Abstract
The gut bacteria of the North American medicinal leech, Macrobdella decora, were characterized. Biochemical tests and DNA sequences indicated that Aeromonas jandaei is the dominant culturable symbiont in leeches from a broad geographic area. In this work we identified a new habitat for A. jandaei, and here we suggest that there is unexpected specificity between leeches and Aeromonas species.
Symbiotic bacteria of leeches recently have been the subject of studies to determine both the specific identities of microbes and the nature of the symbiotic relationships (9). A particularly interesting model symbiosis is the symbiosis between European medicinal leeches and their gut symbiont, Aeromonas veronii biovar sobria (8, 9), which was once considered a unique species, Pseudomonas hirudinis (4). A. veronii biovar sobria is the single cultured symbiont with clinical significance residing in the crop of the digestive tract with an uncultured member of the Bacteroidetes (30). It is unclear how widespread Aeromonas species are in the guts of different leech species.
The potential for aeromonad infections due to postoperative leech use was quickly recognized (29), and concern for appropriate prophylaxis with third-generation cephalosporins soon followed this recognition (11). Meanwhile, following the use of medicinal leeches, the digestive tract symbiont has been implicated in cellulitis and loss of replanted tissue (7, 15), as well as septicemia and meningitis (6, 19). The incidence of such infections can be reduced by preemptive antibiotic treatment.
European medicinal leeches in the genus Hirudo are not the only leeches being used for the relief of venous congestion. In Asia, Hirudinaria manillensis is more commonly encountered, whereas Aliolimnatis michaelseni is the leech of choice in South Africa (2, 27). The dominant gut symbiont of leeches was reported by Mackay et al. to be Aeromonas caviae, not A. veronii biovar sobria (16), but considering the difficulty of accurately identifying environmental Aeromonas isolates to the species level with biochemical tests, this result should be considered preliminary. Here, we investigated the aeromonad gut flora of the common North American medicinal leech, Macrobdella decora, a species often encountered in freshwater environments by swimmers and anglers across North America (14).
Isolates.
Leeches (M. decora) were collected from four localities using the traditional method of wading into water bare legged and retrieving them either with a dip net as they approached or after they attached to bare skin but prior to the onset of blood feeding. These localities were Broadwing Lake, Ontario, Canada (45°35′50"N, 78°31′42"W); Douglas Lake, MI (45°34′49" N, 84°40′12"W); a pond in Storrs, CT (41°49′2.80"N, 72°15′32.12"W); and Horseshoe Pond in Chester, VT (43°14′19.27"N, 72 34′22.39"W). European medicinal leeches were obtained from LeechesUSA (Westbury, NY). Leeches were rinsed with distilled water and washed with bleach. A longitudinal incision was made in the ventral surface, and intraluminal fluid of the crop was collected and serially diluted in saline (0.85% NaCl). Intraluminal fluid dilutions were streaked onto sheep blood agar (Becton-Dickinson, Sparks, MD) using sterile swabs and incubated aerobically at 30°C. Multiple colonies were subcultured on blood agar for isolation.
Amplification, sequencing, and phylogenetic analyses.
DNA was isolated from luminal contents of the crop ceca of leeches using a DNeasy tissue kit (QIAGEN Inc., Valencia, CA). A portion of the 16S rRNA gene was amplified using universal primers AGAGTTTGATCCTGGCTCAG and ATTACCGCGGCTGCTGGC and a cycling program consisting of 94°C for 4 min and then 35 cycles of 94°C for 15 s, 57°C for 15 s, and 72°C for 30 s, followed by 72°C for 7 min. A portion of the gyrB locus was amplified using specific primers TGTTGCTGACCATTCGTCGTAAC and TTGGCATCGCTCGGGTTTTC and a cycling program consisting of 94°C for 4 min and then 35 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 30 s, followed by 72°C for 7 min. For all amplifications we used Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, NJ), 0.5 μl of each primer at a concentration of 10 μM, 1 μl DNA template, and 23 μl RNase-free H2O. Products were sequenced in both directions. Each sequencing reaction mixture contained 1 μl BigDye (Applied Biosystems, Perkin-Elmer Corporation), 1 μl of primer at a concentration of 1 μM (a single primer was used for each direction), and 3 μl of DNA template, and the reaction was performed by using 40 cycles consisting of 96°C for 15 s, 50°C for 30 s, and 60°C for 4 min. Sequences were purified by ethanol precipitation and electrophoresed in an ABI Prism 3730 sequencer (Applied Biosystems). Sequences of complementary strands were edited and reconciled using CodonCode Aligner (CodonCode, Dedham, MA). In addition to sequences obtained from direct sequencing of DNA from freshly collected specimens, gyrB data were obtained from GenBank for taxa as described in previous analyses (25, 31) and for three outgroup taxa. The gyrB sequences required 12-nucleotide insertion/deletion sites. These sites corresponded to one amino acid insertion for Aeromonas simiae, another amino acid insertion shared by nine Aeromonas species, and two amino acid indels for only the outgroup taxa.
Parsimony analyses were conducted with PAUP* (26). ModelTest (21) suggested a GTR+I+Γ nucleotide substitution model for gyrB. Maximum likelihood analyses were conducted with the separate data sets using PhyML (10). The Bayesian method was employed with MrBayes (12) for 1,000,000 generations (the last 500,000 generations of which were used for clade credibility values).
Amplification of the 16S rRNA gene and gyrB generated sequenced fragments up to 603 and 717 bp long, respectively. PCR amplicons obtained from M. decora were identical to each other at both loci regardless of the geographic origin. The 16S rRNA genes of the isolates were identical to 16S rRNA gene of Aeromonas jandaei strain ATCC 49568 (GenBank accession no. X74678). The gyrB sequence obtained from the crop contents of M. decora corroborated this identification by most closely matching the A. jandaei sequence (GenBank accession no. AJ868391). In contrast, the gyrB sequence obtained for isolates from European medicinal leeches most closely matched that obtained for A. veronii strain MTCC 3249 substrain SH (GenBank accession no. AY130993; originally described as Aeromonas culicicola before more recent phylogenetic work [22]).
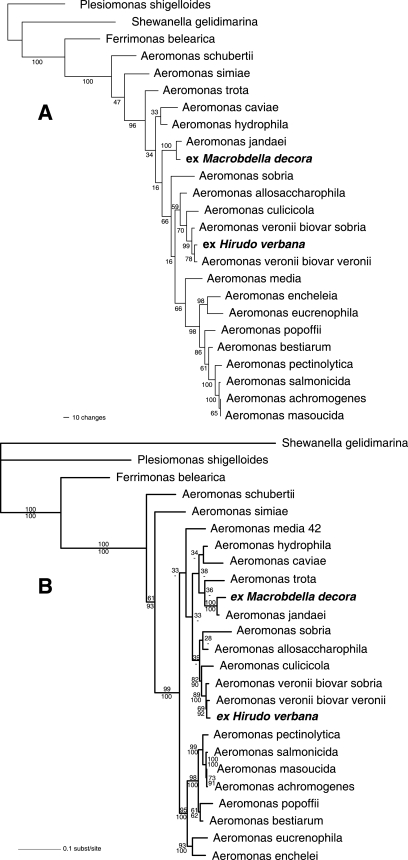
Parsimony analysis of gyrB yielded one tree with a length of 1,136 steps and a retention index of 0.591 (Fig. 1A). Maximum likelihood analysis also generated a single tree with a log(L) value of −6397.199 for gyrB (Fig. 1B). Notably, the isolates from M. decora grouped together with A. jandaei with high support values in all analyses. As expected, the isolates from the European medicinal leech were identified as A. veronii.
FIG. 1.
Results of phylogenetic analyses. (A) Tree with bootstrap support values resulting from parsimony analysis. (B) Tree with the highest likelihood, including both bootstrap support values (upper numbers at the nodes) and Bayesian clade credibility values (lower numbers at the nodes).
Phenotypic tests.
Bacteria were successfully cultured from two M. decora individuals, and five isolates were characterized further using biochemical tests as described previously (1, 8). Only colonies resembling Aeromonas colonies were observed after 48 h. The sensitivities of three isolates to antibiotics were evaluated using Sensi-Discs (Becton-Dickinson and Company, Sparks, MD). All isolates were sensitive to cefotaxime (30 μg), cefuroxime (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), nalidixic acid (30 μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg). One of the isolates was resistant to cephalothin (30 μg), and another isolate exhibited intermediate resistance, indicating that cephalothins are not an appropriate choice for antibiotic therapy. The results of biochemical tests for five isolates were identical. Consistent results (positive or negative for >90% of colonies) favored identification as either Aeromonas hydrophila, A. jandaei, or A. caviae (Table 1). Overall, the inability to utilize citrate was shared only with Aeromonas eucrenophila. Overall, the failure to ferment sucrose was shared only with A. jandaei. Together, our data are consistent with the hypothesis that A. jandaei is the dominant culturable symbiont of the North American medicinal leech.
TABLE 1.
Biochemical tests distinguishing M. decora crop isolates from selected Aeromonas spp.
| Test | M. decora isolates | Aeromonas hydrophilaa | Aeromonas trotaa | Aeromonas eucrenophilaa | Aeromonas jandaeia | Aeromonas veronii bv. sobriaa | Aeromonas caviaea |
|---|---|---|---|---|---|---|---|
| Voges-Proskauer | −b | + | − | − | d | d | − |
| Lysine decarboxylase | − | + | + | − | + | + | − |
| Orthinine decarboxlyase | − | − | − | − | − | − | − |
| Arginine dehydrolase | + | + | + | d | + | + | + |
| Esculin hydrolysis | + | + | − | d | − | − | d |
| Gas from d-glucose | + | + | d | d | + | + | − |
| l-Arabanose fermentation | + | d | − | d | − | − | + |
| Sucrose fermentation | − | + | d | d | − | + | + |
| d-Mannitol fermentation | + | + | d | + | + | + | + |
| Citrate utilization | − | d | + | − | d | d | d |
| Hemolysis | + | + | d | d | + | + | d |
Biochemical tests for Aeromonas species (1).
−, <10% positive; +, >90% positive; d, 11 to 89% positive.
It seems remarkable that two ecologically similar leech species contain distinct Aeromonas species as the dominant culturable bacterial symbionts in the gut lumen, with consistency across the geographic ranges of the leeches. Both A. jandaei and A. veronii are ubiquitous and global in terms of their known distributions (5, 28) and even have been found coinfecting the same wound (13). There seems to be little reason to contemplate that there is a geographic or ecological barrier excluding either species of symbiont from either species of leech. Like the European leech symbiont, A. jandaei has been implicated in several pathological cases, usually involving the exposure of wounds to a freshwater environment (13, 23, 28), although its involvement is less common than the involvement of other aeromonads.
The phylogenetic results obtained here are in agreement (where support values are strong) with those obtained previously for this group of bacteria (25, 31). Whereas genetic and phylogenetic characterization of the gut symbiont of M. decora was clear, like the previous characterization of Aeromonas from Hirudo species, the biochemical results did not agree unambiguously with the previously published biochemical results for any single Aeromonas species. Such difficulties in identifying environmental Aeromonas strains have been reported previously (18) and may reflect the source of most of the characterized isolates or perhaps the possibility that a different subset of strains inhabits leeches.
M. decora, although not yet used clinically, produces a useful platelet aggregation inhibitor, decorsin (24), and it belongs to an evolutionary lineage that is distinct from the Old World medicinal leeches (3). Moreover, it is a widespread, commonly encountered species in freshwater environments, where its typical hosts, besides the occasional human, are frogs and fish. Different species of Aeromonas have different susceptibilities to available antimicrobial agents (17, 28). Notably, A. jandaei may be the most resistant species of Aeromonas both in terms of the extent of multiple-antibiotic resistance and in terms of the frequency with which such resistance is found in clinical isolates (20). The spectrum of Aeromonas species dominating the gut lumen of various leeches commonly encountered around the world deserves closer scrutiny, particularly for the leech species that are used locally for the relief of venous congestion or simple hematomas.
Acknowledgments
This research was supported by grants from NSF (grant DEB 0119329) and NIH (grant NIGMS 5R01GM062351) to M.E.S. and by grant MCB 0448052 from NSF to J.G.
Footnotes
Published ahead of print on 17 November 2006.
REFERENCES
- 1.Abbott, S. L., W. K. Cheung, and J. M. Janda. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41:2348-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bickel, K. D., W. C. Lineaweaver, S. Follansbee, R. Feibel, R. Jackson, and H. J. Buncke. 1994. Intestinal flora of the medicinal leech Hirudinaria manillensis. J. Reconstr. Microsurg. 10:83-85. [DOI] [PubMed] [Google Scholar]
- 3.Borda, E., and M. E. Siddall. 2004. Arhynchobdellida (Annelida: Oligochaeta: Hirudinida): phylogenetic relationships and evolution. Mol. Phylog. Evol. 30:213-225. [DOI] [PubMed] [Google Scholar]
- 4.Büsing, K. H. 1951. Pseudomonas hirudinis, ein bakterieller Darmsymbiont des Blutegels (Hirudo officinalis). Zentrbl. Bakteriol. 157:478-484. [PubMed] [Google Scholar]
- 5.Carnahan, A. M., and S. W. Joseph. 1991. Aeromonas update: new species and global distribution. Experientia 47:402-403. [PubMed] [Google Scholar]
- 6.Evans, J., P. J. Lunnis, P. N. Gaunt, and D. J. Hanley. 1990. A case of septicaemia due to Aeromonas hydrophila. Br. J. Plast. Surg. 43:371-372. [DOI] [PubMed] [Google Scholar]
- 7.Fenollar, F., P. E. Fournier, and R. Legre. 1999. Unusual case of Aeromonas sobria cellulitis associated with the use of leeches. Eur. J. Clin. Microbiol. Infect. Dis. 18:72-73. [DOI] [PubMed] [Google Scholar]
- 8.Graf, J. 1999. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect. Immun. 67:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graf, J., K. Yoshitomo, and R. V. M. Rio. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 10.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 11.Hermansdorfer, J., W. Lineaweaver, S. Follansbee, F. A. Valauri, and H. J. Buncke. 1988. Antibiotic sensitivities of Aeromonas hydrophila cultured from medicinal leeches. Br. J. Plast. Surg. 41:649-651. [DOI] [PubMed] [Google Scholar]
- 12.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 13.Joseph, S. W., A. M. Carnahan, P. R. Brayton, G. R. Fanning, R. Almazan, C. Drabick, E. W. Trudo, Jr., and R. R. Colwell. 1991. Aeromonas jandaei and Aeromonas veronii dual infection of a human wound following aquatic exposure. J. Clin. Microbiol. 29:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm, D. J. 1982. The leeches (Annelida: Hirudinea) of North America. Aquatic Biology Section, Environmental Monitoring and Support Laboratory, Office of Research and Development, U.S. Environmental Protection Agency. Cincinnati, OH.
- 15.Lineaweaver, W. C., M. K. Hill, G. M. Buncke, S. Follansbee, H. J. Buncke, R. K. Wong, E. K. Manders, J. C. Grotting, J. Anthony, and S. J. Mathes. 1992. Aeromonas hydrophila infections following use of medicinal leeches in replantation and flap surgery. Ann. Plast. Surg. 29:238-244. [DOI] [PubMed] [Google Scholar]
- 16.Mackay, D. R., E. K. Manders, G. C. Saggers, D. R. Banducci, J. Prinsloo, and K. Klugman. 1999. Aeromonas species isolated from medicinal leeches. Ann. Plast. Surg. 42:275-279. [DOI] [PubMed] [Google Scholar]
- 17.Nonomura, H., N. Kato, Y. Ohno, M. Itokazu, T. Matsunaga, and K. Watanabe. 1996. Indigenous bacterial flora of medicinal leeches and their susceptibilities to 15 antimicrobial agents. J. Med. Microbiol. 45:490-493. [DOI] [PubMed] [Google Scholar]
- 18.Ormen, O., P. E. Granum, J. Lassen, and M. J. Figueras. 2005. Lack of agreement between biochemical and genetic identification of Aeromonas spp. Acta Pathol. Microbiol. Immunol. Scand. 113:203-207. [DOI] [PubMed] [Google Scholar]
- 19.Ouderkirk, J. P., D. Bekhor, G. S. Turett, and R. Murali. 2004. Aeromonas meningitis complicating medicinal leech therapy. Clin. Infect. Dis. 38:e36-e37. [DOI] [PubMed] [Google Scholar]
- 20.Overman, T. L., and J. M. Janda. 1999. Antimicrobial susceptibility patterns of Aeromonas jandaei, A. schubertii, A. trota, and A. veronii biotype veronii. J. Clin. Microbiol. 37:706-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 22.Saavedra, M. J., M. J. Figueras, and A. J. Martínez-Murcia. 2006. Updated phylogeny of the genus Aeromonas. Int. J. Syst. Evol. Microbiol. 56:2481-2487. [DOI] [PubMed] [Google Scholar]
- 23.Sarma, P. S. 2002. Aeromonas jandaei cellulitis and bacteremia in a man with diabetes. Am. J. Med. 112:325. [DOI] [PubMed] [Google Scholar]
- 24.Seymour, J. L., W. J. Henzel, B. Nevins, J. T. Stults, and R. A. Lazarus. 1990. Decorsin. A potent glycoprotein IIb-IIIa antagonist and platelet aggregation inhibitor from the leech Macrobdella decora. J. Biol. Chem. 265:10143-10147. [PubMed] [Google Scholar]
- 25.Soler, L., M. A. Yanez, M. R. Chacon, M. G. Aguilera-Arreola, V. Catalan, M. J. Figueras, and A. J. Martinez-Murcia. 2004. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int. J. Syst. Evol. Microbiol. 54:1511-1519. [DOI] [PubMed] [Google Scholar]
- 26.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 27.Van Wingerden, J. J., and J. H. Oosthuizen. 1997. Use of the local leech Hirudo michaelseni in reconstructive plastic and hand surgery. S. Afr. J. Surg. 35:29-31. [PubMed] [Google Scholar]
- 28.Vila, J., J. Ruiz, F. Gallardo, M. Vargas, L. Soler, M. J. Figueras, and J. Gascon. 2003. Aeromonas spp. and traveler's diarrhea: clinical features and antimicrobial resistance. Emerg. Infect. Dis. 9:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitlock, M. R., P. M. O'Hare, R. Sanders, and N. C. Morrow. 1983. The medicinal leech and its use in plastic surgery: a possible cause for infection. Br. J. Plast. Surg. 36:240-244. [DOI] [PubMed] [Google Scholar]
- 30.Worthen, P. L., C. J. Gode, and J. Graf. 2006. Culture-independent characterization of the digestive tract microbiota of the medicinal leech, a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanez, M. A., V. Catalan, D. Apraiz, M. J. Figueras, and A. J. Martinez-Murcia. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol Microbiol. 53:875-883. [DOI] [PubMed] [Google Scholar]