Abstract
Two primer sets for automated ribosomal intergenic spacer analysis (ARISA) were used to assess the bacterial community composition (BCC) in Lake Mendota, Wisconsin, over 3 years. Correspondence analysis revealed differences in community profiles generated by different primer sets, but overall ecological patterns were conserved in each case. ARISA is a powerful tool for evaluating BCC change through space and time, regardless of the specific primer set used.
Studies conducted within an ecological framework often involve broad-scale research questions whose answers depend less on obtaining a complete census of microbial populations than on the ability to sample a system of interest with high spatial or temporal resolution. Current molecular microbial ecology methods have allowed environmental microbiologists to ask important ecological questions that previously could not be addressed through culture-based approaches. Culture-independent methods also suffer from well-established limitations (7), however, and as such are useful but always imperfect. This is particularly true for PCR-based methods, since PCR amplification of microbial DNA can be subject to bias for a variety of reasons (10). In spite of these limitations, great advances in the study of microbial ecology have been made through the use of molecular methods.
A key component of any PCR-based method is the PCR primer set. Ideally, a universal primer set would amplify the target DNA from all taxonomic groups with equal efficiencies. No known primer sets meet this criterion. As a result, inconsistencies in DNA amplification are likely to be observed between particular primer sets.
The purpose of this study was to compare two recently evaluated automated ribosomal intergenic spacer analysis (ARISA) primer sets (3) for use in aquatic microbial ecology. ARISA distinguishes microbial populations based on the length heterogeneity in the ribosomal intergenic spacer region and provides a sensitive analysis of microbial communities at a relatively high level of taxonomic resolution with significant reproducibility (3, 6). In addition, the automated nature of the method enables rapid analysis of a large number of samples collected over space or time. The efficacy of this method for ecological research has been demonstrated by our research group and others (8, 9, 11). Here we sought to answer two important questions: (i) do different ARISA primer sets provide dissimilar snapshots of bacterial community composition (BCC), and (ii) do these primer sets result in different conclusions about the ecology of an aquatic system?
Sampling and environmental data.
Integrated water samples were taken from Lake Mendota (Dane County, WI) to a depth of 12 m approximately every 2 weeks during the ice-off season from April 2002 to October 2004. Microbial communities from 250 to 500 ml of lake water were captured on 0.2-μm filters (Pall Life Sciences; catalog no. 63025). Filters were stored at −80°C until DNA was extracted using a FastPrep DNA purification kit (Bio 101). Environmental data were collected by the North Temperate Lakes Long Term Ecological Research program (http://lter.limnology.wisc.edu). Complete methodology and data are available at http://lterquery.limnology.wisc.edu.
ARISA.
ARISA was conducted on all samples using two primer sets. Primer set 1406f/23Sr consisted of 5′-TGYACACACCGCCCGT-3′ (forward primer sequence) and 5′-GGGTTBCCCCATTCRG-3′ (reverse primer sequence). Primer set ITSF/ITSFReub consisted of 5′-GTCGTAACAAGGTAGCCGTA-3′ (forward primer sequence) and 5′-GCCAAGGCATCCACC-3′ (reverse primer sequence). The conditions for ARISA PCR using the 1406f/23Sr primer set were described previously (9). ARISA PCR conditions for the ITSF/ITSReub primer set were previously described by Cardinale et al. (3). Denaturing capillary electrophoresis was carried out for each PCR using an ABI 3730 genetic analyzer (PE Biosystems) as described previously (9).
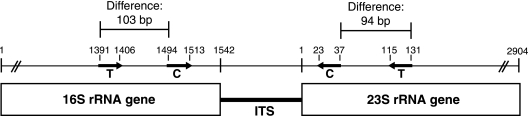
ARISA profiles were analyzed using a 100- to 2,000-bp custom internal size standard (Bioventures), Genescan 3.1.2 (Applied Biosystems), and Genotyper 2.5 (Applied Biosystems), as described by Kent et al. (9). To include the maximum number of peaks while excluding background fluorescence, a threshold of 50 fluorescence units greater than the baseline was used. Profiles obtained with each primer set were independently analyzed and compared for each sample. In addition, profiles generated using ITSF/ITSReub were converted to the fragment length that would theoretically be detected using 1406f/23Sr by adding 197 base pairs and aligned using Matlab 5.0 (The Mathworks). The 197-bp addition corresponds to the difference in length obtained by amplifying the 16S-23S intergenic spacer region using primer set ITSF/ITSReub versus that obtained with primer set 1406f/23Sr, based on Escherichia coli numbering (1) (Fig. 1). This “converted” data set will be referred to as cITSF/ITSReub.
FIG. 1.
The positions of the primers used in this study are shown on the prokaryotic rRNA operon. Bold lines indicate primer annealing sites using E. coli numbering. T, 1406f/23Sr primers utilized by Fisher and Triplett (6); C, ITSF/ITSReub primers introduced by Cardinale et al. (3); ITS, 16S-23S rRNA operon internal transcribed spacer. Length heterogeneity in the ITS sequence is used to distinguish microbial populations in ARISA. The 1406f/23Sr primer set amplifies an additional 197 bases compared to that amplified by the ITS/ITSReub primer set (based upon standard E. coli numbering). This value was added to ITSF/ITSReub data to generate the cITSF/ITSReub data set and allowed direct comparison of data produced from the two primer sets.
Multivariate statistical analyses.
Correspondence analysis (CA) was carried out on the 1406f/23Sr, ITSF/ITSReub, and cITSF/ITSReub data sets. This approach is useful for summarizing and graphically representing the multivariate data collected over time and enables comparisons of ecological patterns apparent in the data generated by each primer set. Samples with similar community structures plot close together in a CA ordination, and axes represent theoretical environmental gradients, which can be compared to measured environmental variables. Axes were selected to explain the greatest variability in the community data. CA biplots and correlation coefficients were generated using Canoco for Windows, version 4.51 (Biometris-Plant Research International; 1997 to 2003).
Analysis of similarity (ANOSIM) is a method to test for differences among defined groups in multivariate data sets that is analogous to analysis of variance in univariate statistics (4). ANOSIM was used to directly compare Lake Mendota BCC data produced by the 1406f/23Sr primer set with the cITSF/ITSReub profiles, testing the hypothesis that community profiles produced by the same primer set are more similar to each other than to profiles produced by different primer sets. Successive ANOSIM analyses were conducted with the data set after peaks falling below a threshold of 1, 2, 3, 4, or 5% of total fluorescence were removed. ANOSIM produces a statistic, R, which indicates the magnitude of difference among groups of samples. An R of 1 indicates that the communities completely differ among defined groups, and an R of 0 indicates no difference among groups (4, 5). The statistical significance of R was tested by Monte Carlo randomization.
Profile composition.
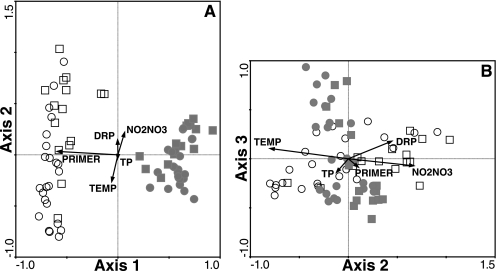
BCC data generated by the two primer sets (1406f/23Sr and cITSF/ITSReub) were combined in a single correspondence analysis (Fig. 2). Samples amplified by the two primer sets strongly separate along axis 1 (Fig. 2A). The “primer” vector nearly parallel to axis 1 illustrates the strong correlation between axis 1 and the primer set used for ARISA (correlation coefficient, 0.975), indicating that the primer set best describes the difference observed among the samples despite correcting for the difference in amplicon length. These differences could be important at the molecular and the methodological level and may significantly influence conclusions about the presence of taxa or their relative abundances.
FIG. 2.
Correspondence analyses biplots directly comparing ARISA data sets generated by using two different primer sets. Axes 1 and 2 (A) and axes 2 and 3 (B) of the ordination comparing 1406f/23Sr and converted ITSF/ITSReub ARISA data sets. Because of the length differences in the amplicons generated by the two primer sets, 197 bp was added to each fragment generated by ITSF/ITSReub PCR to allow direct comparison of community compositions by correspondence analysis. Filled symbols represent bacterial community data generated from ITSF/ITSReub ARISA, and open symbols represent community data generated using 1406f/23Sr. Squares, samples collected in spring and fall; circles, samples collected in summer. Vectors represent environmental variables and the dummy variable “PRIMER” (1, 1406f/23Sr; 0, ITSF/ITSReub). TP, total phosphorus; TEMP, water temperature; DRP, dissolved reactive phosphorus; NO2NO3, nitrite and nitrate.
To determine if the strong differences in BCC patterns due to the primer set were driven by dominant or less-abundant populations (based upon relative areas of ARISA peaks), we gradually increased the minimum relative fluorescence threshold for inclusion in the analysis. ANOSIM R values consistently decreased as the threshold was increased from 0% (all peaks included) to 5% (only dominant peaks included), indicating that when minor community members are excluded the primer effect is reduced (Table 1) .
TABLE 1.
ANOSIM R values for comparisons between ARISA profiles generated from primer set 1406f/23Sr and converted primer set ITSF/ITSReub
| Data seta | Rb | P | nc |
|---|---|---|---|
| Full | 0.941 | 0.001 | 38 |
| 1% | 0.925 | 0.001 | 38 |
| 2% | 0.869 | 0.001 | 38 |
| 3% | 0.770 | 0.001 | 38 |
| 4% | 0.638 | 0.001 | 38 |
| 5% | 0.379 | 0.001 | 36 |
Comparisons of ARISA profiles were performed by analyses using various thresholds (1 to 5%) for minimum relative fluorescence contributions by a single “taxon.” See text for details.
An ANOSIM R value of 1 indicates complete dissimilarity between samples analyzed with the two primer sets; 0 corresponds to complete similarity among these groups of samples.
n, number of samples included in the ANOSIM analysis.
The observed differences between BCC determined using 1406f/23Sr and ITSF/ITSReub appear to be driven largely by amplicons that do not contribute significantly to overall ARISA profile fluorescence. If we assume that relative fluorescence is proportional to relative abundance (supported by (2), this observation could be the result of differential amplification of rare taxa. If each primer set preferentially amplifies different groups of rare taxa, the simple conversion of the ITSF/ITSReub profiles to 1406f/23Sr bins by adding 197 bp would generate dissimilar fingerprints, as we observe in Fig. 2A. Even if relative fluorescence is not correlated with relative abundance, the same subset of strongly amplified taxa is represented in profiles generated by both primer sets. The ANOSIM sensitivity analysis (Table 1) demonstrates that this subset of taxa dictates our interpretation of the Lake Mendota bacterial community dynamics with either primer set.
Ecological conclusions and seasonal patterns.
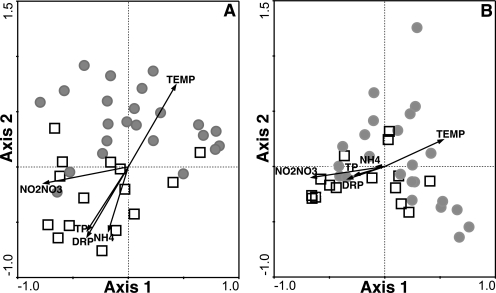
Within the combined analysis, axes 2 and 3 of the CA ordination combine to best explain the variance in BCC driven by environmental parameters and reveal substantial overlap between the two data sets (Fig. 2B). In addition, sample ordination along axes 2 and 3 demonstrates good agreement between the 1406f/23Sr and cITSF/ITSReub data in seasonal separation and correlation with environmental variables. Individual CA ordinations of ARISA profiles generated by each primer set show similar seasonal patterns and relationships to chemical and physical variables (Fig. 3). The visual patterns in the ordinations are supported by correlation coefficients between the axis 1 sample scores from the independent correspondence analyses and environmental drivers that are shared among these data sets (Table 2). The signs and strengths of correlation were similar for bacterial community data generated by each primer set.
FIG. 3.
Correspondence analysis biplots of bacterial community compositions in Lake Mendota. Seasonal dynamics of bacterioplankton communities were assessed from April 2002 to October 2004 using ARISA relative fluorescence values. ARISA data were generated with two different primer sets: (A) 1406f/23Sr (6) and (B) ITSF/ITSReub (3). Squares, samples collected in spring and fall; circles, samples collected in summer. Vectors represent environmental variables; the direction of increase for each variable and the length of each arrow indicate the degree of correlation with the ordination axes. TP, total phosphorus; TEMP, water temperature; DRP, dissolved reactive phosphorus; NH4, ammonium; NO2NO3, nitrite and nitrate.
TABLE 2.
Correlation coefficients between sample scores on the first ordination axis and environmental vectors calculated for community data generated by each primer set
| Factor | Correlation coefficient
|
|
|---|---|---|
| ITSF/ITSReub | 1406f/23Sr | |
| Temperature | 0.617 | 0.376 |
| Nitrite/nitrate | −0.776 | −0.667 |
| Dissolved reactive phosphorus | −0.391 | −0.326 |
| Total phosphorus | −0.318 | −0.325 |
The predominant ecological patterns in BCC were evident regardless of the choice of primer set used for ARISA. Our two primer sets lead to similar conclusions about seasonal patterns in BCC dynamics in Lake Mendota over 3 years and the environmental factors that drive these patterns (Fig. 3).
A recent study evaluated three primer sets for use with ARISA, comparing the richnesses of ARISA profiles generated by each primer set from six environments (3). The results of this study indicated dramatic differences in measured bacterial richness among the three primer sets, but we suggest that this method of evaluation has limited ecological relevance, given the well-known limitations of molecular microbial ecology methods (7). The purpose of ARISA is not to evaluate richness and composition of single samples. ARISA provides rapid, reproducible community fingerprints that alone contain little information but, when applied in the context of an ecological question and compared to other fingerprints through time or across space (9, 11), offer insight about drivers of community composition change. Our study suggests that different primer sets provide dissimilar fingerprints from a given environmental sample, likely due to differential amplification of taxa with a small contribution to ARISA total fluorescence. However, when fingerprints from different primer sets are compared along some ecological gradient (time, space, etc.) the pattern of change inferred by ARISA is remarkably robust with respect to primer set.
Acknowledgments
We thank Jed Fuhrman, Eric Triplett, and Ian Hewson for helpful discussions and comments on the manuscript. We are also grateful to the University of Wisconsin Center for Limnology and the North Temperate Lakes Long Term Ecological Research program for environmental data and logistical support.
This research was supported in part by National Science Foundation Microbial Observatories grant MCB-9977903 and by funding supplied by the University of Wisconsin—Madison Graduate School to K.D.M.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. V., M. S. Schwalbach, I. Hewson, and J. A. Fuhrman. 2005. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 7:1466-1479. [DOI] [PubMed] [Google Scholar]
- 3.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke, K. R. 1993. Nonparametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 5.Clarke, K. R., and R. H. Green. 1988. Statistical design and analysis for a biological effects study. Mar. Ecol. Prog. Ser. 46:213-226. [Google Scholar]
- 6.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 8.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent, A. D., S. E. Jones, A. C. Yannarell, J. M. Graham, G. H. Lauster, T. K. Kratz, and E. W. Triplett. 2004. Annual patterns in bacterioplankton community variability in a humic lake. Microb. Ecol. 48:550-560. [DOI] [PubMed] [Google Scholar]
- 10.von Wintzingerode, F. V., U. B. Goebel, and E. Stackebrandt. 1997. Determinations of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 11.Yannarell, A. C., and E. W. Triplett. 2005. Geographic and environmental sources of variation in lake bacterial community composition. Appl. Environ. Microbiol. 71:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]