericson. 1998. Phylogenetic analysis of cultivable oral treponemes from the Smibert collection. Int. J. Syst. Bacteriol. 48(Pt. 3):713-722. [DOI] [PubMed] [Google Scholar]
- 47.Quinn, S. M., J. B. Zhang, J. C. Gunsolley, J. G. Schenkein, H. A. Schenkein, and J. G. Tew. 1996. Influence of smoking and race on immunoglobulin G subclass concentrations in early-onset periodontitis patients. Infect. Immun. 64:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riviere, G. R., K. S. Smith, E. Tzagaroulaki, S. L. Kay, X. Zhu, T. A. DeRouen, and D. F. Adams. 1996. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J. Periodontol. 67:109-115. [DOI] [PubMed] [Google Scholar]
- 49.Sakamoto, M., Y. Huang, M. Ohnishi, M. Umeda, I. Ishikawa, and Y. Benno. 2004. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J. Med. Microbiol. 53:563-571. [DOI] [PubMed] [Google Scholar]
- 50.Sakamoto, M., Y. Huang, M. Umeda, I. Ishikawa, and Y. Benno. 2002. Detection of novel oral phylotypes associated with periodontitis. FEMS Microbiol. Lett. 217:65-69. [DOI] [PubMed] [Google Scholar]
- 51.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seymour, G. J. 1991. Importance of the host response in the periodontium. J. Clin. Periodontol. 18:421-426. [DOI] [PubMed] [Google Scholar]
- 53.Shackelford, P. G., D. M. Granoff, M. H. Nahm, M. G. Scott, B. Suarez, J. P. Pandey, and S. J. Nelson. 1985. Relation of age, race, and allotype to immunoglobulin subclass concentrations. Pediatr. Res. 19:846-849. [DOI] [PubMed] [Google Scholar]
- 54.Slots, J., D. Feik, and T. E. Rams. 1990. Prevalence and antimicrobial susceptibility of Enterobacteriaceae, Pseudomonadaceae and Acinetobacter in human periodontitis. Oral Microbiol. Immunol. 5:149-154. [DOI] [PubMed] [Google Scholar]
- 55.Slots, J., T. E. Rams, and S. E. Schonfeld. 1991. In vitro activity of chlorhexidine against enteric rods, pseudomonads and acinetobacter from human periodontitis. Oral Microbiol. Immunol. 6:62-64. [DOI] [PubMed] [Google Scholar]
- 56.Slots, J., and M. Ting. 2000. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontology 20:82-121. [DOI] [PubMed] [Google Scholar]
- 57.Smith, G. T., C. J. Greenbaum, B. D. Johnson, and G. R. Persson. 1996. Short-term responses to periodontal therapy in insulin-dependent diabetic patients. J. Periodontol. 67:794-802. [DOI] [PubMed] [Google Scholar]
- 58.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 59.Socransky, S. S., C. Smith, and A. D. Haffajee. 2002. Subgingival microbial profiles in refractory periodontal disease. J. Clin. Periodontol. 29:260-268. [DOI] [PubMed] [Google Scholar]
- 60.Socransky, S. S., C. Smith, L. Martin, B. J. Paster, F. E. Dewhirst, and A. E. Levin. 1994. “Checkerboard” DNA-DNA hybridization. BioTechniques 17:788-792. [PubMed] [Google Scholar]
- 61.Takeuchi, Y., M. Umeda, M. Sakamoto, Y. Benno, Y. Huang, and I. Ishikawa. 2001. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J. Periodontol. 72:1354-1363. [DOI] [PubMed] [Google Scholar]
- 62.Tangada, S. D., J. V. Califano, K. Nakashima, S. M. Quinn, J. B. Zhang, J. C. Gunsolley, H. A. Schenkein, and J. G. Tew. 1997. The effect of smoking on serum IgG2 reactive with Actinobacillus actinomycetemcomitans in early-onset periodontitis patients. J. Periodontol. 68:842-850. [DOI] [PubMed] [Google Scholar]
- 63.Thoden van Velzen, S. K., L. Abraham-Inpijn, and W. R. Moorer. 1984. Plaque and systemic disease: a reappraisal of the focal infection concept. J. Clin. Periodontol. 11:209-220. [DOI] [PubMed] [Google Scholar]
- 64.Tonetti, M. S., and A. Mombelli. 1999. Early-onset periodontitis. Ann. Periodontol. 4:39-53. [DOI] [PubMed] [Google Scholar]
- 65.Torresyap, G., A. D. Haffajee, N. G. Uzel, and S. S. Socransky. 2003. Relationship between periodontal pocket sulfide levels and subgingival species. J. Clin. Periodontol. 30:1003-1010. [DOI] [PubMed] [Google Scholar]
- 66.Tran, S. D., and J. D. Rudney. 1999. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J. Clin. Microbiol. 37:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tresse, O., M. J. Lorrain, and D. Rho. 2002. Population dynamics of free-floating and attached bacteria in a styrene-degrading biotrickling filter analyzed by denaturing gradient gel electrophoresis. Appl. Microbiol. Biotechnol. 59:585-590. [DOI] [PubMed] [Google Scholar]
- 68.van der Gast, C. J., A. S. Whiteley, A. K. Lilley, C. J. Knowles, and I. P. Thompson. 2003. Bacterial community structure and function in a metal-working fluid. Environ. Microbiol. 5:453-461. [DOI] [PubMed] [Google Scholar]
- 69.van Winkelhoff, A. J., C. J. Bosch-Tijhof, E. G. Winkel, and W. A. van der Reijden. 2001. Smoking affects the subgingival microflora in periodontitis. J. Periodontol. 72:666-671. [DOI] [PubMed] [Google Scholar]
- 70.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willis, S. G., K. S. Smith, V. L. Dunn, L. A. Gapter, K. H. Riviere, and G. R. Riviere. 1999. Identification of seven Treponema species in health- and disease-associated dental plaque by nested PCR. J. Clin. Microbiol. 37:867-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ximénez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722-732. [DOI] [PubMed] [Google Scholar]
- 73.Yang, C. H., D. E. Crowley, J. Borneman, and N. T. Keen. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 98:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zijnge, V., H. J. Harmsen, J. W. Kleinfelder, M. E. van der Rest, J. E. Degener, and G. W. Welling. 2003. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol. Immunol. 18:59-65. [DOI] [PubMed] [Google Scholar]