Abstract
Cyanide is a serious environmental pollutant and a biocontrol metabolite in plant growth-promoting Pseudomonas species. Here we report on the presence of multiple sulfurtransferases in the cyanogenic bacterium Pseudomonas aeruginosa PAO1 and investigate in detail RhdA, a thiosulfate:cyanide sulfurtransferase (rhodanese) which converts cyanide to less toxic thiocyanate. RhdA is a cytoplasmic enzyme acting as the principal rhodanese in P. aeruginosa. The rhdA gene forms a transcriptional unit with the PA4955 and psd genes and is controlled by two promoters located upstream of PA4955 and rhdA. Both promoters direct constitutive RhdA expression and show similar patterns of activity, involving moderate down-regulation at the stationary phase or in the presence of exogenous cyanide. We previously observed that RhdA overproduction protects Escherichia coli against cyanide toxicity, and here we show that physiological RhdA levels contribute to P. aeruginosa survival under cyanogenic conditions. The growth of a ΔrhdA mutant is impaired under cyanogenic conditions and fully restored upon complementation with rhdA. Wild-type P. aeruginosa outcompetes the ΔrhdA mutant in cyanogenic coculture assays. Hence, RhdA could be regarded as an effector of P. aeruginosa intrinsic resistance to cyanide, insofar as it provides the bacterium with a defense mechanism against endogenous cyanide toxicity, in addition to cyanide-resistant respiration.
Cyanide is extremely toxic to several forms of life because of its inhibitory activity on a variety of key enzymes. Cyanide toxicity results from its reaction with keto compounds and Schiff base intermediates to give cyanohydrins and stable nitrile derivatives, respectively, and from chelation of di- and trivalent metal ions in the prosthetic groups of several metalloenzymes. A case in point is cyanide binding to the ferric heme iron of cytochrome c oxidase, which inhibits aerobic respiration (36).
An apparent paradox is that several organisms synthesize, degrade, or metabolize cyanide compounds. Cyanogenesis has been documented to occur in a variety of microorganisms (25), among which the genus Pseudomonas provides a paradigmatic example (7). Many Pseudomonas species release in culture up to 300 μM cyanide generated by oxidative decarboxylation of glycine (3). Pseudomonas cyanogenesis has the typical features of secondary metabolism, occurring at the end of exponential growth and being influenced by oxygen, iron, and phosphate concentrations (6). It has been shown that cyanide released by Pseudomonas fluorescens suppresses the growth of microorganisms (e.g., phytopathogenic bacteria and fungi) sharing the same ecological niche (e.g., the rhizosphere), thereby acting as a biocontrol metabolite (41). Hence, cyanide production would increase the biological fitness by providing cyanogenic species with a selective advantage over competitors (22, 41). Cyanide biosynthesis has also been associated with Pseudomonas aeruginosa pathogenesis in the Caenorhabditis elegans infection model (16, 20).
Microorganisms adopt different strategies to face cyanide toxicity. In P. aeruginosa, a cyanide-insensitive oxidase (CIO) is induced at the stationary phase, enabling aerobic respiration even in the presence of 1 mM cyanide (14, 15). Exogenous cyanide is a potent inducer of cioAB genes during exponential growth, while endogenous cyanide is not the physiological stimulus for CIO expression (14). In addition, P. aeruginosa can rely upon fermentation for energy generation in the presence of cyanide (44), while P. fluorescens is able to couple scavenging of cyanide with its utilization as a nitrogen source (19). Besides these effective alternative metabolic pathways, it is plausible that enzymatic conversion of cyanide into a less toxic compound(s) may aid in protection (29, 44).
Rhodaneses (thiosulfate:cyanide sulfurtransferases; EC 2.8.1.1) catalyze the transfer of a sulfane sulfur atom from a suitable donor (e.g., thiosulfate) to cyanide, yielding the less toxic thiocyanate (43). Rhodaneses have been purified from a number of sources and found to consist of two structurally similar domains, each one characterized by a RHOD module containing either a catalytic Cys or an inactive Asp residue (4). The function(s) of rhodaneses is not conclusively defined, though these enzymes are deemed to act as effectors of cyanide detoxification in animals (1, 2, 39). Given the high conservation of rhodaneses in the three evolutionary lineages (4), it seems likely that these enzymes contribute to cyanide detoxification also in bacteria. Rhodaneses have been identified in a variety of bacterial species, including Escherichia coli (30), Azotobacter vinelandii (13), and P. aeruginosa (11, 32). The high sequence homology between A. vinelandii and P. aeruginosa RhdA rhodaneses predicts that these two enzymes belong to the same subfamily (11), sharing structural features with mitochondrial bovine rhodanese (4, 9), whose role in cyanide detoxification has been established (39).
We previously showed that heterologous expression of P. aeruginosa RhdA increases cyanide tolerance in E. coli (12), and here we report on the role of RhdA in cyanide detoxification by P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids are listed in Table 1. Bacteria were routinely grown at 37°C with shaking in LB medium (33). Cyanogenic cultures were performed at 37°C in cyanogenic synthetic medium (C-MM) under microaerophilic conditions in static rubber-stoppered flasks (6). Noncyanogenic cultures were performed at 37°C with shaking in noncyanogenic synthetic medium (NC-MM) (40 mM glutamic acid, 10 mM methionine, 2 mM MgSO4, 50 mM NaH2PO4, 50 mM K2HPO4, pH 7.0), which was obtained by varying the concentrations of glycine, iron, and phosphate in C-MM to abrogate cyanogenesis (7, 8). Unless otherwise stated, antibiotics were added at the following concentrations (μg ml−1): carbenicillin (Cb), 500; gentamicin (Gm), 200; nalidixic acid (Nal), 250; rifampin (Rif), 200; and tetracycline (Tc), 100.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristics | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 (ATCC 15692) | Prototroph | American Type Culture Collection |
| PAO1 ΔrhdA | rhdA null mutant | This study |
| PAO1-Nal | Nalr | This study |
| PAO1-Rif | Rifr | This study |
| PAO1-Rif ΔrhdA | PAO1 ΔrhdA, Rifr | This study |
| Plasmids | ||
| pEX18Tc | PMB1 replicon, oriT sacB lacZα, Mob+ Tcr, allelic exchange vector | 23 |
| pFLP2 | AprsacB; broad-host-range recombination system | 23 |
| pPS858 | Apr; source of Gmr-GFP cassette | 23 |
| pEXΔrhdA | pEX18Tc derivative carrying the Gmr-GFP cassette between rhdA flanking regions | This study |
| pUCP18 | E. coli-Pseudomonas shuttle vector derived from pUC18/19; ColE1, pRO1600 replicon, lacZ bla, Apr Cbr | 34 |
| pUCPrhdA | 1,002-bp PCR product, encompassing the rhdA coding sequence and its putative promoter, ligated to the BamHI-HindIII sites of pUCP18 | This study |
| pUCP4955-rhdA | 2,645-bp PCR product encompassing both PA4955 and rhdA coding sequences with their putative promoters, ligated to the BamHI-HindIII sites of pUCP18 | This study |
| pMP220 | Broad-host-range, low-copy-number promoter-probe vector; IncP replicon, lacZ Tcr Tra− | 38 |
| pMP4955 | 322-bp PCR product encompassing the putative PA4955 promoter, ligated to the BglII-PstI sites of pMP220 | This study |
| pMPrhdA | 305-bp PCR product encompassing the putative rhdA promoter, ligated to the BglII-PstI sites of pMP220 | This study |
| pMP4955-rhdA | 1,948-bp PCR product encompassing both PA4955 and rhdA putative promoters with the PA4955 intervening ORF, ligated to the BglII-PstI sites of pMP220 | This study |
| pMP220R | Derivative of pMP220 with polylinker in inverted orientation | This study |
| pMPR4955 | Same insert of pMP4955 cloned in pMP220R | This study |
| pMPRrhdA | Same insert of pMPrhdA cloned in pMP220R | This study |
| pMPR4955-rhdA | Same insert of pMP4955-rhdA cloned in pMP220R | This study |
The susceptibility of P. aeruginosa (ca. 107 cells ml−1) to 300 μM KCN was determined after 40 min of incubation in LB at 37°C. Viability was calculated as the CFU ratio between cyanide-treated and untreated cultures.
For growth comparison under noncyanogenic and cyanogenic conditions, overnight cultures in NC-MM were diluted in NC-MM or in C-MM to an optical density at 600 nm (OD600) of ca. 0.02, followed by periodic OD600 measurements. In coculture assays, cells from overnight cultures in LB were 100-fold diluted in the same medium and grown to and OD600 between 0.90 and 1.10. P. aeruginosa PAO1-Nal and test strains (PAO1-Rif ΔrhdA carrying or not carrying the complementing plasmid pUCP4955-rhdA) were washed and suspended in saline at an OD600 of 1.0 ± 0.03, mixed in different volumetric ratios (1/1, 1/10, and 10/1), and 250-fold diluted in NC-MM or C-MM. In long-term coculture experiments, P. aeruginosa PAO1-Nal and PAO1-Rif ΔrhdA mixtures (1:1 ratio) were serially subcultured in NC-MM and C-MM for nine passages of 12 h each (250-fold dilution at each passage), for a total of 108 h of growth. Bovine methemoglobin (Sigma-Aldrich) was used at 75 μM, equivalent to 300 μM ferric heme. Viable counts were performed at given times on LB agar plates supplemented with either Nal or Rif. The competitive index (CI) was calculated as log(CFU of mutant or complemented mutant/CFU of wild type).
Construction of mutant strains.
Site-specific excision of the rhdA coding sequence was performed using the previously described sacB-based strategy (23). Two regions of ca. 1,000 bp flanking the rhdA gene were generated by PCR with primers rhdA mutup forward (5′-GGGAATTCAGTGGGTCATCCAGGGCT-3′ [EcoRI site underlined]) and reverse (5′-GCCGGATCCGGAGAAAACGGACATGCG-3′ [BamHI site underlined]) or rhdA mutdown forward (5′-GGGGGATCCTGTAGAGGTTTGAGGAATG-3′ [BamHI site underlined]) and reverse (5′-CGGCAAGCTTTCCGAACTCTAGCAGCAT-3′ [HindIII site underlined]). Upstream and downstream fragments were digested with appropriate restriction enzymes and directionally cloned into the pEX18Tc suicide vector (23). The Gmr-green fluorescent protein (GFP) cassette from pPS858 (23) was inserted at the BamHI site between the two cloned fragments, and the resulting pEXΔrhdA construct was conjugally transferred into P. aeruginosa PAO1. Resolution of merodiploids and excision of the Gmr-GFP cassette were achieved as described previously (23). Deletion events were checked by colony PCR using the mutup forward and mutdown reverse primers.
Spontaneous Nalr or Rifr mutants were selected on LB agar plates supplemented with 2,000 μg ml−1 Nal or 500 μg ml−1 Rif, respectively.
Cell fractionation, immunoblot analysis, and biochemical assays.
Protein fractions were obtained by use of a minor modification of a previously described method (10). Briefly, P. aeruginosa cells from LB cultures were treated for 15 min at 25°C with 200 mM MgCl2 in 10 mM Tris-HCl, pH 8.4. The periplasmic fraction was recovered from the supernatant after centrifugation. The pellet was suspended in 10 mM Tris-HCl (pH 8.4), disrupted by sonication, and ultracentrifuged at 50,000 × g for 60 min to recover the soluble cytoplasmic fraction. Isocitrate dehydrogenase (21) and β-lactamase constitutively expressed by the pUCP18 bla gene (42) were used as markers for purity of cytoplasmic and periplasmic fractions, respectively.
The protein concentration was measured according to the method of Bradford (5). Protein fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining. Immunoblotting was performed with a mouse anti-RhdA antiserum (1:500) (12) or a rabbit anti-β-lactamase antiserum (Chemicon), using alkaline phosphatase-conjugate anti-mouse or anti-rabbit immunoglobulin G secondary antibodies (1:7,500) as described previously (33). Recombinant RhdA was prepared as previously reported (11).
Resting cell suspensions (ca. 1011 cells ml−1) were prepared in 10 mM Tris-HCl, 150 mM NaCl (pH 7.3), using late-exponential-phase cultures in LB medium.
RhdA activity was determined by a colorimetric method (37), with or without sodium thiosulfate (68 mM) as a sulfur donor and KCN (68 mM for soluble enzyme and 0 to 300 μM for cell-associated enzyme) as the acceptor. Enzyme activity was assayed in 10 mM Tris-HCl (pH 7.3) at 25°C and expressed as nmol thiocyanate min−1 mg−1 protein (11).
LacZ (β-galactosidase) activity was determined as described by Miller (28).
Cyanogenesis was determined by a qualitative method (18). Reactive paper strips were aseptically placed in the gaseous phase of flasks containing P. aeruginosa cultures, and HCN production was indicated by a white-to-blue color change of the strips.
Plasmid constructions and genetic techniques.
Recombinant DNA procedures have been described elsewhere (33). Different DNA fragments in the rhdA genomic region of P. aeruginosa PAO1 were generated by PCR and sequenced on both strands. The 1,002-bp DNA fragment encompassing the rhdA gene with its putative promoter (from position −182 to position +820 relative to the rhdA start codon) was generated with primers P4955FW (5′-GCGAGATCTCGGCGGCCAGCCGTT-3′ [BglII site underlined]) and rhdARV (5′-GGGCAAGCTTCCTCAAACCTCTACAGGGG-3′ [HindIII site underlined]) and then cloned in pUCP18 (34) to yield pUCPrhdA. The 2,645-bp DNA fragment encompassing both the PA4955 and rhdA genes with their putative promoters (from position −239 upstream of the PA4955 start codon to position +4 downstream of the rhdA stop codon) was generated with primers PmotFW (5′-GCGAGATCTCGAAGGGCTGGATGATG-3′ [BglII site underlined]) and rhdARV and then cloned in pUCP18 to yield pUCP4955-rhdA (Table 1).
Transcriptional fusions were generated by cloning putative promoter fragments at the PstI and BglII sites of pMP220 (38) (Table 1). Plasmid pMP4955, carrying the predicted PA4955 promoter region from position −239 to +83 relative to the PA4955 start codon, was obtained with primers PmotFW and P4955RV (5′-GCGCTGCAGGCCGGCATAGTGG-3′ [PstI site underlined]). Plasmid pMPrhdA, carrying the predicted rhdA promoter region from position −182 to +123 relative to the rhdA start codon, was obtained with primers P4955FW and PrhdARV (5′-GCGCTGCAGATATGCCCTTCGGCGTAG-3′ [PstI site underlined]). Plasmid pMP4955-rhdA, encompassing both the PA4955 and rhdA promoter regions from position −239 upstream of PA4955 to position +123 downstream of rhdA (relative to the start codons), was obtained with primers PmotFW and PrhdARV. The same fragments were also cloned in reverse orientation in pMP220R. Plasmid pMP220 was used as a control.
RT-PCR.
Total RNA was isolated from late-exponential-phase P. aeruginosa PAO1 cultures in LB by using the hot-phenol extraction method (26). Residual DNA was digested with 30 U of RNase-free DNase I (Roche), except for the positive PCR control. Five micrograms of total RNA was subjected to reverse transcription (RT) reactions using 2 pmol of primer rhdARV and the Superscript II kit (Life Technologies), except for the negative RT control. PCR was performed on 2 μl of each RT reaction mixture, using primer pairs rhdAFW (5′-GGGGGGATCCGCATGTCCGTTTTCTCCGA-3′) and rhdARV to amplify the rhdA transcript and P4955FW and PrhdARV to amplify the PA4955-rhdA intergenic transcript.
Computational DNA and protein analysis.
P. aeruginosa PAO1 DNA sequences, genome organization, and protein functional inference were retrieved from the Pseudomonas Genome Project (http://v2.pseudomonas.com). The presence of putative prokaryotic promoter elements was predicted by the Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html) and BPROM (Softberry Inc.) software. Transcriptional terminators were predicted with the TransTerm software (http://www.cbcb.umd.edu/software/TransTerm). Proteins containing RHOD module(s) and domain organization were retrieved using the SMART (simple modular architecture research tool) protein domain database (http://smart.embl-heidelberg.de) and screened by combined searches for rhodanese in the Pfam homology superfamily database (PF00581; http://sanger.ac.uk/cgi-bin/Pfam) and in the Cluster of Orthologous Group database (COG0607, COG1054, COG2603, and COG2897; http://www.ncbi.nlm.nih.gov/COG). Protein sequences were aligned with CLUSTALW (40). Signal peptide searches were performed at http://www.cbs.dtu.dk/services/SignalP.
RESULTS
Identification of RhdA as the most likely rhodanese candidate in P. aeruginosa PAO1.
A simple modular architecture search for the RHOD module in nonredundant Swiss-Prot and Ensembl databases retrieved 738 predicted proteins, 372 of which were encoded by eubacterial genomes. Multiple proteins containing the RHOD module were frequently retrieved from the same genome. Ten open reading frames (ORFs) encoding putative rhodanese-related sulfurtransferases were identified in P. aeruginosa PAO1 (Fig. 1A), all of which were predicted to encode single or multiple RHOD modules (Fig. 1B). The typical double-domain rhodanese structure was predicted only for PA1292 and PA4956 (RhdA), both belonging to COG2897 (Fig. 1A). The amino acid motif surrounding the catalytic Cys residue of PA1292 matches the 3-mercaptopyruvate:cyanide sulfurtransferases signature, whereas the corresponding motif in RhdA is typical of thiosulfate:cyanide sulfurtransferases or rhodaneses sensu stricto (4, 9) (Fig. 1B). Moreover, the three-dimensional model of RhdA reveals a striking similarity to the crystal structures of bovine and A. vinelandii rhodanese (R. Cipollone, unpublished data). Since the rhdA gene product is the most likely P. aeruginosa rhodanese, its contribution to cyanide detoxification was further investigated.
FIG. 1.
Prediction of rhodanese-related proteins encoded by the P. aeruginosa PAO1 genome. (A) Gene products were identified for the presence of RHOD module(s), subdivided according to their COG, and designated according to their PA number (www.pseudomonas.com). Protein organization is reported to scale. Black boxes are potentially active RHOD modules, while white circles denote Cys→Asp substitutions. The ThiF family domain in MoeB, the THUMP domain in ThiI, and the acylphosphatase-like domain together with the antifungal protein (AGAFP) fold in PA0858 are shown with hatched, slashed, white, and gray boxes, respectively. Straight lines denote uncharacterized amino acid sequences. (B) Alignment of partial RHOD modules from P. aeruginosa PAO1 rhodanese-related proteins. Modules are defined by PA number followed by module number. The conserved, potentially catalytic Cys residue in the active domain (upper alignment) and the corresponding Asp residue in the inactive domain (lower alignment) are boxed. Identical residues and conserved and semiconserved substitutions are indicated by asterisks, colons, and periods, respectively. Residues forming the canonical six-amino-acid active-site loop in thiosulfate:cyanide sulfurtransferases (PA4956_2) and 3-mercaptopyruvate:cyanide sulfurtransferases (PA1292_2) are underlined. Amino acid sequences were retrieved from the Swiss-Prot/TrEMBL database and aligned with the program CLUSTALW (40).
RhdA is the main cytoplasmic rhodanese in P. aeruginosa PAO1.
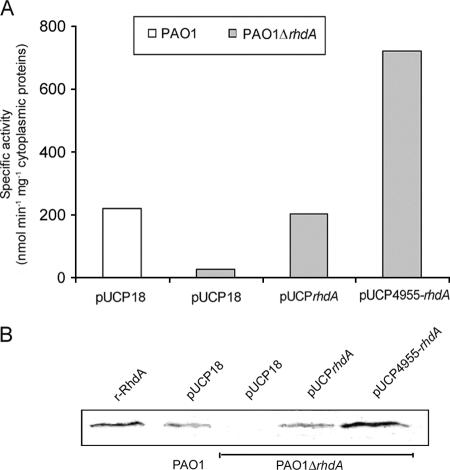
To assess the actual contribution of RhdA to the total rhodanese activity in P. aeruginosa PAO1, a ΔrhdA mutant was generated and rhodanese activities in subcellular fractions of wild-type PAO1 and PAO1 ΔrhdA were compared. The purity of fractions was preliminarily tested; isocitrate dehydrogenase activity in periplasmic preparations was 0.1% of cytoplasmic activity, while β-lactamase was immunodetectable only in periplasmic fractions (data not shown). Rhodanese activity was differently distributed between the periplasm and the cytoplasm of wild-type P. aeruginosa PAO1, attaining 2.8 and 221.3 nmol min−1 mg−1 of whole-cell protein in late exponential phase, respectively. The same rhodanese activity was detected in the periplasmic fraction of both wild-type and ΔrhdA strains (ca. 1,000 nmol min−1 mg−1 of periplasmic protein), while remarkable differences were observed at the cytoplasmic level, with PAO1ΔrhdA showing 12% of wild-type activity (26 versus 219 nmol min−1 mg−1 of cytoplasmic protein, respectively) (Fig. 2A). Accordingly, immunoblot analysis did not detect RhdA in the cytoplasm of PAO1 ΔrhdA (Fig. 2B) or in the periplasm of both parent and mutant strains (data not shown). Complementation of PAO1 ΔrhdA with either rhdA or both rhdA and the preceding PA4955 ORF restored or increased the rhodanese activity relative to the wild-type level, respectively (Fig. 2A). A similar profile of complementation was observed when cytoplasmic RhdA levels were estimated by immunoblotting (Fig. 2B).
FIG. 2.
Effect of the rhdA mutation on rhodanese activity in P. aeruginosa PAO1. (A) Complementation analysis of P. aeruginosa PAO1 ΔrhdA with pUCPrhdA and pUCP4955-rhdA. The pUCP18 vector was used as a control. Bacteria were grown in LB medium at 37°C for 8 h before cell fractionation (see Materials and Methods). Specific rhodanese activity is reported for the cytoplasmic protein fraction. Results are the means from three independent experiments. Standard deviations are <5% of given values. (B) Immunoblot analysis of RhdA levels in cytoplasmic preparations (35 μg of total protein) from P. aeruginosa PAO1, PAO1 ΔrhdA, and the ΔrhdA mutant complemented with either pUCPrhdA or pUCP4955-rhdA. Purified recombinant RhdA (r-RhdA) (150 ng) was used as a control. Blots were hybridized with a mouse anti-RhdA polyclonal antiserum.
Identification of rhdA promoter regions and analysis of RhdA expression in P. aeruginosa PAO1.
Inspection of the rhdA region in the P. aeruginosa genome revealed the presence of four contiguous ORFs (PA4954 to PA4957), three of which (PA4955 to PA4957) were in the same orientation (Fig. 3A). The rhdA gene is preceded by an uncharacterized ORF (PA4955) and is followed by the psd gene (PA4957), which is predicted to encode a phosphatidylserine decarboxylase. Remarkably, a similar organization of the rhdA genomic region is conserved in several rRNA group I Pseudomonas species (http://v2.pseudomonas.com).
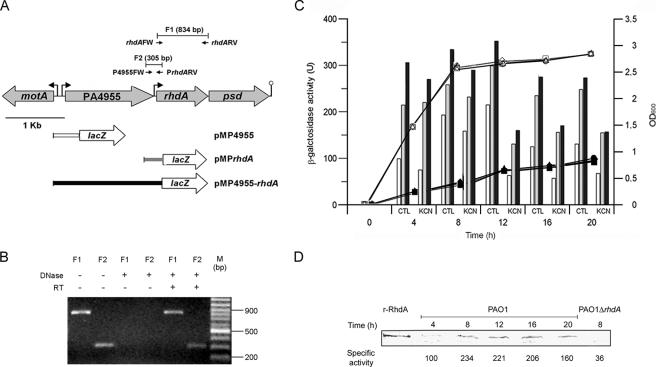
FIG. 3.
Identification of rhdA promoter regions and regulation of rhdA expression. (A) Schematic representation of the rhdA locus. Gray arrows show the orientations of ORFs. Small black arrows show the positions and orientations of RT-PCR primers. The lengths of the predicted RT-PCR products (F1 and F2) are reported. Bent arrows and the lollipop show the locations of putative promoters and the transcription terminator, respectively. Gene fusions with lacZ are shown in the lower part (see Materials and Methods and Table 1). For clarity, inserts of pMP4955 (white), pMPrhdA (gray), and pMP4955-rhdA (black) have the same color as the corresponding bars in the histograms in panel C. (B) Detection of rhdA and PA4955-rhdA transcripts by RT-PCR. Lanes show RT-PCR products amplified from total RNA isolated from P. aeruginosa PAO1 grown in LB for 12 h. PCRs were with or without DNase or reverse transcriptase pretreatment as indicated; F1 and F2 indicate the predicted RT-PCR products (see panel A). M, molecular size standard. (C) Activity of rhdA promoter regions and regulation of rhdA expression. Bacterial growth (OD600 on the right ordinate) was determined at 37°C in LB medium with (black symbols) or without (white symbols) 300 μM KCN. Circles, PAO1(pMP4955); triangles, PAO1(pMPrhdA); squares, PAO1(pMP4955-rhdA). The histogram shows the LacZ activity (on the left ordinate) of pMP4955 (white), pMPrhdA (gray), and pMP4955-rhdA (black) transcriptional fusions in P. aeruginosa PAO1 grown in LB medium at 37°C with (KCN) or without (control [CTL]) 300 μM KCN. The background LacZ activity for PAO1(pMP220) was ca. 20 U. Values are the averages from three independent assays. Standard deviations are <9% of the given values. (D) Time course analysis of RhdA expression by P. aeruginosa PAO1 as revealed by immunoblot analysis of RhdA levels in the cytoplasmic fraction (35 μg of total protein) of bacteria grown in LB at 37°C. The specific rhodanese activity (nmol min−1 mg−1 of cytoplasmic protein) at each time point is also reported. Purified recombinant RhdA (r-RhdA) (150 ng) and cytoplasmic proteins (35 μg) from P. aeruginosa PAO1 ΔrhdA were used as the positive and negative controls, respectively. Blots were hybridized with a mouse anti-RhdA polyclonal antiserum.
In silico analysis predicted the existence of divergent promoter elements in the 140-nucleotide (nt) motA (PA4954)-PA4955 intergenic region, namely, the FliA-dependent promoter (TATAGTCT-N11-ACCGATAA) for motA (35) and an RpoD-dependent promoter oriented like PA4955 (TTatCg-N17-TATAgT), whose putative −35 hexamer diverges (as indicated by lowercase bases) from the P. aeruginosa RpoD consensus (17). No promoter-like sequence was detected in the PA4955-rhdA intergenic region. Since no transcription termination signal can be detected in the 73-nt PA4955-rhdA intergenic spacer and only 3 nt separates rhdA from psd, these three ORFs were predicted to form a single transcriptional unit, ending with a Rho-independent terminator located 145 nt downstream of the psd stop codon (Fig. 3A). To verify this hypothesis, RT-PCR was performed on RNA extracted from late-exponential-phase P. aeruginosa PAO1 cultures, using primer pairs lying within the rhdA coding sequence or flanking the PA4955-rhdA intergenic region (Fig. 3A). Both predicted RT-PCR products (F1 and F2 in Fig. 3A) were observed, providing evidence of readthrough transcription from the upstream PA4955 ORF into rhdA (Fig. 3B).
To confirm the promoter location and investigate the expression profile of rhdA in P. aeruginosa PAO1, three transcriptional fusions linking different regions upstream of rhdA gene with the promoterless lacZ gene were constructed and tested in standard LB medium. These include pMP4955 and pMPrhdA, carrying the motA-PA4955 and PA4955-rhdA intergenic regions, respectively, and pMP4955-rhdA, encompassing both regions (Table 1 and Fig. 3A). Detection of appreciable LacZ activity in P. aeruginosa PAO1(pMP4955) and PAO1(pMPrhdA) substantiated the existence of a promoter element upstream of PA4955 and also made it possible to map an additional promoter in the PA4955-rhdA intergenic spacer. Although lacZ expression was weaker for pMP4955 than for pMPrhdA, the activities of both promoters were maximal from the late exponential to the early stationary phase and progressively decreased thereafter (Fig. 3C). The coexistence of both PA4955 and rhdA promoter regions in pMP4955-rhdA resulted in increased LacZ expression compared with both PAO1(pMP4955) and PAO1(pMPrhdA). Moreover, similar time-dependent expression profiles of the three promoter-probe constructs were observed. These expression data can be interpreted as a cumulative effect due to the existence of tandemly arranged PA4955 and rhdA promoters, both contributing to rhdA transcription (Fig. 3C). Promoter activity was also detected for pMPR4955 and pMPR4955-rhdA reverse constructs (ca. 250 U and ca. 650 U in exponential and stationary phases, respectively), likely resulting from the divergently oriented motA promoter. No activity was observed for pMPRrhdA (data not shown).
The time course analysis of promoter activity correlates well with both rhodanese activity and the intracellular RhdA content. Specific rhodanese activity increased from 98 to 226 nmol min−1 mg−1 of cytoplasmic protein in the first 8 h and then progressively decreased to 162 nmol min−1 mg−1 at 20 h. In accordance, immunoblot analysis of the cytoplasmic protein fraction from P. aeruginosa PAO1 showed that RhdA levels increased during the exponential phase to attain a maximum in the 8- to 16-h time interval and then slightly decreased in the late stationary phase (Fig. 3D).
Effect of RhdA substrates on rhdA expression.
To investigate the influence of RhdA substrates (i.e., thiosulfate and cyanide) on rhdA expression, LacZ activity was determined in cultures of P. aeruginosa PAO1 carrying either pMP4955, pMP4955-rhdA, or pMPrhdA transcriptional fusions and grown in LB medium supplemented with either thiosulfate or cyanide. The presence of up to 10 mM thiosulfate had no effect on bacterial growth and LacZ expression (data not shown). Conversely, 300 μM KCN inhibited P. aeruginosa growth and the activity of all three promoters, particularly at late growth stages (12 to 20 h) (Fig. 3C).
Involvement of RhdA in the protection of P. aeruginosa from cyanide toxicity.
To investigate the role of RhdA as a scavenger of exogenous cyanide, thiocyanate production was compared between resting cells of PAO1 and PAO1 ΔrhdA exposed to 300 μM KCN. Under these conditions, both the wild type and the ΔrhdA mutant showed negligible thiocyanate-forming capability compared with the corresponding clear lysates (0.38 and 0.31 versus 248.5 and 65.1 nmol min−1 mg−1 of total protein, respectively), even after addition of thiosulfate in a 200-fold molar excess over cyanide (data not shown). This could plausibly be explained by the impossibility for thiosulfate to cross the cytoplasmic membrane at sufficient rate (24) and by the inability of a cytoplasmic sulfurtransferase(s) to perform multiple persulfuration cycles. To investigate the involvement of RhdA in protection against exogenous cyanide, exponential-phase cultures of P. aeruginosa PAO1 and PAO1 ΔrhdA carrying or not carrying the complementing plasmid pUCP4955-rhdA were challenged with 300 μM KCN in LB medium. The resulting viability of PAO1 ΔrhdA at 40 min postexposure was on average 26% and 37% lower than those of wild-type PAO1 and PAO1(pUCP4955-rhdA), respectively (data not shown). However, such minor differences were biased by poor statistical significance due to the intrinsic high variability of the viable-count assay.
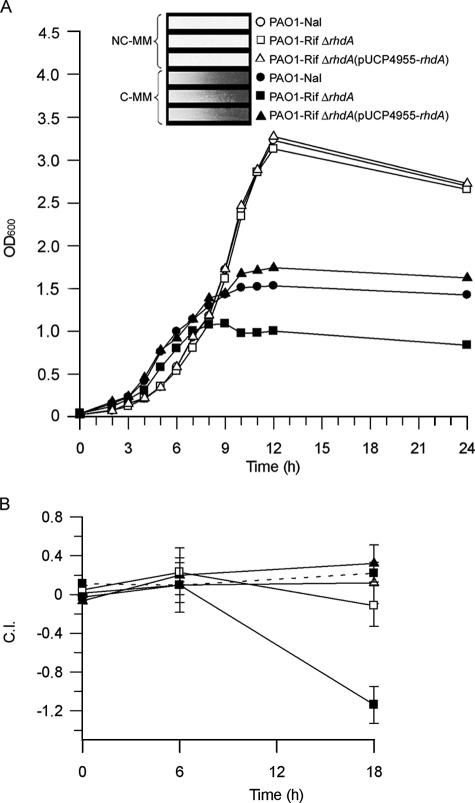
The ability of RhdA to act as a scavenger of endogenous cyanide was assessed by comparing the growth rates of wild-type P. aeruginosa PAO1 and PAO1 ΔrhdA in NC-MM and C-MM synthetic media. P. aeruginosa PAO1, the isogenic mutant PAO1 ΔrhdA, and the mutant complemented with pUCP4955-rhdA showed similar growth rates in NC-MM and attained equally high cell densities at the stationary phase (Fig. 4A), while remarkable differences were observed under cyanogenic conditions. Apart from the overall poor growth in C-MM, the ΔrhdA mutant reproducibly displayed 35% and 50% growth reduction compared to the wild type and the complemented mutant, respectively, resulting from impaired growth at the transition from exponential to stationary phase (Fig. 4A). Under these conditions, cyanogenesis was confirmed for all three strains (Fig. 4A, inset).
FIG. 4.
Effect of the rhdA mutation on P. aeruginosa growth under cyanogenic conditions. (A) Growth of wild-type P. aeruginosa PAO1-Nal (circles), PAO1-Rif ΔrhdA (squares), and PAO1-Rif ΔrhdA(pUCP4955-rhdA) (triangles) in NC-MM (white symbols) or C-MM (black symbols) at 37°C. Results are the means from triplicate growth assays. Standard deviations are <6% of the given values. The colorimetric detection of cyanide produced by individual strains at 12 h is shown in the inset. (B) CIs of P. aeruginosa PAO1-Rif ΔrhdA (squares) and PAO1-Rif ΔrhdA(pUCP4955-rhdA) (triangles) calculated at exponential (6 h) and stationary (18 h) phases in NC-MM (white symbols), C-MM (black symbols), and C-MM supplemented with 75 μM bovine methemoglobin (dashed line). CI values were calculated relative to wild-type P. aeruginosa PAO1-Nal (see Materials and Methods). Results are the means from at least three independent experiments. Standard deviations are shown.
To determine the influence of the ΔrhdA mutation on the biological fitness of P. aeruginosa PAO1 under cynogenic conditions, short- and long-term coculture experiments with wild-type PAO1-Nal and PAO1-Rif ΔrhdA (1:1 ratio) were performed in both NC-MM and C-MM. In the short-term coculture assay, samples for CFU counts were taken at 6 and 18 h, corresponding to the exponential and stationary phases (Fig. 4B), and results were expressed as CI relative to PAO1-Nal. The CI of PAO1-Rif was within the ±0.1 range at both sampling times, irrespective of the culture medium (data not shown). The PAO1-Rif ΔrhdA/PAO1-Nal ratio was essentially constant between 6 and 18 h in NC-MM (CI of between −0.1 and 0.2) (Fig. 4B), consistent with the similar growth rates of these strains under noncyanogenic conditions (Fig. 4A). Conversely, the CI of PAO1-Rif ΔrhdA was severely affected by coculturing in C-MM (Fig. 4B), as shown by a dramatic reduction between 6 and 18 h. Complementation of PAO1-Rif ΔrhdA with pUCP4955-rhdA restored wild-type CI values. A similar effect was obtained with the wild type and the ΔrhdA mutant upon supplementation of C-MM with 75 μM bovine methemoglobin (equivalent to 300 μM ferric heme) as a cyanide scavenger (Fig. 4B). The low CI of PAO1-Rif ΔrhdA in C-MM was further confirmed by coculture experiments performed with different initial PAO1-Rif ΔrhdA/PAO1-Nal ratios, all showing a 1-log difference between the wild type and the mutant after 18 h of coculture (data not shown).
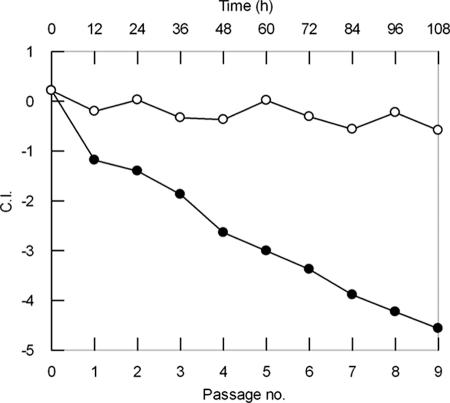
To gain further insight into the dominance of PAO1-Nal over PAO1-Rif ΔrhdA, a long-term coculture experiment in NC-MM and C-MM was performed, involving nine serial passages at 12-h intervals, starting from a CI of 0.2. The results (Fig. 5) show that wild-type PAO1-Nal and the PAO1-Rif ΔrhdA mutant coexist in similar proportions for up to nine passages in NC-MM, attaining ca. 95 generations in 108 h of semicontinuous growth (data not shown). In contrast, wild-type PAO1-Nal showed a marked advantage over PAO1-Rif ΔrhdA in C-MM, progressively outcompeting the mutant during subcultures. At the first passage in C-MM, the mean generation times of PAO1-Nal and PAO1-Rif ΔrhdA were 124 and 450 min, respectively. However, with subsequent passages the growth of both strains in C-MM improved significantly (mean generation times were 108 and 140 min for PAO1-Nal and PAO1-Rif ΔrhdA, respectively).
FIG. 5.
Outcompetition of the ΔrhdA mutant by wild-type P. aeruginosa PAO1. The mean CI of the PAO1-Rif ΔrhdA mutant, relative to PAO1-Nal, was calculated at indicated times during 108 h of semicontinuous growth in NC-MM (white circles) and C-MM (black circles), corresponding to nine serial passages of 12 h each. Each inoculum consisted of a 250-fold dilution from the preceding culture.
DISCUSSION
Rhodaneses have been historically regarded as the main enzymes responsible for cyanide detoxification in eukaryotes, while their function in prokaryotes remains obscure. In P. aeruginosa, rhodanese activity was first documented in 1977 (32), and its genetic basis has recently been unraveled (11). Here, genome-wide analysis of P. aeruginosa PAO1 revealed a redundancy of ORFs characterized by the occurrence of RHOD modules within a variable protein scaffold. Among these, the rhdA gene product is the only protein endowed with features typical of two-domain rhodaneses (4, 11).
While rhodanese activity has previously been localized in the P. aeruginosa periplasm (31), the detection of similar rhodanese levels in periplasmic preparations of both wild-type and ΔrhdA strains excludes a contribution of RhdA to such activity. Cell fractionation unambiguously localized RhdA in the cytoplasm, where it accounts for ca. 90% of total rhodanese activity. The occurrence of six additional ORFs encoding putatively functional sulfurtransferases in P. aeruginosa PAO1 (Fig. 1) could explain the residual activity detected in PAO1ΔrhdA (Fig. 2) and/or the presence of periplasmic rhodanese activity. However, sequence analysis and a recent computational study did not reveal any export signal for any of these proteins (27).
Several lines of evidence suggest that rhdA forms a transcriptional unit with the flanking PA4955 and psd genes (Fig. 3). At the transcriptional level, rhdA expression appears to be governed by two promoters: an upstream element preceding PA4955 and a downstream one in the PA4955-rhdA intervening region. Partially overlapping rhdA transcripts originate either upstream or downstream of PA4955, cumulatively contributing to the extent of rhdA expression (Fig. 2 and 3).
At the regulatory level, RhdA is constitutively expressed during the entire growth cycle, and both rhdA promoter regions show similar time course activities. Promoter fusion assays, rhodanese activity measurements, and immunoblot analyses concur to demonstrate that rhdA expression increases during exponential growth to attain a maximum level at the onset of the stationary phase, slightly decreasing thereafter. Moreover, growth in the presence of exogenous cyanide reduces rhdA expression during the entire growth cycle, with a more pronounced effect in stationary phase (Fig. 3C). While rhdA regulation deserves more in-depth investigation, one could speculate that the synchronism between RhdA down-regulation and cyanogenesis, which is maximal at the stationary phase, would in principle prevent a futile cycle leading to the loss of an ecologically relevant metabolite (cyanide), while keeping its concentration at permissive levels for growth. In fact, comparative analysis of P. aeruginosa PAO1 and PAO1 ΔrhdA growth under cyanogenic and noncyanogenic conditions highlights the involvement of RhdA in protection from endogenous cyanide. Competitive growth assays under cyanogenic conditions result in ca. 10-fold disproportion of stationary-phase yields of the ΔrhdA mutant and impaired competition in long-term coculture assays (Fig. 4B and 5, respectively). Although to different extents, the generation times of both the wild type and the ΔrhdA mutant in cyanogenic medium were higher at the first passage than at following passages, suggesting that adaptive mechanisms take place during cyanogenesis. However, such mechanisms do not fully compensate for the growth defect due to loss of rhdA.
CIO is regarded as the most effective system for protection of P. aeruginosa against cyanide toxicity. Although CIO is physiologically induced after the entry into stationary phase or following exposure to exogenous cyanide, its expression is apparently insensitive to endogenous cyanide (14). Thus, while CIO provides an efficient response to exogenous cyanide, it is rational that P. aeruginosa has evolved additional strategies to face cyanide toxicity while producing this compound, including transition to fermentative metabolism (44) and RhdA-dependent cyanide scavenging. Notably, active breakdown of endogenous cyanide has recently been reported as a means for ensuring the stationary-phase viability of a cio mutant during cyanogenesis (44), and here we show that RhdA facilitates P. aeruginosa survival under cyanogenic conditions. The low growth yield of the ΔrhdA mutant under cyanogenic conditions results from bacteriostasis at the passage from exponential to stationary phase (Fig. 4A). Being concomitant with cyanide production (6, 14), this effect is likely to reflect the inability of the ΔrhdA mutant to face endogenous cyanide. Conversely, when P. aeruginosa cultures were treated with exogenous cyanide, only minor differences in viability between the ΔrhdA mutant and the wild type were observed. Since exogenous cyanide is a potent inducer of CIO expression (14), our results would argue for a marginal role of RhdA following induction of cyanide-insensitive respiration.
While the temporal and functional relationships between the different P. aeruginosa strategies used to face cyanide remain to be established, it is interesting that rhdA orthologues could be retrieved from all genomes of plant growth-promoting Pseudomonas spp. sequenced so far. Since RhdA alleviates the toxic effects of endogenous cyanide, it could facilitate the growth of cyanogenic rhizobacteria under natural conditions of cyanogenicity, ultimately improving their biocontrol performances. Moreover, the modest activity of cytoplasmic RhdA against cyanide from the extracellular milieu would avoid consumption of this compound once it is released in the environment.
Acknowledgments
This work was supported by grants from ISPESL, MIUR-COFIN 2004, and Ministero della Salute-Ricerca Corrente INMI Lazzaro Spallanzani 2005 to P.V.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Aminlari, M., and M. Shahbazi. 1994. Rhodanese (thiosulfate:cyanide sulfurtransferase) distribution in the digestive tract of chickens. Poult. Sci. 73:1465-1469. [DOI] [PubMed] [Google Scholar]
- 2.Aminlari, M., A. Li, V. Kunanithy, and C. H. Scaman. 2002. Rhodanese distribution in porcine (Sus scrofa) tissues. Comp. Biochem. Physiol. 132:309-313. [DOI] [PubMed] [Google Scholar]
- 3.Blumer, C., and D. Haas. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173:170-177. [DOI] [PubMed] [Google Scholar]
- 4.Bordo, D., and P. Bork. 2002. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 3:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Castric, P. A. 1975. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21:613-618. [DOI] [PubMed] [Google Scholar]
- 7.Castric, P. A. 1977. Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 130:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castric, P. A. 1983. Hydrogen cyanide production by Pseudomonas aeruginosa at reduced oxygen levels. Can. J. Microbiol. 29:1344-1349. [DOI] [PubMed] [Google Scholar]
- 9.Cereda, A., F. Forlani, S. Iametti, R. Bernhardt, P. Ferranti, G. Picariello, S. Pagani, and F. Bonomi. 2003. Molecular recognition between Azotobacter vinelandii rhodanese and a sulfur acceptor protein. Biol. Chem. 384:1473-1481. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, K. J., J. M. Ingram, and J. W. Costerton. 1970. Release of alkaline phosphatase from cells of Pseudomonas aeruginosa by manipulation of cation concentration and of pH. J. Bacteriol. 104:748-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cipollone, R., M. G. Bigotti, E. Frangipani, P. Ascenzi, and P. Visca. 2004. Characterization of a rhodanese from the cyanogenic bacterium Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 325:85-90. [DOI] [PubMed] [Google Scholar]
- 12.Cipollone, R., P. Ascenzi, E. Frangipani, and P. Visca. 2006. Cyanide detoxification by recombinant bacterial rhodanese. Chemosphere 63:942-949. [DOI] [PubMed] [Google Scholar]
- 13.Colnaghi, R., S. Pagani, C. Kennedy, and M. Drummond. 1996. Cloning, sequence analysis and overexpression of the rhodanese gene of Azotobacter vinelandii. Eur. J. Biochem. 236:240-248. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, M., G. R. Tavankar, and H. D. Williams. 2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 149:1275-1284. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham, L., and H. D. Williams. 1995. Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J. Bacteriol. 177:432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Argenio, D. A. 2004. The pathogenic lifestyle of Pseudomonas aeruginosa in model systems of virulence, p. 477-503. In J. L. Ramos (ed.), Pseudomonas: genomics, life style and molecular architecture, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 17.Domínguez-Cuevas, P., and S. Marqués. 2004. Compiling sigma-70-dependent promoters, p. 319-343. In J. L. Ramos (ed.), Pseudomonas: virulence and gene regulation, vol. 2. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 18.Feigl, F., and V. Anger. 1966. Replacement of benzidine by copper ethylacetoacetate and tetra base as spot-test reagent for hydrogen cyanide and cyanogen. Analyst 91:282-284. [DOI] [PubMed] [Google Scholar]
- 19.Fernández, R. F., and D. A. Kunz. 2005. Bacterial cyanide oxygenase is a suite of enzymes catalyzing the scavenging and adventitious utilization of cyanide as a nitrogenous growth substrate. J. Bacteriol. 187:6396-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg, D. M., and G. Ellis. 1983. Isocitrate dehydrogenase, p. 183-190. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, vol. 3. Verlag Chemie, Deerfield Beach, Fla. [Google Scholar]
- 22.Haas, D., and G. Défago. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307-319. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kuthma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of cromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 24.Kertesz, M. A. 2004. Metabolism of sulphur-containing organic compounds, p. 323-357. In J. L. Ramos (ed.), Pseudomonas: biosynthesis of macromolecules and molecular metabolism, vol. 3. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 25.Knowles, C. J. 1976. Microorganisms and cyanide. Bacteriol. Rev. 40:652-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leoni, L., A. Ciervo, N. Orsi, and P. Visca. 1996. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J. Bacteriol. 178:2299-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Pessi, G., and D. Haas. 2004. Cyanogenesis, p. 671-687. In J. L. Ramos (ed.), Pseudomonas: biosynthesis of macromolecules and molecular metabolism, vol. 3. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 30.Ray, W. K., G. Zeng, M. B. Potters, A. M. Mansuri, and T. J. Larson. 2000. Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J. Bacteriol. 182:2277-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan, R. W., M. P. Gourlie, and R. C. Tilton. 1979. Release of rhodanese from Pseudomonas aeruginosa by cold shock and its localization within the cell. Can. J. Microbiol. 25:340-351. [DOI] [PubMed] [Google Scholar]
- 32.Ryan, R. W., and R. C. Tilton. 1977. The isolation of rhodanese from Pseudomonas aeruginosa by affinity chromatography. J. Gen. Microbiol. 103:197-199. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 35.Simpson, D. A., and D. P. Speert. 2000. RpmA is required for nonopsonic phagocytosis of Pseudomonas aeruginosa. Infect. Immun. 68:2493-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomonson, L. P. 1981. Cyanide as a metabolic inhibitor, p. 11-28. In B. Vennesland, E. E. Conn, C. J. Knowles, J. Westley, and F. Wissing (ed.), Cyanide in biology. Academic Press, London, United Kingdom.
- 37.Sörbo, B. H. 1953. Crystalline rhodanese. I. Purification and physicochemical examination. Acta Chem. Scand. 7:1129-1136. [Google Scholar]
- 38.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 39.Sylvester, M., and C. Sander. 1990. Immunohistochemical localization of rhodanese. Histochem. J. 22:197-200. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voisard, C., C. Keel, D. Haas, and G. Dèfago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8:351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voulhoux, R., A. Filloux, and I. J. Schalk. 2006. Pyoverdine-mediated iron uptake in Pseudomonas aeruginosa: the Tat system is required for PvdN but not for FpvA transport. J. Bacteriol. 188:3317-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westley, J., H. Adler, L. Westley, and C. Nishida. 1983. The sulfurtransferases. Fundam. Appl. Toxicol. 3:337-382. [DOI] [PubMed] [Google Scholar]
- 44.Zlosnik, J. E., G. R. Tavankar, J. G. Bundy, D. Mossialos, R. O'Toole, and H. D. Williams. 2006. Investigation of the physiological relationship between the cyanide-insensitive oxidase and cyanide production in Pseudomonas aeruginosa. Microbiology 152:1407-1415. [DOI] [PubMed] [Google Scholar]