Abstract
Here we describe the diversity and activity of sulfate-reducing bacteria (SRB) in sulfidogenic bioreactors by using the simultaneous analysis of PCR products obtained from DNA and RNA of the 16S rRNA and dissimilatory sulfite reductase (dsrAB) genes. We subsequently analyzed the amplified gene fragments by using denaturing gradient gel electrophoresis (DGGE). We observed fewer bands in the RNA-based DGGE profiles than in the DNA-based profiles, indicating marked differences in the populations present and in those that were metabolically active at the time of sampling. Comparative sequence analyses of the bands obtained from rRNA and dsrB DGGE profiles were congruent, revealing the same SRB populations. Bioreactors that received either ethanol or isopropanol as an energy source showed the presence of SRB affiliated with Desulfobulbus rhabdoformis and/or Desulfovibrio sulfodismutans, as well as SRB related to the acetate-oxidizing Desulfobacca acetoxidans. The reactor that received wastewater containing a diverse mixture of organic compounds showed the presence of nutritionally versatile SRB affiliated with Desulfosarcina variabilis and another acetate-oxidizing SRB, affiliated with Desulfoarculus baarsii. In addition to DGGE analysis, we performed whole-cell hybridization with fluorescently labeled oligonucleotide probes to estimate the relative abundances of the dominant sulfate-reducing bacterial populations. Desulfobacca acetoxidans-like populations were most dominant (50 to 60%) relative to the total SRB communities, followed by Desulfovibrio-like populations (30 to 40%), and Desulfobulbus-like populations (15 to 20%). This study is the first to identify metabolically active SRB in sulfidogenic bioreactors by using the functional gene dsrAB as a molecular marker. The same approach can also be used to infer the ecological role of coexisting SRB in other habitats.
Certain industrial waste streams, such as effluents from paper mills, potato starch factories, and edible oil production plants, contain high concentrations of sulfate (6, 42). Sulfate as such may not be any problem to the environment, because it is chemically inert and nontoxic. However, under anaerobic conditions, dissimilatory sulfate-reducing bacteria (SRB) use sulfate as a terminal electron acceptor in the degradation of organic matter, resulting in the production of sulfide (17). In contrast to sulfate, sulfide is a highly reactive, corrosive, and a very toxic compound, causing numerous environmental problems (19). Therefore, the removal of sulfate from wastewater is often required. However, sulfide produced by SRB can also be used beneficially, such as in the removal of toxic heavy metals through precipitation (6), as demonstrated with the groundwater treatment system of the Zinifex Budel Company in The Netherlands, which removes sulfate, zinc, and cadmium (47).
The major metabolic processes that take place in anaerobic bioreactors, such as methanogenesis, sulfidogenesis, and acetogenesis, are nowadays well understood. However, our knowledge of the diversity and dynamics of the microbial communities responsible for these processes is still limited. This is because microbial communities in large-scale biotechnological processes, such as wastewater treatment, are often treated as a black box (18). This is not due to the underestimation of the significance of the biological component but is due to our inability to isolate most of the microorganisms in pure cultures. Fortunately, molecular techniques have provided alternative approaches to overcome the problems associated with culture-dependent analysis of complex microbial communities (2).
Different molecular techniques, such as fluorescence in situ hybridization (FISH) (52), PCR-denaturing gradient gel electrophoresis (DGGE) (9), and DNA microarrays (25), have been used to study the diversity of SRB in natural and engineered ecosystems. Most of these studies have focused on the presence rather than on the activity of SRB in the samples. There have been only a few studies (64, 65) in which the metabolically active populations were monitored by targeting the mRNA of the [NiFe] hydrogenase gene fragments of SRB. However, the limited distribution of this gene among the SRB has restricted its use to the study of Desulfovibrio species only (62).
dsrAB encodes the α and β subunits of an enzyme that catalyzes the six-electron reduction of sulfite to sulfide (57). Due to a remarkably high degree of conservation observed in dsrAB across sulfate-reducing bacteria and archaea (30), it is a potential candidate for phylogenetic studies of these organisms. Previous studies using partial sequences of dsrAB to evaluate the phylogeny of different SRB lineages has revealed topology congruent with 16S rRNA gene-based phylogenetic tree (68). The dsrAB gene-based molecular approach has been used to discriminate among SRB in diverse environments (4, 12, 15, 34, 43). Recently, Geets and coworkers described DGGE of PCR-amplified dsrB gene fragments to specifically follow population dynamics of SRB (16).
This paper describes the results of a comparative study of SRB in lab- and full-scale sulfidogenic wastewater treatment reactors using 16S rRNA and dsrB gene fragments as molecular markers. The goals of our research were to obtain insight into the diversity of sulfate-reducing bacteria in different lab- and full-scale reactors treating sulfate-rich industrial wastewater and to identify the metabolically active community members by comparative DGGE analysis of PCR products obtained from both DNA and RNA of the 16S rRNA and the dsrAB genes. Apart from DGGE analysis, hybridization probes were designed to validate the presence and to determine the abundance of dominant SRB by using FISH.
MATERIALS AND METHODS
Reactor samples.
Reactor samples were obtained from different lab-scale (7 liters) and full-scale (>100 m3) sulfidogenic wastewater treatment reactors. The sulfate-rich wastewater fed to most of the reactors did not contain organic compounds, so an external electron donor, i.e., ethanol (reactors A, B, C, and D) or a 1:1 mixture of isopropanol and butanol (reactor E), was added. The wastewater fed to reactor F contained a diverse mixture of organic compounds, and hence no external electron donor was added. The lab-scale reactors, A and B, were fed with synthetic mineral medium based on the medium of Vishniac and Santer (60) and ethanol as electron donor. The ratio between the amount of electron donor added and the amount of sulfate in all reactors was always less than 0.7 kg/kg, thus making the reactors electron donor limited. The sulfide produced in the reactors was either converted to elemental sulfur through biological sulfide oxidation or precipitated with toxic heavy metals, thus keeping the sulfide concentrations in the reactors below toxic levels. Sludge from reactor C, which was started in 1995, was used as the inoculum for all the other reactors. Other operational details of the reactors are given in Table 1.
TABLE 1.
Characteristics of the reactors
| Reactor | Type | Scale | Wastewater source | Key component in wastewater | Electron donor | Temp (°C) | pH | Conductivity (mS/cm) | SO42− (mg/liter)
|
|
|---|---|---|---|---|---|---|---|---|---|---|
| In | Out | |||||||||
| A | Expanded bed | 7 dm3 | Mineral medium | Na2SO4 | Ethanol | 35 | 7.0-7.5 | 5 | 2,500 | 1,000 |
| B | Expanded bed | 7 dm3 | Mineral medium | Na2SO4 | Ethanol | 35 | 7.0-7.5 | 5 | 2,500 | 1,000 |
| C | UASBa | >100 m3 | Fiber-producing chemical industry | Na2SO4 | Ethanol | 30 | 7.0-7.5 | 3 | 1,500 | 100 |
| D | UASB | >100 m3 | Fiber-producing chemical industry | Na2SO4 | Ethanol | 30 | 7.0-7.5 | 3 | 1,500 | 100 |
| E | BESTb | >100 m3 | Acid mine drainage from coal mine | Ca2SO4 | Isopropanol-butanol (1:1) | 25 | 7.0-7.5 | 3 | 2,500 | 200 |
| F | BEST | >100 m3 | Pulp- and fiber-producing chemical industry | Na2SO4 and a mixture of organic compounds | None | 35 | 7.0-7.5 | 18 | 10,000 | 8,000 |
UASB, upflow anaerobic sludge bed.
BEST, bioreactor enhanced by separation technology.
Nucleic acid extraction.
Bacterial biomass was concentrated from the reactor samples by centrifugation. Genomic DNA was extracted directly from the concentrated biomass by using the Ultra Clean Soil DNA extraction kit (MOBIO Laboratories, Inc., CA) according to the manufacturer's protocol. Extracted DNA was stored at −20°C until further use. Total RNA was extracted from 500 μl concentrated biomass by using the RNeasy minikit (QIAGEN GmbH, Hilden, Germany). A volume of 450 μl lysis buffer containing 4.5 μl β-mercaptoethanol and 0.1-mm-diameter autoclaved glass beads was added to the tubes. Cells were disrupted by vortexing for 10 min at maximum speed. From the cell lysate, total RNA was isolated according to the manufacturer's protocol. Although the RNA extraction kit protocol included a DNase treatment step, the extracted RNA was subjected to an additional DNase treatment using Ambion's Turbo DNA-free kit (Ambion Inc., Austin, TX). DNA contamination was removed according to the protocol recommended by the manufacturer. The absence of DNA was confirmed by a direct PCR on the RNA samples, using primers and PCR conditions as described below.
Reverse transcription of RNA and PCR amplification.
Reverse transcription of isolated RNA into cDNA was carried out using the iScript cDNA synthesis kit (Bio-Rad, CA) according to the protocol provided by the manufacturer, using 1 μl (80 to 100 ng) of the RNA template. Amplification of 16S rRNA and dsrB gene fragments was performed using the primer pairs 341F-GC (5′ CCT ACG GGA GGC AGC AG 3′)/907R (5′ CCG TCA ATT CMT TTG AGT TT 3′) (33) and DSRp2060F-GC (5′ CAA CAT CGT YCA YAC CCA GGG 3′) (16)/DSR4R (5′GTG TAG CAG TTA CCG CA 3′) (63), respectively. We used 1 μl of genomic DNA and 2 μl of cDNA as templates for the amplification reactions. The protocol used for the amplification of 16S rRNA gene fragments or cDNA was same as described previously (33). However, the protocol for amplification of the dsrB gene fragment as described by Geets et al. (16) was modified slightly to increase the specificity of the amplification reaction. A “touchdown” protocol was used, wherein the annealing temperature was decreased from 65°C to 55°C in 20 cycles. Thermal cycling was carried out as follows: 5 min of initial denaturation of DNA/cDNA at 95°C, followed by 20 cycles of denaturation at 95°C for 40 s, a “touchdown”-annealing step for 40 s, and elongation at 72°C for 1 min. This was followed by another 30 cycles of denaturation at 95°C for 40 s, annealing at 55°C for 40 s, and elongation at 72°C for 1 min. Amplification was completed by a final elongation step at 72°C for 10 min. DNA from Desulfobulbus propionicus was used as a positive control and water as a negative control in all PCR amplifications. The quality of the PCR products was examined on 1% (wt/vol) agarose gels, and the yield was quantified by absorption spectrophotometry using the NanoDrop ND-1000 (NanoDrop Technologies, DE).
DGGE of 16S rRNA and dsrB gene fragments.
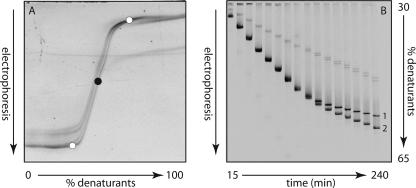
DGGE was performed as described by Schäfer and Muyzer (56) using the D-Code system (Bio-Rad Laboratories, CA). Electrophoresis was performed with 1-mm-thick 6% polyacrylamide gels (ratio of acrylamide to bisacrylamide, 40:1) submerged in 1× TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA, pH 7.5) at a constant temperature of 60°C. PCR product in an amount ranging from 300 to 500 ng was applied to the individual lanes on the gel. The electrophoresis conditions for the 16S rRNA gene fragment were the same as described previously (56): 16 h at 100 V in a linear 20 to 80% denaturant gradient (100% denaturant is a mixture of 7 M urea and 40% [vol/vol] formamide). However, the conditions used for dsrB gene fragments were based on the results of a perpendicular DGGE (see Fig. 2A) and a “time travel” experiment (see Fig. 2B): 6 h at 150 V in a linear 30% to 65% denaturant gradient. After electrophoresis, the gels were incubated for 30 min in Milli-Q water containing ethidium bromide (0.5 μg/ml), rinsed for 20 min in Milli-Q water, and photographed using a Bio-Rad GelDoc station (Bio-Rad, CA). Individual bands were excised, resuspended in 20 μl of Milli-Q water, and stored overnight at 4°C. A volume of 3 to 5 μl of the supernatant was used for reamplification with the original primer sets. The reamplified PCR products were run again on a denaturing gradient gel to check their purity. Prior to sequencing, the PCR products were purified using the Qiaquick PCR purification kit (QIAGEN GmbH, Hilden, Germany).
FIG. 2.
(A) Negative image of a perpendicular denaturing gradient gel of PCR-amplified dsrB fragments from Desulfobulbus propionicus obtained with primer pair DSRp2060F-GC and DSR4R. The black dot indicates a urea-formamide concentration of 46%. The white dots indicate the urea-formamide concentrations (30% and 65%) used for the “time travel” experiment. (B) “Time travel” experiment with PCR-amplified dsrB fragments from Desulfobulbus propionicus (1) and Desulfomicrobium escambience (2). A mixture of the two fragments was loaded onto the gel every 15 min for a total of 240 min. The electrophoresis conditions were 150 V for 4 h.
Phylogenetic analysis.
The 16S rRNA gene sequences obtained were first compared to sequences stored in GenBank by using the BLAST algorithm. Subsequently, the sequences were imported into the ARB software program (26) and aligned using the automatic aligner function. The alignment was further corrected manually, and an optimized tree was calculated using the neighbor-joining algorithm with the Felsenstein correction.
The partial dsrB gene sequences and the deduced amino acids were first analyzed and aligned using the BioEdit (version 7.0.5) sequence alignment editor (www.mbio.ncsu.edu/bioedit/bioedit.html). Subsequently, the sequences were imported into the ARB software program (26), in which the alignment was further corrected manually. The nucleotide sequences were aligned according to the alignment of deduced amino acid sequences. The alignment regions of insertions and deletions were omitted using a suitable alignment mask (indel filter). A full-length dsrAB consensus tree was constructed after comparing the topologies of phylogenetic trees calculated by maximum parsimony, neighbor-joining, and maximum-likelihood analyses. For tree reconstruction, only nearly full-length sequences were considered. Partial sequences were then inserted into the reconstructed tree by applying parsimony criteria, without allowing changes in the overall tree topology.
Design of oligonucleotide probes.
Specific probes for the dominant sulfate-reducing bacteria were designed using the Probe Design function in the ARB software (26). The complete 16S rRNA sequences of Desulfobacca acetoxidans and Desulfovibrio sulfodismutans and the partial sequences obtained from the excised DGGE bands were used for designing probes. The probes were named with a number that indicates the position of the first base in the target sequence (by Escherichia coli numbering). The following fluorescently labeled oligonucleotide probes were used in this study: (i) DSV827, a probe for members of the genus Desulfovibrio (5′ GGT CGC CCC CCG ACA CCT 3′) (this study); (ii) DSBA1017, a probe for Desulfobacca acetoxidans (5′ GTT GCC AGG CAC CCC CAT 3′) (this study); and (iii) DSR660, a probe for members of the genus Desulfobulbus (5′ GAA TTC CAC TTT CCC CTC TG 3′) (11). Desulfobacca acetoxidans DSM11109 and Desulfovibrio sulfodismutans DSM3696 were used as reference strains to check the specificity of the designed probes. Hybridization stringencies were determined by performing hybridizations with increasing formamide concentrations of 10% (vol/vol), 20% (vol/vol), 30% (vol/vol), and 40% (vol/vol) using the reference strains.
Whole-cell hybridization.
Samples from the reactors were washed with 10 mM sodium phosphate buffer (pH 7.2) containing 130 mM NaCl and were resuspended in the same buffer. The cells were fixed with 4% (wt/vol) paraformaldehyde in potassium phosphate buffer for at least 1 h. The cell suspension was subsequently immobilized on Teflon-coated multiwell microscopic slides. Hybridization was carried out according to the protocol described by Pernthaler et al. (45), using 30% (vol/vol) formamide for probes DSV827 and DSBA1017 and 40% (vol/vol) formamide for probe DSR660. Following hybridization, the slides were washed in washing buffer (5 mM EDTA, 20 mM Tris [pH 8.0], 112 mM NaCl, and 0.01% [wt/vol] sodium dodecyl sulfate) at 48°C for 20 min and then rinsed with Milli-Q water. The slides were embedded in Vectashield (Vector Laboratories, Burlingame, CA) and observed with a Zeiss Axioplan 2 epifluorescence microscope. Images were acquired with Leica FW4000 software. Cell counts of dominant SRB were determined as described by Neef et al. (36). The hybridized cells were analyzed by two independent observers for determining the fraction of positive signal from each probe relative to the signal visualized with general probes for bacteria (i.e., probe EUB338 [5′ GCT GCC TCC CGT AGG AGT 3′] [1]) and for SRB (i.e., probes SRB385 [5′ CGG CGT CGC TGC GTC AGG 3′] and SRB385Db [5′ CGG CGT TGC TGC GTC AGG 3′] [1, 50]) or with the general DNA stain DAPI (4′,6′-diamidino-2-phenylindole). The hybridization experiments were done in triplicate using different fluorochromes for each probe; different microscopic fields on each slide were analyzed to confirm the results.
Nucleotide sequence accession numbers.
The sequences determined in this study were submitted to GenBank under accession numbers DQ514554 to DQ514564 for the dsrB sequences and DQ514565 to DQ514583 for the 16S rRNA sequences.
RESULTS
DGGE analysis of 16S rRNA gene fragments.
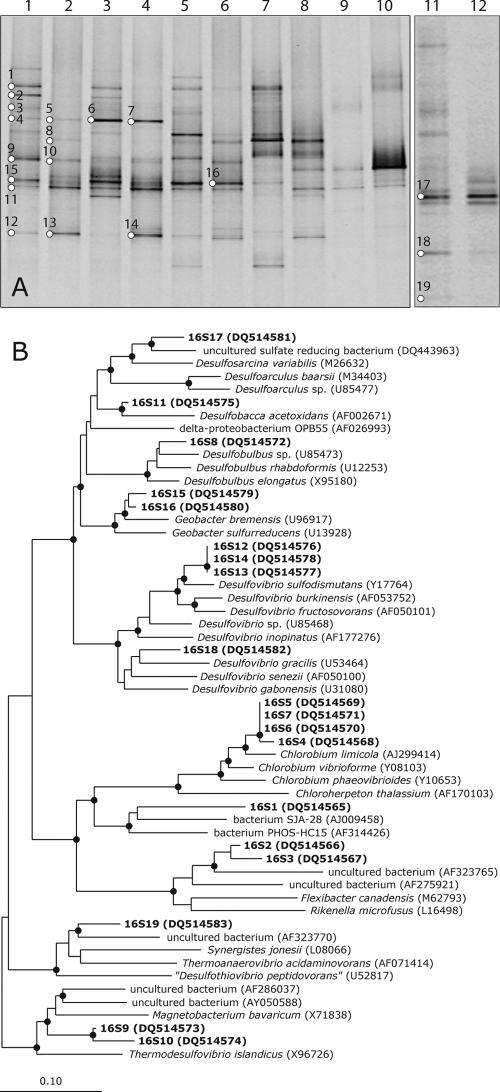
We studied the microbial communities of different lab- and full-scale sulfidogenic reactors (Table 1) by DGGE profiling of 16S rRNA gene fragments, which were amplified either from genomic DNA or from reverse-transcribed RNA, i.e., cDNA. The number of DNA-derived bands relates to the presence of bacterial populations that are above the detection limit of DGGE, while the RNA-derived bands reflect the predominantly active populations. DGGE profiles of the PCR-amplified bacterial 16S rRNA gene differed markedly from those of the reverse-transcribed PCR amplified 16S rRNA (Fig. 1A). In general, patterns obtained with PCR products from genomic DNA showed a higher number of bands than patterns obtained from rRNA. For instance, the DNA-derived pattern of reactor A resulted in approximately 11 bands, while the RNA-derived profile from the same reactor showed only 7 bands. Similar trends were observed for the other reactors, except for reactor E. Several bands (i.e., bands 9, 15, 16, 11, and 12 in Fig. 1A) in the DGGE profile from DNA-derived PCR products showed increased or decreased intensities relative to the corresponding similar bands in the profiles from RNA-derived PCR products. Approximately 25 prevalent bands were excised and sequenced, of which 4 gave ambiguous sequences that were not included in the phylogenetic analysis. Bands showing similar mobility gave identical sequences. Therefore, only a few representative sequences of these bands were used in the phylogenetic analysis. The DGGE profiles of all reactor samples indicated the presence of a highly diverse bacterial population, with some bands present in most of the reactors although at different intensities (e.g., bands 8, 11, 12, 13, and 14 in Fig. 1A). Bands 11, 12, 13, and 14 (Fig. 1A) were more intense in RNA-derived profiles, suggesting that these bacteria were metabolically active. Some bands (i.e., bands 4, 5, 6, and 7 in Fig. 1A) occurred only in the DGGE profiles obtained from samples of the lab-scale reactors A and B.
FIG. 1.
(A) DGGE analysis of 16S rRNA gene fragments using DNA and RNA samples from different sulfidogenic anaerobic bioreactors as templates. Lane 1, DNA sample from reactor A; lane 2, RNA sample from reactor A; lane 3, DNA sample from reactor B; lane 4, RNA sample from reactor B; lane 5, DNA sample from reactor C; lane 6, RNA sample from reactor C; lane 7, DNA sample from reactor D; lane 8, RNA sample from reactor D; lane 9, DNA sample from reactor E; lane 10, RNA sample from reactor E; lane 11, DNA sample from reactor F; lane 12, RNA sample from reactor F. Bands indicated with numbers were excised from the gels and sequenced. (B) Phylogenetic tree based on 16S rRNA gene sequences obtained from the DGGE bands. Sequences determined in this study are in boldface; the band number is preceded by 16S. The sequence accession numbers are shown in parentheses. The sequence of Archaeoglobus fulgidus was used as an outgroup but was pruned from the tree. A black dot indicates a bootstrap value of between 90 and 100%. The scale bar indicates 10% sequence difference.
The phylogenetic affiliation of 16S rRNA gene sequences is presented in Fig. 1B. A neighbor-joining tree was generated using the sequences of the DNA fragments excised from the denaturing gel (Fig. 1A). Band 1 occurred only in the DGGE profile of DNA-derived PCR products from most of the reactor samples. It showed high similarity to uncultured bacteria. Bands 4, 5, 6, and 7 were present only in the profiles obtained from samples of lab-scale reactors and showed high sequence similarity to the green sulfur bacteria, in particular to the sequence of Chlorobium limicola. Band 8 was observed in the DGGE profiles of reactors A, C, and D but not in those of reactors B, E, and F. The band was relatively intense in the RNA-derived pattern of reactor D. Comparative sequence analysis showed that band 8 was closely related to members of the genus Desulfobulbus. The sequence of the most dominant fragment (i.e., band 11), present in the DGGE patterns of all reactors except reactor F, was most closely affiliated to Desulfobacca acetoxidans, a sulfate-reducing bacterium that can use acetate as the sole electron donor. Another fragment (bands 12, 13, and 14) that was significantly present, especially in the profiles obtained from the RNAs of reactor samples, was related to members of the genus Desulfovibrio. The Fe(III)- and Mn(IV)-reducing deltaproteobacterium Geobacter bremensis, represented by bands 15 and 16, was also present in the lab-and full-scale reactors. A reactor F sample resulted in a markedly different DGGE profile, with most of the bands being unique to this reactor. Band 17 was closely related to an uncultured sulfate-reducing bacterium detected in different bioreactors fed with the same seed sludge. The closest relative among the cultured sulfate reducing bacteria was Desulfosarcina variabilis. Band 18 clustered well with members of the genus Desulfovibrio. Band 19 grouped with an uncultured bacterium that was detected in a benzoate-degrading methanogenic consortium.
Optimization of the dsr DGGE.
A PCR product of approximately 390 bp was amplified using the primers DSRp2060F-GC and DSR4R. The optimal gradient of the denaturant concentration for the amplified product was determined by performing a perpendicular DGGE. We loaded 600 ng of amplified PCR product from a pure culture of Desulfobulbus propionicus into a single large well on a polyacrylamide gel, in which the denaturant gradient was perpendicular to the direction of electrophoresis (65). After running through the perpendicular gel, the dsrB gene fragment appeared as a sigmoid curve (Fig. 2A). At low concentrations of denaturants (i.e., from 0% to approximately 40% urea-formamide), the fragment runs as a double-stranded molecule with no denaturation of different melting domains. At a concentration higher than 55% urea-formamide, the fragment undergoes direct melting and is held together only by the GC clamp. At a denaturant concentration of between 40 and 50%, the fragment displays a reduced mobility with transitional denaturation of the melting domain, with a melting temperature corresponding to 46% denaturants. From the perpendicular gradient analysis, a denaturant concentration gradient of 30 to 65% was defined to resolve different dsrB sequence variants in parallel denaturing gradient gels.
We also performed a “time travel” experiment with dsrB fragments obtained from Desulfobulbus propionicus and Desulfomicrobium escambiense to determine the optimal electrophoresis time (Fig. 2B). PCR fragments were loaded at 15-min intervals for up to 240 min (4 h) onto a polyacrylamide gel containing a 30% to 65% linear gradient of denaturants. After ca. 120 min the individual fragments from the two pure culture samples started to separate, and they were clearly distinguished after 4 h. Even after 4 h of electrophoresis the fragments were still migrating through the gel, although at a considerably reduced mobility. We thus decided on an electrophoresis time of 6 h at a constant voltage of 150 V to obtain a good separation between dsrB gene fragments from different sulfate reducers.
DGGE analysis of dsrB gene fragments.
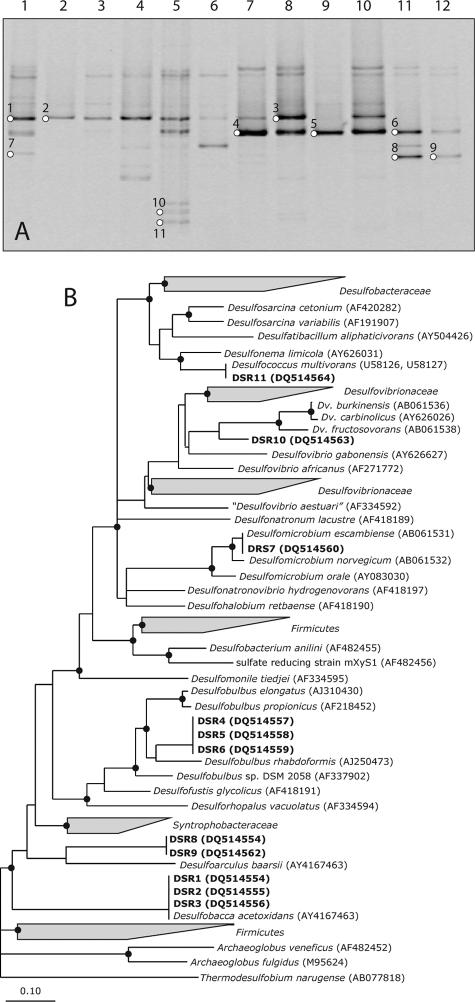
The presence of dormant and metabolically active members of SRB in the sulfidogenic bioreactors was also studied using DGGE of dsrB gene fragments, amplified in parallel from genomic DNA as well as mRNA. The DGGE analysis of the PCR products is shown in Fig. 3A. A maximum of two dominant bands were observed in most of the reactor samples, with an additional one to three less-intense bands appearing in the DNA-derived profiles of reactors A and C. The DNA- and mRNA-derived patterns in reactor B were similar and showed the presence of one dominant band. The DGGE profile of the mRNA-derived PCR product from a reactor C sample was markedly different from the corresponding DNA-derived pattern. mRNA amplification products showed very weak bands corresponding to bands at similar positions in the DNA-derived profile. On the other hand, one intense band was also observed in the mRNA profile, which after repeated attempts gave an ambiguous sequence and hence could not be included in the phylogenetic studies. Reactors D and E gave similar patterns for both the DNA- and mRNA-based PCR products; the only difference is in the intensity of band 3. The intensity of band 3 was higher in the RNA pattern than in the DNA-derived profile, suggesting a highly active population of SRB represented by band 3. The DNA- and RNA-derived patterns in reactor F were similar, although the bands in the RNA-derived profile were less intense, suggesting less-active SRB populations in reactor F. Bands 8 and 9 were found in the profiles of reactor F only, which suggests a different SRB population dominating this reactor. The dominant bands were excised and sequenced, and their phylogenetic affiliations were analyzed and depicted in a phylogenetic tree (Fig. 3B). Bands with similar melting behaviors had the same sequences. In general, the sequencing results were consistent with the results obtained from the 16S rRNA gene sequences. All lab- and full-scale reactors, except for reactor F, showed the presence of Desulfobacca acetoxidans-like sequences (i.e., bands 1, 2, and 3), as observed in the 16S rRNA DGGE profiles. These bands were found in both the DNA- and the RNA-derived dsrB profiles. The second most prominent group of bands (i.e., bands 4, 5, and 6) that occurred in all the reactors, except reactor B, were closely related to Desulfobulbus rhabdoformis. One of the sequences from reactor F (i.e., bands 8 and 9) was closely affiliated with Desulfoarculus baarsii. Furthermore, sequences of bacteria closely affiliated with the genera Desulfovibrio, Desulfomicrobium, and Desulfococcus were found, but the bands (bands 7, 10, and 11) were low in intensity.
FIG. 3.
(A) DGGE patterns of dsrB gene fragments using DNA and RNA samples from different sulfidogenic anaerobic bioreactors as templates (see the legend to Fig. 1A for specifications of the samples). Bands indicated with numbers were excised from the gels and sequenced. (B) Phylogenetic consensus tree for dsrAB amino acid sequences deduced from nearly full-length dsrAB sequences. Branching orders that were not supported by all treeing methods are shown as multifurcations. Partial sequences were individually added to the reconstructed consensus tree by applying parsimony criteria without allowing changes in the overall tree topology. Sequences determined in this study are in boldface; the band number is preceded by DSR. The sequence accession numbers are shown in parentheses. The sequences of Thermodesulfovibrio islandicus and Thermodesulfovibrio yellowstonii were used as an outgroup but were pruned from the tree. A black dot indicates a bootstrap value of between 90 and 100%. Bootstrap values were calculated only for nearly full-length dsrAB sequences. The scale bar indicates 10% sequence difference.
Whole-cell hybridization of sulfate-reducing bacteria.
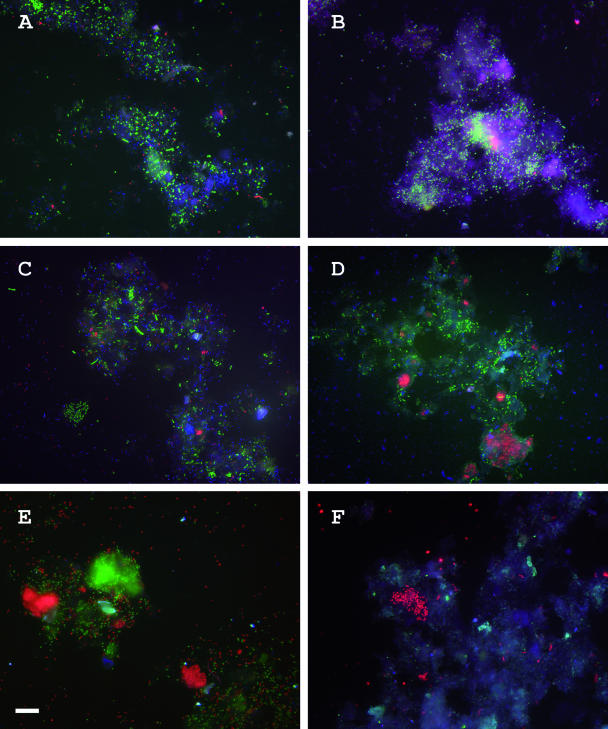
Two 16S rRNA probes, namely, DSBA1017 and DSV827, were designed to target community members that were closely affiliated to Desulfobacca acetoxidans and Desulfovibrio spp., respectively. For detection of Desulfobulbus-like community members, probe DSR660 (11) was chosen. The probes were used to confirm the presence of the dominant sulfate reducers as identified by DGGE and to make an estimate of the abundances of three sulfate-reducing bacterial strains in the reactors. Members of the phylogenetic groups of SRB for which the probes were designed were consistently present in the reactors but varied in relative percentages. The percent abundances of the specific probes relative to the general probes are summarized in Table 2. The Desulfobacca-specific probe, DSBA1017, gave a positive signal with cells of all reactor samples except those from reactor F (Fig. 4). The relative percentage of cells that hybridized with this probe was 50 to 60% of the total SRB385-positive cells and 20 to 30% of the total EUB338-positive cells. The probe specific for members of the genus Desulfovibrio, probe DSV827, detected between 25 to 35% of the total SRB385-positive cells in reactors A, C, and D, whereas in reactor B and F the probe detected ca. 40% (Fig. 4). Compared to EUB338-positive cells, the signal was between 10 and 20% in reactors A, C, and D and ca. 27% in reactor F. In reactor E, no positive signal could be detected with the Desulfovibrio-specific probe (Fig. 4). Probe DSR660, specific for members of the genus Desulfobulbus, gave a positive hybridization signal with most of the reactor samples (Fig. 4). The signal was approximately 10% or less in the lab-scale reactors and one of the full-scale reactors (reactor C), whereas the same probe accounted for about 20 to 30% of the signal in reactors D, E, and F. The probe detected fewer than 5 to 8% of the EUB338-positive cells in reactors A and C but accounted for a signal of approximately 10 to 20% of the EUB338-positive cells in reactors D, E, and F. A large majority of the cells detected by probes DSR660 and DSBA1017 were present as aggregates, although scattered cells were detected as well.
TABLE 2.
Relative abundances of SRB in different sulfidogenic reactors
| Reactor | Probe | SRB abundance (%) relative to detection witha:
|
||
|---|---|---|---|---|
| SRB385 | EUB338 | DAPI | ||
| A | DSBA1017 | 50-60 | 30-35 | 25-30 |
| DSR660 | <10 | <5 | <4 | |
| DSV827 | 30-35 | 15-20 | 8-10 | |
| B | DSBA1017 | 60-65 | 25-30 | 15-20 |
| DSR660 | 0 | 0 | 0 | |
| DSV827 | 30-40 | 15-20 | 10-15 | |
| C | DSBA1017 | >50 | 20-25 | 15-18 |
| DSR660 | <10 | <8 | <5 | |
| DSV827 | 25-30 | 10-15 | 5-8 | |
| D | DSBA1017 | 50-55 | 30-35 | 20-25 |
| DSR660 | 15-20 | 10-15 | 5-10 | |
| DSV827 | 30-35 | 15-20 | 8-10 | |
| E | DSBA1017 | 50-60 | >30 | 20-25 |
| DSR660 | 25-30 | 15-20 | 10-15 | |
| DSV827 | 0 | 0 | 0 | |
| F | DSBA1017 | 0 | 0 | 0 |
| DSR660 | 20-25 | 15-20 | 10-15 | |
| DSV827 | 30-40 | 25-30 | 15-20 | |
Abundances of probe-positive cells relative to cells detected by probe SRB385 (specific for SRB), to cells detected by probe EUB338 (specific for Bacteria), and to cells stained with the DNA stain DAPI.
FIG. 4.
Whole-cell hybridization of reactor samples A to F with probe DSBA1017 (specific for Desulfobacca acetoxidans and labeled with Fluos) (green), probe DSR660 (specific for Desulfobulbus and labeled with Cy3) (red), and probe DSV827 (specific for Desulfovibrio and labeled with Cy5) (blue). Bar, 20 μm.
DISCUSSION
In this study, a comparative analysis of the microbial communities in different lab- and full-scale sulfidogenic reactors was performed using a combined approach with PCR-DGGE and FISH. The combination of these molecular techniques provided us with a detailed and consistent description of the SRB populations within the reactors.
Microbial community analysis using 16S rRNA as a molecular marker.
DGGE analysis of the PCR-amplified 16S rRNA gene was used to obtain an estimate of the microbial diversity within the reactors. To obtain an overview of the metabolically active members in the microbial communities, reverse transcription-PCR amplification of 16S rRNA molecules was used. The analysis of rRNA as a means of inferring the composition of the active members in the total bacterial community has been proposed for many years (46, 66). The rRNA content as a function of growth rate in E. coli (10) is well established. Poulsen et al. (49) found nearly a linear relation between rRNA content and growth rate of a specific SRB population in anaerobic biofilms. Since then an increasing number of studies have been conducted to characterize the metabolically active bacterial and archaeal populations in diverse environments (20, 28, 29, 31, 38). Comparative analysis of PCR products obtained by amplification of rRNA genes and reverse-transcribed rRNA resulted in different DGGE profiles, indicating a marked difference in the microorganisms present and those that are metabolically active. This difference in patterns may be attributed in part to the difference in the ribosome content between the metabolically active and the dormant populations, assuming that actively growing cells contain increased levels of rRNA as has been shown in previous studies (10, 21, 49). A more complex and diverse pattern was obtained from 16S rRNA gene-derived PCR amplicons than from 16S rRNA-derived PCR products, indicating a lesser number of metabolically active populations. DGGE bands 11, 12, 13, and 14 (Fig. 1A) were of low intensity in the DNA-derived profiles compared to the RNA-derived profiles. Assuming that the intensity of DGGE bands is an indirect measure of the relative abundance (13, 32), bands 11, 12, 13, and 14 could be attributed to relatively smaller but metabolically active populations.
Considering the sulfidogenic nature of both the lab- and full-scale reactors, with high sulfate reduction and low electron donor-to-sulfate ratios, most of the 16S rRNA gene/rRNA sequences obtained from the reactor samples, as expected, showed high sequence identity to those of sulfate-reducing bacteria. Sequences were obtained from both completely and incompletely oxidizing sulfate reducers. Sequences of the bands 16S8, 16S12, 16S13, 16S14, and 16S18 grouped with two genera of incompletely oxidizing sulfate reducers, Desulfobulbus and Desulfovibrio (Fig. 1B). The sequence of band 16S8 was closely related to Desulfobulbus rhabdoformis (23), a gram-negative, mesophilic sulfate-reducing bacterium that can oxidize a broad range of substrates, such as propionate, lactate, pyruvate, and ethanol, to acetate and CO2.
The sequences of 16S12, 16S13, and 16S14 closely grouped with members of the genus Desulfovibrio, in particular with Desulfovibrio sulfodismutans, which was isolated from freshwater mud (3). Desulfovibrio sulfodismutans is characterized by the ability to obtain energy for growth by disproportionation of thiosulfate or sulfite to sulfate and sulfide. Growth by dissimilatory sulfate reduction has also been reported (3); however, it is slower than growth by disproportionation. The presence of Desulfovibrio sulfodismutans and Desulfobulbus species in wastewater treatment systems has often been reported in the literature (35, 39, 51, 53, 55, 64). The detection of Desulfovibrio and Desulfobulbus species suggests their fundamental role in the oxidation of carbon through sulfate reduction or through disproportionation of sulfite and thiosulfate in these reactors. Because, Desulfovibrio has a higher growth rate than Desulfobulbus (22), it may be assumed that Desulfovibrio species are mainly responsible for the oxidation of carbon in most of these reactors, although oxidation of some carbon compounds by Desulfobulbus species cannot be ruled out. However, the fermentation product propionate is degraded by Desulfobulbus spp. only, since members of genus Desulfovibrio cannot use propionate as an electron donor. In reactor F, the sequence of 16S18 also clustered with members of the genus Desulfovibrio, with Desulfovibrio gracilis (27) as the closest relative. Desulfovibrio gracilis has been described as moderately halophilic SRB, with optimal NaCl concentrations for growth being 50 to 60 g liter−1. The higher salinity in reactor F (Table 1) may explain the presence of a Desulfovibrio gracilis-like SRB in this reactor. Sequence 16S17, obtained from the most dominant band in both the DNA- and RNA-derived DGGE patterns of reactor F, indicates the presence of an active population. Phylogenetic analysis revealed that this organism was closely related to an uncultured sulfate-reducing bacterium. The closest known sulfate-reducing bacterium was found to be Desulfosarcina variabilis. The availability of a variety of complex organic compounds in reactor F, rather than the input of one specific substrate as an energy source, might be responsible for the predominance of nutritionally versatile Desulfosarcina-like SRB (67).
Sequence 16S11, which could be detected in samples from all reactors except reactor F, showed close similarity to Desulfobacca acetoxidans, which oxidizes its carbon source completely in sulfate reduction (Fig. 1B). First isolated from a sulfidogenic bioreactor (41), Desulfobacca acetoxidans has been described as a mesophilic, gram-negative, completely oxidizing sulfate-reducing bacterium that can utilize acetate as the only source of organic carbon and electron donor. The specific growth rate of Desulfobacca acetoxidans (μmax = 0.013 to 0.017 h−1) has been reported to be higher than those of acetate-degrading methanogenic archaea, such as Methanosaeta spp. (μmax = 0.003 to 0.012 h−1), which dominate methanogenic bioreactors (41). Although archaea were not the focus of this study, it was expected that because of the high sulfate concentrations and limited electron donor quantity in these reactors and the better growth kinetics of Desulfobacca acetoxidans, it would outcompete the acetate-degrading methanogenic archaea.
The sequences 16S4, 16S5, 16S6, and 16S7 were detected in the lab-scale reactors only. The presence of the band was more prominent in reactor B, indicating the presence of a more abundant population. Phylogenetically the sequences grouped with the sequence of Chlorobium limicola (Fig. 1B), an anaerobic, photosynthetic green sulfur bacterium that can use sulfide or sulfur as an electron donor (14). The presence of these bacteria can be explained because the lab-scale reactors were made from glass and were exposed to light. The greenish color of the culture from the lab-scale reactor B at the time of sampling may be attributed to the presence of this bacterium. A likely syntrophic association of this bacterium with sulfate-reducing bacteria, oxidizing sulfide to sulfur and/or sulfate, may be assumed to be present in these reactors. Although present in high numbers in reactor B, the recycling of sulfur does not seem to significantly influence the sulfate reduction, as the percentages of sulfate removal in the two lab reactors were the same (Table 1).
Sequences 16S15 and 16S16 were closely related to the Fe(III)- and Mn(IV)-reducing deltaproteobacterium Geobacter bremensis (58). In the absence of ferric iron, Geobacter species can use alternate electron acceptors, such as nitrate, fumarate, and elemental sulfur, for the oxidation of variety of organic compounds. A coculture of Geobacter spp. in syntrophic association with sulfate-reducing bacteria in the presence of sulfate as an electron acceptor has been described before (7, 54). However, this culture grows very slowly, with a doubling time of more than 7 days.
Microbial community analysis using dsrB as a molecular marker.
We used the dissimilatory sulfite reductase (dsrAB) gene as a molecular marker to elucidate the community composition of sulfate-reducing bacteria in the reactors. For this purpose, the primers originally designed as internal sequencing primers by Pérez-Jiménez et al. (44) and later modified for DGGE analysis by Geets et al. (16) were used. A 350-bp dsrB gene fragment was amplified and used for DGGE analysis; individual bands were sequenced and used for identification of the SRB. Previous dsr gene-based studies to evaluate SRB communities used primers DSR1F/DSR4R (63), amplifying a 1.9-kb dsr gene fragment. The subsequent phylogenetic studies were then based either on sequencing the cloned 1.9-kb dsr gene fragment (12) or on restriction analysis of the cloned fragment (24, 43). Although our study was based on a much shorter sequence of 350 bp, previous studies have shown that the general topology of the dsrAB trees based on sequences of different lengths remains consistent (44), and other studies (5, 8) have indicated that shorter sequences of the dsrAB gene can be successfully used for the phylogenetic analysis of the SRB. We are well aware that short sequences may not be suitable for detailed phylogenetic inferences or for tracing the evolutionary history of sulfate-reducing bacteria, but short sequences are suitable for identification of SRB in environmental samples.
Detection of dsrB sequences indicated the presence of SRB in these reactors, but this does not provide evidence for metabolically active populations. Therefore, the expression of the dsrAB gene was studied by targeting mRNA, indicating the presence of active populations of sulfate reducers. Validation of this approach was given by Neretin and coworkers (37), who quantified dsr gene expression in Desulfobacterium autotrophicum by using real-time reverse transcription-PCR. They found a strong positive correlation between dsr mRNA concentration and cell-specific sulfate reduction rate. Our DGGE results with dsrB gene fragments indicated a relatively simple sulfate-reducing bacterial community, with similar populations present in all reactors except reactor F. This similarity is probably due to the similar environmental conditions (i.e., pH and temperature) or to the same carbon source being fed to most of the reactors. The complex organic compounds and higher salinity in reactor F (Table 1) might have contributed to the presence of a different SRB community in this reactor.
In most reactors, identification of SRB based on dsrB sequences showed a similarity to the SRB identified by comparative 16S rRNA gene sequence analysis. The prominent dsrB sequences (DSR1/DSR2/DSR3 and DSR4/DSR5/DSR6) were phylogenetically affiliated to Desulfobacca acetoxidans and Desulfobulbus rhabdoformis, respectively. The absence of Desulfobulbus rhabdoformis-like sequences in reactor B was consistent with the results obtained by 16S rRNA gene analysis. Desulfobacca acetoxidans-like sequences were not detected in reactor F, which again was in agreement with 16S rRNA gene analysis. The sequence of bands DSR8/DSR9 grouped with Desulfoarculus baarsii, which is capable of complete oxidation of organic compounds to CO2. The oxidation of acetate by Desulfoarculus baarsii is reported to take place at a low rate, with no substantial formation of biomass (6). This might explain the small amount of sulfate reduction (ca. 20%) in reactor F relative to the amount of sulfate reduction in other reactors.
Although a phylogenetically similar Desulfovibrio-like sequence (i.e., DSR10) was detected in reactor C, the result was in contrast to the 16S rRNA gene/rRNA DGGE, in which Desulfovibrio sulfodismutans-like sequences were found in all the reactors except reactor E. This may be attributed to PCR bias (48, 59), as two different primer sets amplifying two different genes, one targeting the whole bacterial population and the other specific to dissimilatory sulfate reducers, may result in different PCR biases. The dsrB DGGE resulted in the detection of two other SRB-like sequences (DSR7 and DSR11), which were closely related to Desulfomicrobium escambiense and Desulfococcus multivorans, respectively. These results may suggest an increased sensitivity in detection of sulfate-reducing bacteria by dsrB DGGE, which seems possible because the primers are specific for microorganism that are capable of dissimilatory sulfate reduction.
Whole-cell hybridization of SRB.
Since PCR-based approaches for the analysis of microbial diversity in mixed populations can be influenced by several constraints (61), our results based on PCR-DGGE do not necessarily reflect the abundance of target sequences in the original sample. We therefore tried to confirm the relevance of the sequence data by whole-cell hybridization using fluorescently labeled oligonucleotide probes. Based on the comparative analysis of the sequences retrieved and those of reference organisms, three probes, DSBA1017, DSV827, and DSR660, were used in the hybridization analysis to validate the results obtained by DGGE. The hybridization results (Fig. 4) not only confirmed the presence of dominant SRB sequences retrieved by DGGE but also gave an estimate of the abundance of these bacteria relative to SRB or total bacterial communities. FISH experiments indicated that Desulfobacca-like sulfate-reducing bacteria made up a significant part of the SRB community in the reactors, with the exception of reactor F, in which no positive hybridization was observed (Fig. 4 and Table 2). This is consistent with the DGGE results. Again in accordance with the results obtained by DGGE, the probe specific for members of the genus Desulfobulbus showed that Desulfobulbus spp. were more abundant in reactors D, E, and F than in reactors A and C (Fig. 4 and Table 2). In the latter reactors, Desulfobulbus spp. do not seem to contribute significantly to the overall SRB population. Cells detected with DSR660 were observed mainly as aggregates of a few hundred cells. Similar clusters of cells of Desulfobulbus spp. have been observed previously in wastewater biofilms (40).
The detection of 30 to 40% of Desulfovibrio cells in most of the reactor samples with probe DSV827 (Table 2) implies that they are important in sulfate reduction in these reactors. The results are in general in agreement with the 16S rRNA gene DGGE, although dsrB DGGE failed to identify Desulfovibrio-like sequences. This might be due to PCR bias such as the preferential amplification of dsrB gene fragments from Desulfobacca- and Desulfobulbus-like species.
So far, most microbial ecology studies have focused on the diversity of microorganisms. However, more important for the cycling of chemical elements, such as sulfur, are the microorganisms that are active. In this study, we compared the structures and functions of sulfate-reducing bacterial communities in different lab- and full-scale wastewater treatment reactors by targeting both DNA and RNA of two different molecular markers, i.e., 16S rRNA and dsrB. Detection of the gene indicates the presence of microorganisms; however, detection of rRNA or the mRNA of the dsrB ensured that the corresponding SRB was metabolically active at the time of sampling. In general, congruent results were obtained with the two genes. Whole-cell hybridization with oligonucleotide probes targeting the 16S rRNAs of the dominant SRB populations confirmed the results obtained with PCR-DGGE and showed the relative abundances of these populations. The approach is an important step forward to gain insight into the niche differentiation of coexisting sulfate-reducing bacteria in different habitats.
Acknowledgments
We are grateful to the Dutch Science Foundation-Earth and Life Sciences (NWO-ALW) for supporting this work financially.
We thank Paques B.V. (Balk, The Netherlands) for providing the samples for this study. We thank Miriam Foti for help in construction of the dsrAB phylogenetic tree and Marzia Miletto and Dimitry Sorokin for helpful discussions.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bak, F., and N. Pfennig. 1987. Chemolithotrophic growth of Desulfovibrio sulfodismutans sp. nov. by disproportionation of inorganic sulfur compounds. Arch. Microbiol. 147:184-189. [Google Scholar]
- 4.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colleran, E., S. Finnegan, and P. Lens. 1995. Anaerobic treatment of sulphate-containing waste streams. Antonie Leeuwenhoek 67:29-46. [DOI] [PubMed] [Google Scholar]
- 7.Cord-Ruwisch, R., D. R. Lovley, and B. Schink. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64:2232-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and S. C. Cary. 1999. Diversity of dissimilatory bisulfite reductase genes of bacteria associated with the deep-sea hydrothermal vent polychaete annelid Alvinella pompejana. Appl. Environ. Microbiol. 65:1127-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dar, S. A., J. G. Kuenen, and G. Muyzer. 2005. Nested PCR-denaturing gradient gel electrophoresis approach to determine the diversity of sulfate-reducing bacteria in complex microbial communities. Appl. Environ. Microbiol. 71:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 11.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 12.Dhillon, A., A. Teske, J. Dillon, D. A. Stahl, and M. L. Sogin. 2003. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 69:2765-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilly, O., J. Bloem, A. Vos, and J. C. Munch. 2004. Bacterial diversity in agricultural soils during litter decomposition. Appl. Environ. Microbiol. 70:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueras, J. B., L. J. Garcia-Gil, and C. A. Abella. 1997. Phylogeny of the genus Chlorobium based on 16S rDNA sequence. FEMS Microbiol. Lett. 152:31-36. [DOI] [PubMed] [Google Scholar]
- 15.Fukuba, T., M. Ogawa, T. Fujii, and T. Naganuma. 2003. Phylogenetic diversity of dissimilatory sulfite reductase genes from deep-sea cold seep sediment. Mar. Biotechnol. 5:458-468. [DOI] [PubMed] [Google Scholar]
- 16.Geets, J., B. Borremans, L. Diels, D. Springael, J. Vangronsveld, D. van der Lelie, and K. Vanbroekhoven. 2006. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 66:194-205. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, G. R. 1990. Physiology and ecology of the sulphate-reducing bacteria. J. Appl. Bacteriol. 69:769-797. [DOI] [PubMed] [Google Scholar]
- 18.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 19.Hulshoff Pol, L. W., P. N. Lens, A. J. Stams, and G. Lettinga. 1998. Anaerobic treatment of sulphate-rich wastewaters. Biodegradation 9:213-224. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki, F., Y. Sakihama, A. Inoue, C. Kato, and K. Horikoshi. 2002. Molecular phylogenetic analyses of reverse-transcribed bacterial rRNA obtained from deep-sea cold seep sediments. Environ. Microbiol. 4:277-286. [DOI] [PubMed] [Google Scholar]
- 21.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 22.Laanbroek, H. J., H. J. Geerligs, L. Sijtsma, and H. Veldkamp. 1984. Competition for sulfate and ethanol among Desulfobacter, Desulfobulbus, and Desulfovibrio species isolated from intertidal sediments. Appl. Environ. Microbiol. 47:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lien, T., M. Madsen, I. H. Steen, and K. Gjerdevik. 1998. Desulfobulbus rhabdoformis sp. nov., a sulfate reducer from a water-oil separation system. Int. J. Syst. Bacteriol. 48:469-474. [DOI] [PubMed] [Google Scholar]
- 24.Liu, X., C. E. Bagwell, L. Wu, A. H. Devol, and J. Zhou. 2003. Molecular diversity of sulfate-reducing bacteria from two different continental margin habitats. Appl. Environ. Microbiol. 69:6073-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magot, M., O. Basso, C. Tardy-Jacquenod, and P. Caumette. 2004. Desulfovibrio bastinii sp. nov. and Desulfovibrio gracilis sp. nov., moderately halophilic, sulfate-reducing bacteria isolated from deep subsurface oilfield water. Int. J. Syst. Evol. Microbiol. 54:1693-1697. [DOI] [PubMed] [Google Scholar]
- 28.Martinez, R. J., H. J. Mills, S. Story, and P. A. Sobecky. 2006. Prokaryotic diversity and metabolically active microbial populations in sediments from an active mud volcano in the Gulf of Mexico. Environ. Microbiol. 8:1783-1795 [DOI] [PubMed] [Google Scholar]
- 29.Mills, H. J., R. J. Martinez, S. Story, and P. A. Sobecky. 2004. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities from Gulf of Mexico cold-seep sediments. Appl. Environ. Microbiol. 70:5447-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miskin, I. P., P. Farrimond, and I. M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977-1987. [DOI] [PubMed] [Google Scholar]
- 32.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa, T., J. Ishibashi, A. Maruyama, T. Yamanaka, Y. Morimoto, H. Kimura, T. Urabe, and M. Fukui. 2004. Analysis of dissimilatory sulfite reductase and 16S rRNA gene fragments from deep-sea hydrothermal sites of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific. Appl. Environ. Microbiol. 70:393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanninga, H. J., and J. C. Gottschal. 1987. Properties of Desulfovibrio carbinolicus sp. nov. and other sulfate-reducing bacteria isolated from an anaerobic-purification plant Appl. Environ. Microbiol. 53:802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neef, A., A. Zaglauer, H. Meier, R. Amann, H. Lemmer, and K. H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neretin, L. N., A. Schippers, A. Pernthaler, K. Hamann, R. Amann, and B. B. Jorgensen. 2003. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5:660-671. [DOI] [PubMed] [Google Scholar]
- 38.Nogales, B., E. R. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 39.Okabe, S., T. Ito, and H. Satoh. 2003. Sulfate-reducing bacterial community structure and their contribution to carbon mineralization in a wastewater biofilm growing under microaerophilic conditions. Appl. Microbiol. Biotechnol. 63:322-334. [DOI] [PubMed] [Google Scholar]
- 40.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oude Elferink, S. J., W. M. Akkermans-van Vliet, J. J. Bogte, and A. J. Stams. 1999. Desulfobacca acetoxidans gen. nov., sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol. 49:345-350. [DOI] [PubMed] [Google Scholar]
- 42.Oude Elferink, S. J. W. H., A. Visser, L. W. H. Pol, and A. J. M. Stams. 1994. Sulfate reduction in methanogenic bioreactors. FEMS Microbiol. Rev. 15:119-136. [Google Scholar]
- 43.Perez-Jimenez, J. R., and L. J. Kerkhof. 2005. Phylogeography of sulfate-reducing bacteria among disturbed sediments, disclosed by analysis of the dissimilatory sulfite reductase genes (dsrAB). Appl. Environ. Microbiol. 71:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Jimenez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsrAB) genes. FEMS Microbiol. Ecol. 35:145-150. [DOI] [PubMed] [Google Scholar]
- 45.Pernthaler, J., F.-O. Glockner, W. Schonhuber, and R. Amman. 2001. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 46.Pichard, S. L., and J. H. Paul. 1993. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl. Environ. Microbiol. 59:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pol, L. W., P. N. Lens, J. Weijma, and A. J. Stams. 2001. New developments in reactor and process technology for sulfate reduction. Water Sci. Technol 44:67-76. [PubMed] [Google Scholar]
- 48.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabus, R., M. Fukui, H. Wilkes, and F. Widdle. 1996. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raskin, L., D. Zheng, M. E. Griffin, P. G. Stroot, and P. Misra. 1995. Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Leeuwenhoek 68:297-308. [DOI] [PubMed] [Google Scholar]
- 52.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jorgensen, and R. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roest, K., H. G. Heilig, H. Smidt, W. M. de Vos, A. J. Stams, and A. D. Akkermans. 2005. Community analysis of a full-scale anaerobic bioreactor treating paper mill wastewater. Syst. Appl. Microbiol. 28:175-185. [DOI] [PubMed] [Google Scholar]
- 54.Rozanova, E., A. Galushko, and T. Nazina. 1990. An acetate-decomposing sulphidogenic syntrophic association, p. 469-470. In J.-L. Garcia (ed.), Microbiology and biochemistry of strict anaerobes involved in interspecies hydrogen transfer. Plenum Press, New York, NY.
- 55.Santegoeds, C. M., L. R. Damgaard, G. Hesselink, J. Zopfi, P. Lens, G. Muyzer, and D. de Beer. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol. 65:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schäfer, H., and G. Muyzer. 2001. Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 30:425-468. [Google Scholar]
- 57.Stahl, D. A., S. Fishbain, M. Klein, B. J. Baker, and M. Wagner. 2002. Origins and diversification of sulfate-respiring microorganisms. Antonie Leeuwenhoek 81:189-195. [DOI] [PubMed] [Google Scholar]
- 58.Straub, K. L., and B. E. Buchholz-Cleven. 2001. Geobacter bremensis sp. nov. and Geobacter pelophilus sp. nov., two dissimilatory ferric-iron-reducing bacteria. Int. J. Syst. Evol. Microbiol. 51:1805-1808. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vishniac, W., and M. Santer. 1957. The thiobacilli. Bacteriol. Rev. 21:195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 62.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 56:3748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wawer, C., M. S. Jetten, and G. Muyzer. 1997. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl. Environ. Microbiol. 63:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [NiFe] hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weller, R., and D. M. Ward. 1989. Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl. Environ. Microbiol. 55:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, NY.
- 68.Zverlov, V., M. Klein, S. Lucker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]