Abstract
The effects of mineral fertilizer (NPK) and organic manure on the community structure of soil ammonia-oxidizing bacteria (AOB) was investigated in a long-term (16-year) fertilizer experiment. The experiment included seven treatments: organic manure, half organic manure N plus half fertilizer N, fertilizer NPK, fertilizer NP, fertilizer NK, fertilizer PK, and the control (without fertilization). N fertilization greatly increased soil nitrification potential, and mineral N fertilizer had a greater impact than organic manure, while N deficiency treatment (PK) had no significant effect. AOB community structure was analyzed by PCR-denaturing gradient gel electrophoresis (PCR-DGGE) of the amoA gene, which encodes the α subunit of ammonia monooxygenase. DGGE profiles showed that the AOB community was more diverse in N-fertilized treatments than in the PK-fertilized treatment or the control, while one dominant band observed in the control could not be detected in any of the fertilized treatments. Phylogenetic analysis showed that the DGGE bands derived from N-fertilized treatments belonged to Nitrosospira cluster 3, indicating that N fertilization resulted in the dominance of Nitrosospira cluster 3 in soil. These results demonstrate that long-term application of N fertilizers could result in increased soil nitrification potential and the AOB community shifts in soil. Our results also showed the different effects of mineral fertilizer N versus organic manure N; the effects of P and K on the soil AOB community; and the importance of balanced fertilization with N, P, and K in promoting nitrification functions in arable soils.
Ammonia oxidation is the primary step in the oxidation of ammonia to nitrate via nitrite and is thought to be the rate-limiting step of nitrification in most systems; it is therefore central to the global nitrogen cycle (22). In arable soils, most ammonia oxidation is carried out by autotrophic ammonia-oxidizing bacteria (AOB) (7). All known terrestrial AOB belong to a monophyletic group within the β-subclass of Proteobacteria, and the currently accepted classification recognizes only two genera within this group, Nitrosospira and Nitrosomonas (41). However, due to their low growth rates, low biomass yields, and limited number of distinguishing phenotypic characteristics, AOB are difficult to isolate and study in culture (36). In the last decade, it has become possible to study terrestrial AOB communities without culturing by targeting the genes encoding 16S rRNA and the α subunit of ammonia monooxygenase (amoA) (38, 41).
The phylogeny of the amoA gene was found to correspond largely to that of the 16S rRNA gene in AOB (2, 22, 37). The 16S rRNA gene sequence similarities among different AOB are so high that only limited phylogenetic information can be obtained using this gene as a molecular marker (1, 37). Aakra et al. (2) pointed out that, within the genus Nitrosospira, the similarity values of 16S rRNA gene sequences are very high (97% to 99.8%), whereas those of amoA range from 82.6% to 100%. The amoA primer set is highly specific for AOB and is suitable for assessing community shifts (3, 18, 19, 38). Denaturing gradient gel electrophoresis (DGGE) has been used to study the community structure of AOB by targeting the amoA gene (3, 4, 6, 14, 31, 32, 34). Avrahami et al. (6) gave a tentative definition of amoA clusters 1, 2, 3a, 3b, 4, and 8 that will have to be redefined in the future, when more pure cultures and clones are available (4). Clusters 2, 3, and 4 can be related to corresponding 16S rRNA gene clusters as defined by Stephen et al. (41), although clusters 2 and 4 cannot be clearly distinguished. Nitrosospira species of clusters 2, 3, and 4 have been frequently observed in soils (10, 15, 20, 23, 24, 28, 35, 40). Various studies have found that the community structure of AOB in soil is influenced by selective factors such as pH, gravimetric water content, and fertilizer treatment (22); temperature (3, 4); and net nitrogen mineralization (11).
Mineral fertilizers and organic manure could affect the community structure of AOB in soils. Several studies have shown that Nitrosospira cluster 3 dominated in early successional soils with relatively high ammonium concentrations while Nitrosospira clusters 2 and 4 dominated in old successional soils with low ammonium concentrations (10, 24, 25). Fields to which manure or compost had been added contained, in addition, significant Nitrosomonas species (16, 21). Under incubation conditions, the community structure of AOB in soil did not change with different ammonium concentrations in a short period (<4 weeks) (6), while obvious community shifts occurred over a longer period (>16 weeks), indicating that ammonium was a selective factor for different AOB populations (3). Until now, however, there has been little information on the effects of long-term application of fertilizer on the AOB community in soil.
In this study, we report the effects of mineral fertilizer and organic manure on soil AOB community structure as well as nitrification potential in a long-term fertilizer experiment that has been carried out since 1989.
MATERIALS AND METHODS
Field site.
The long-term field fertilizer experiment was carried out in the Fengqiu Ecological Experimental Station (35o00′N, 114o24′E) of the Chinese Academy of Sciences, Henan Province, China. The soil, with a sandy loam texture, was derived from alluvial sediments of the Yellow River and classified as aquic inceptisol. The soil contained 5.8 g kg−1 of organic C, 0.45 g kg−1 of total N, 0.50 g kg−1 of total P, and 18.6 g kg−1 of total K and had a pH (H2O) of 8.6 at the beginning of the experiment in September 1989. Seven treatments (four replicates of each) were established in completely randomized blocks in 28 plots (9.5 by 5 m2) under a rotation of winter wheat (Triticum aestivum L.) and summer maize (Zea mays L.): organic manure (OM), half organic manure N plus half mineral N fertilizer (1/2 OMN), mineral NPK fertilizer (NPK), mineral NP fertilizer (NP), mineral NK fertilizer (NK), mineral PK fertilizer (PK), and the control (without fertilization). For NPK treatment, N, P, and K were applied in the form of urea (300 kg N ha−1 per year), super phosphate (150 kg P2O5 ha−1 per year), and potassium sulfate (300 kg K2O ha−1 per year), respectively, while no K, P, or N was applied for the NP, NK, and PK treatments, respectively. The organic manure was a composted mixture of wheat straw, oil cake, and cotton cake in a ratio of 100:40:45. These materials were ground to achieve lengths of 3 to 5 mm, mixed completely with limited water, and composted for 2 months. The oil cakes and cotton cakes were the machine-dried residues of oil-harvested rapeseeds and cottonseeds, respectively. Detailed information on the organic manure has been given before (29). The OM and 1/2 OMN treatments were designed to give the same application rates of N, P, and K as those given with the NPK treatment. For the OM treatment, N was applied as organic manure, while for the 1/2 OMN treatment, half of the N was applied as organic manure and the other half as urea. Because the amounts of P and K contained in the organic manure were generally less than the prescribed doses, supplemental super phosphate and potassium sulfate were added to the OM and 1/2 OMN treatments to equal the amounts given in the NPK treatment. Detailed information on the experimental design and field management has been described by Meng et al. (29). Each plot had received the same fertilizer management every year since 1989. All phosphorus, potassium, and organic manure were applied as basal fertilizers, whereas urea was added in two applications as both basal and supplementary fertilizers. Basal fertilizers were evenly broadcast onto the soil surface and immediately incorporated into the plowed layer before sowing in June for maize and in October for wheat. Supplementary fertilizer urea was also applied to the soil surface and incorporated into the plowed layer. On 3 March 2006, soil samples were collected at a depth of 0 to 15 cm in the wheat season. For each plot, soil samples were collected from 16 points and then mixed and sieved (<2 mm), with aboveground plant materials, roots, and stones being removed. The fresh soil samples were used for the analysis of nitrification potential and AOB community structure as well as soil mineral N content, while dried and ground samples were used for the analysis of other soil chemical properties.
Soil property and nitrification potential analysis.
Soil pH was determined with a glass electrode using a soil-to-water ratio of 1:1. Soil organic C and total N were determined by dichromate oxidation (27) and Kjeldahl digestion (9), respectively. Mineral N in soil was extracted with 2 mol liter−1 KCl in a 1:4 soil-to-solution ratio for 1 h. NH4+-N and NO3−-N in the extracts were determined by an automated procedure (Skalar SANplus segmented flow analyzer). Available P in soil was extracted by sodium bicarbonate and determined using the molybdenum blue method (33). Available K in soil was extracted by ammonium acetate and determined by flame photometry (12).
Soil nitrification potential was determined according to the method of Hayatsu and Kosuge (17) with slight modifications. Briefly, moist soil (10 g) was amended with ammonium sulfate solution at a rate of 40 mg NH4+-N 100 g−1 dry soil, and then the soil water content was adjusted to 60% of the maximum water-holding capacity. Soils without added ammonium sulfate served as the control. The soils were incubated at 28°C for 48 h in darkness. At the end of the incubation, NO3−-N in the soils was extracted with 40 ml of 2 mol liter−1 KCl for 1 h and determined by an automated procedure (Skalar SANplus segmented flow analyzer).
All results were expressed on an oven-dried soil weight basis (105°C, 24 h). The data were subjected to analysis of variance, and the means and standard deviations for four replicates were calculated. Significant differences of means for all treatments were judged by least significant difference multiple-comparison tests.
DNA extraction from soil.
Soil total DNA was extracted using a FastDNA spin kit for soil (Qbiogene, Inc., Irvine, CA) according to the manufacturer's instructions. Cell lysis was performed by vigorous shaking in a bead beater at an intensity of 5.5 for 30 s (FastPrep DNA extractor; Qbiogene, Inc.). DNA was finally eluted with 50 μl of the DNA elution solution included in the kit. The extracted soil DNA was then purified using an UltraClean 15 DNA purification kit (MO BIO Labs, Solana Beach, CA) and stored at −20°C until ready for PCR-DGGE analysis.
PCR amplification of amoA.
The primer pair amoA-1F (forward) and amoA-2R (reverse) was used for the amplification of the ammonia monooxygenase gene amoA (38). For the DGGE analysis, a guanosine-cytosine clamp was added to the 5′ end of the forward primer (30). PCR amplification was performed with an iCycler thermocycler (Bio-Rad Laboratories, Hercules, CA). PCR mixtures were prepared with a 0.25 mM concentration of each deoxynucleoside triphosphate, 5 μl of 10× PCR buffer, 2.0 mM MgCl2, 0.4 μM of each primer, 2.5 U of Ex Taq HS polymerase (Takara, Japan), and 1 μl of soil DNA template, in a final volume of 50 μl. Amplification was always started by placing the PCR tubes into the preheated (94°C) thermal block of the thermocycler. The thermal profile for amplification was as follows: 2 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; and 5 min at 72°C. The PCR products were electrophoresed on 1.0% agarose to ascertain their size and quality, with the amoA fragment exhibiting a size close to what we expected (531 bp).
DGGE.
The PCR products were separated by using DGGE with a D-Code universal mutation detection system (Bio-Rad Laboratories) according to the instruction manual. The PCR products (20 μl of each) were loaded on 8% (wt/vol) polyacrylamide (acrylamide-bisacrylamide [37.5:1]) gels with a denaturing gradient of 45% to 60% (100% denaturant contains 7 M urea and 40% formamide). The gels were run in 1.0× Tris-acetate-EDTA buffer at 80 V and 60°C for 18 h. After DGGE, the gels were stained with 1:10,000 SYBR green I (Molecular Probes, Leiden, The Netherlands) and scanned with a Molecular Imager FX (Bio-Rad Laboratories). The DGGE image was analyzed with Quantity One software (Bio-Rad Laboratories).
Cloning and sequencing.
The dominant bands in the DGGE gel were excised, and each excised piece was washed twice with 1 ml of sterilized distilled water. A small chip (less than 1 mm3) of each piece was used as a direct template for PCR to recover the DNA fragment. The PCR conditions were the same as for the original PCR, except that the duration of initial denaturation at 94°C was extended to 5 min and the number of cycles in the second step was increased to 40. The fragments recovered from the PCR were subjected to DGGE again to confirm the equality of their mobilities in comparison with that of the soil samples. If a single band appeared in the DGGE gel for one sample, the PCR products were purified with PCR Clear-Up (MO BIO Labs, Solana Beach, CA) and used as a template for direct sequencing. When multiple bands appeared in one sample, the reamplified bands were cloned using the TOPO cloning kit (pCR 2.1 vector for Escherichia coli, TOP 10F′; Invitrogen) according to the manufacturer's instructions. Clones containing a correct insert were reamplified and screened by DGGE and always compared with their original soil samples. The sequencing reactions were performed with a DNA sequencing kit, BigDye Terminator v3.0 (Applied Biosystems, Foster City, CA), and the reaction products were analyzed with an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Phylogenetic analysis.
The nucleotide sequences determined in this study or obtained from the DNA Data Bank of Japan were aligned, and the neighbor-joining trees were constructed using MEGA version 3.1 (Molecular Evolutionary Genetics Analysis [http://www.megasoftware.net]).
Nucleotide sequence accession numbers.
The sequences generated in this study have been deposited in the DNA Data Bank of Japan under accession numbers AB259696 to AB259708.
RESULTS
Soil pH and nutrient contents.
Soil pH and nutrient contents under long-term application of mineral fertilizer and organic manure are shown in Table 1. Soil pH slightly declined with all fertilizer treatments. Soil organic C and total N were greatly increased by the application of organic manure. Organic C and total N were also significantly increased (P < 0.05) by the application of mineral fertilizers, except in the case of the NK treatment. The NH4+-N content in soil was very low, and there were no significant differences in content between the treatments. Since the last fertilization was conducted in October 2005, about 4.5 months before soil sampling, the NH4+-N in the fertilizers had been nitrified to NO3−-N. The accumulation of NO3−-N was observed in all N-fertilized soils, with higher accumulation in mineral N soils. Available P and K in soil were significantly increased (P < 0.05) due to the application of P and K fertilizers, respectively.
TABLE 1.
Soil pH and nutrient contents after long-term application of mineral fertilizer and organic manurea
| Treatment | pHb | Organic C (g kg−1) | Total N (g kg−1) | NH4+-N (mg kg−1) | NO3−-N (mg kg−1) | Available P (mg kg−1) | Available K (mg kg−1) |
|---|---|---|---|---|---|---|---|
| Control | 8.4 (0.1)B | 4.7 (0.5)A | 0.43 (0.03)A | 0.91 (0.17)A | 10.6 (0.6)B | 1.8 (0.3)A | 64 (5)B |
| OM | 8.1 (0.1)A | 12.6 (1.0)E | 1.13 (0.04)E | 1.00 (0.55)A | 19.1 (3.7)C | 25.1 (2.9)C | 154 (8)C |
| 1/2 OMN | 8.1 (0.1)A | 9.8 (0.5)D | 0.92 (0.05)D | 0.89 (0.45)A | 19.3 (1.0)C | 22.2 (4.0)C | 159 (21)C |
| NPK | 8.0 (0.1)A | 7.0 (0.7)C | 0.64 (0.04)C | 0.84 (0.22)A | 32.4 (1.6)D | 14.3 (2.7)B | 146 (5)C |
| NP | 7.9 (0.1)A | 6.6 (0.6)C | 0.65 (0.01)C | 0.96 (0.34)A | 47.9 (6.5)E | 14.3 (3.4)B | 48 (11)A |
| NK | 8.0 (0.1)A | 4.7 (0.5)A | 0.48 (0.04)AB | 0.66 (0.13)A | 52.5 (7.6)E | 2.4 (0.5)A | 243 (9)E |
| PK | 8.1 (0.1)A | 5.6 (0.3)B | 0.54 (0.05)B | 0.73 (0.36)A | 6.5 (0.5)A | 30.5 (2.0)D | 216 (4)D |
Standard deviations are given in parentheses. Values within the same column not followed by the same letter differ significantly (P < 0.05).
pH in water.
Soil nitrification potential.
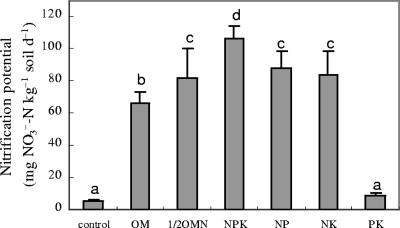
Nitrification potentials in the N-fertilized soils were 13 to 21 times greater than in the control, and the nitrification potential in the PK treatment was not significantly different from that in the control (Fig. 1). Nitrification potentials in mineral N-fertilized soils were significantly higher (P < 0.05) than in soils that received the OM treatment. The NPK treatment led to a significantly higher (P < 0.05) nitrification potential than did the P- and K-deficient treatments. These results indicate that long-term application of N fertilizers (especially mineral N) could greatly increase the nitrification potential of soils and that balanced fertilization (NPK) is important in promoting the nitrification functions in arable soils.
FIG. 1.
Soil nitrification potentials under long-term application of mineral fertilizer and organic manure. Vertical T bars indicate standard deviations. Bars topped by the same letter indicate a significant difference in values (P < 0.05).
Nondegenerate amoA reverse primers.
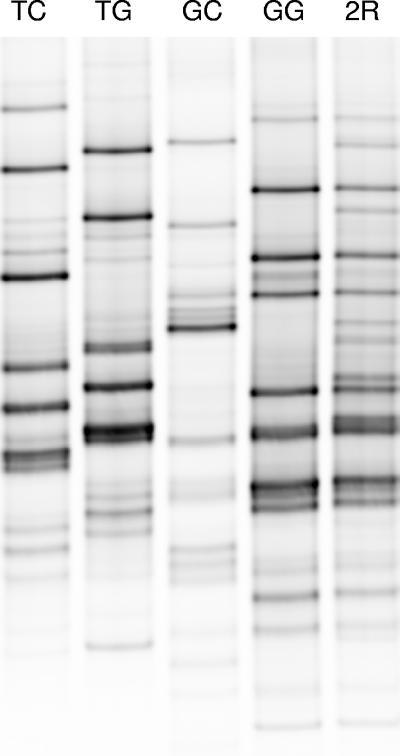
The reverse primer amoA-2R has two degeneracies: K (G or T) and S (G or C) at seven and nine nucleotides from the 5′ end, respectively. Nicolaisen and Ramsing (31) reported that multiple DGGE bands were the result of the two degeneracies in the amoA-2R primer and that the apparent complexity of the DGGE pattern produced by the primer set amoA-1F/amoA-2R was, therefore, a side effect of using a degenerate primer and did not reflect the original diversity. Nondegenerate reverse primers have also been used for PCR-DGGE analysis of AOB in several studies (3, 4, 14, 31, 32). In this study, we used the degenerate amoA-2R primer and four nondegenerate primers, amoA-2R-GG, amoA-2R-GC, amoA-2R-TG, and amoA-2R-TC (the last two letters in the designations indicate the nucleic acids at seven and nine nucleotides from the 5′ end of amoA-2R, respectively), to check which primer gave better results. The DGGE banding patterns in the NPK treatment obtained by using five different reverse primers are shown in Fig. 2. We observed that the results obtained with amoA-2R-GG had a banding pattern similar to that obtained with the degenerate amoA-2R primer and exhibited the dominant bands seen in the DGGE gel with amoA-2R. We also observed that the amoA-2R-GG results exhibited higher AOB diversity than those of the other three nondegenerate primers in our DGGE gels. Therefore, we chose the amoA-2R-GG primer for further study.
FIG. 2.
Comparison of DGGE banding patterns obtained using five different reverse primers: TC, amoA-2R-TC; TG, amoA-2R-TG; GC, amoA-2R-GC; GG, amoA-2R-GG; and 2R, amoA-2R. See the text for explanation of primer names.
Soil AOB community structure.
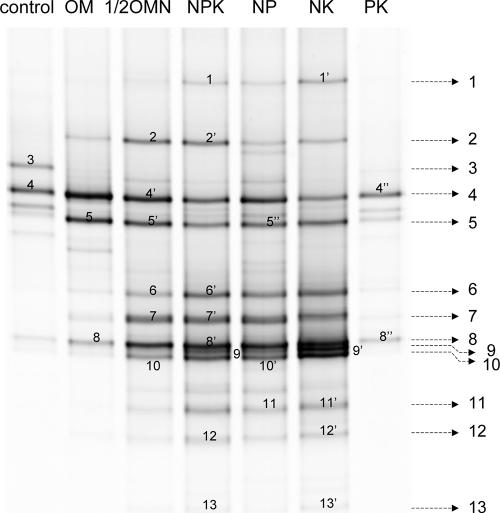
The AOB community structure in soils was analyzed by DGGE. The analysis of four replicates for each treatment showed good reproducibility of DGGE banding patterns (data not shown); therefore, the results for only one replicate are shown in the DGGE patterns (Fig. 3). The numbered bands in the DGGE gel were excised for sequencing. After sequencing, we found that bands (e.g., bands 4, 4′, and 4") with the same mobilities in the DGGE gel had the same nucleotide sequences; thus, the same band names (e.g., band 4) were used to describe the sequences used for phylogenetic analysis (Fig. 4). N fertilization generally increased the number of bands in the DGGE patterns, indicating an increased diversity of the AOB community with N amendment. Mineral N-fertilized treatments exhibited more-diverse banding patterns, with the additional bands 1, 9, and 11 to 13, than the OM treatment, suggesting that mineral N fertilizer and organic manure had different effects on soil AOB community. Band 3 was unique to the control and could not be detected in any of the fertilized soils, including the PK treatment. This result also indicated the significant effect of PK treatment on the AOB community, in addition to N fertilizer. Comparing the DGGE patterns among these three mineral N fertilizer treatments, we observed that, although the dominant bands were the same for all three, the intensities of these bands differed. For example, band 4 had the highest intensity in the NP treatment, while bands 9 and 10 had the highest intensities in the NK treatment. This result further indicated the possible effect of P and K on the soil AOB community. We concluded that long-term N fertilization could greatly affect the soil AOB community. We assumed the possibility that the different forms of N in mineral fertilizers and organic manure could affect the soil AOB community in different ways and that P and K could have a secondary effect in addition to the decisive factor N.
FIG. 3.
DGGE analysis of amoA fragments retrieved from soils after long-term application of mineral fertilizer and organic manure. The excised bands are numbered.
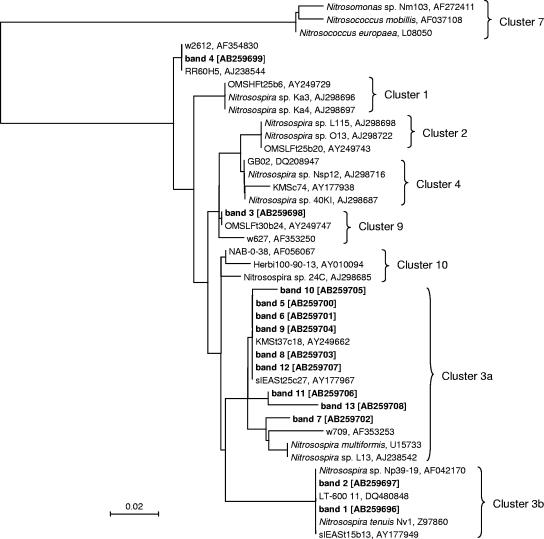
FIG. 4.
Phylogenetic tree based on partial amoA sequences (150 amino acids) retrieved from the DGGE bands (shown in bold) in this study and the DNA Data Bank of Japan. The accession number for each sequence is enclosed in brackets. The scale bar indicates two changes per 100 amino acid positions.
A neighbor-joining tree was constructed using the translated amino acid sequences of DGGE bands and the related sequences obtained from the DNA Data Bank of Japan (Fig. 4). We used the nomenclature for Nitrosospira amoA clusters as defined by Avrahimi et al. (4, 6). All the band sequences belong to the genus Nitrosospira, and no Nitrosomonas species were detected in our soils. The main body(5-13) of these bands grouped in cluster 3a. Bands 1 and 2, which were detected only in N fertilizer treatments, grouped in Nitrosospira cluster 3b, indicating that Nitrosospira cluster 3b had a higher N demand than Nitrosospira cluster 3a. Band 3, which was unique to the control, grouped in cluster 9. Band 4, the dominant common band observed for all the treatments, did not group in some clusters. These results demonstrated that soil AOB community shifts occurred after long-term fertilization.
DISCUSSION
Our study showed that soil nitrification potential responded positively to the application of N fertilizer, indicating that long-term N application could greatly promote soil nitrification functions. The increased nitrification potentials can be ascribed mainly to the mineral N derived from N fertilizers. Increased nitrification rates and potentials along with N amendment have been widely reported in other studies (13, 28, 39, 42, 43). We observed that mineral N (NH4+-forming) fertilizers resulted in higher nitrification potentials than organic manure but with the same application rates of N. This result supports the observation that available NH4+ limits both the nitrification rates and the nitrifier population size (39). We also observed that nitrification potential in the NPK treatment was significantly (P < 0.05) higher than that in the P- and K-deficient treatments, indicating that balanced fertilization with N, P, and K resulted in the highest nitrification potential in our study.
In the present study, we observed that the results obtained with amoA-2R-GG had a banding pattern similar to that obtained with the degenerate amoA-2R primer and exhibited higher AOB diversity than was obtained with the other three nondegenerate primers in our DGGE gels. Nicolaisen and Raming (31) observed that the mobility of clones related to Nitrosospira was much greater than that of Nitrosomonas in the DGGE gel, producing bands within a denaturant concentration gradient of 50% to 60%, whereas clones related to Nitrosomonas ceased to move at much lower concentrations of denaturant (30% to 46%). The DGGE bands were within a denaturant concentration gradient of 50% to 60% in our soils, suggesting that the AOB species in our soils were related to the genus Nitrosospira, which was confirmed by sequence determination (Fig. 4). Avrahami et al. (3) examined a mixture of Nitrosospira clones from different clusters using amoA-2R-GG and found out that this primer was able to detect them all. They also observed that the DGGE patterns obtained with the primer amoA-2R-GG exhibited the highest diversity in their soils (mainly Nitrosospira species). Therefore, the reverse primer amoA-2R-GG is suitable in our soil system.
N fertilizers resulted in clear shifts of the DGGE patterns, indicating a strong influence of long-term (16-year) application of N fertilizer on the soil AOB community. In a short-term incubation experiment (4 weeks), Avrahami et al. (6) in fact obtained no evidence for an AOB community shift with NH4+ concentrations, though community shifts with denitrifiers were observed. Mendum et al. (28) arrived at a similar conclusion after studying soil AOB populations over a period of 6 weeks. Obviously, AOB grow so slowly that changes in their community cannot be detected within 4 to 6 weeks of incubation. AOB community shifts did occur after 16 weeks of incubation (3) and even after just 8 weeks of incubation at 30°C (4). Webster et al. (42) observed significant AOB community shifts in grassland by N amendments in acidic soils. Horz et al. (18) also reported that increased N deposition significantly altered the structure of soil AOB and that the community shifts were associated with an increase in nitrification. In the present study, the DGGE pattern in mineral N-fertilized soil was more diverse than that in the organic manure-fertilized soil, indicating that the forms of N in mineral fertilizer and organic manure could affect AOB community shifts in different ways.
In our study, one dominant DGGE band (band 3) observed in the control could not be detected in all fertilized treatments, including the PK treatment. Although N fertilization was generally beneficial to the increase in AOB community diversity, it might have a negative effect on some resident species. This result also indicated that community shifts could occur not only in N-fertilized soil with high nitrification potential but also in soil receiving the N-deficient (PK) treatment with low nitrification potential. The difference in DGGE banding patterns among the NPK, NP, and NK treatments further indicated the possible effects of P and K on the soil AOB community. To our knowledge, our study is the first to describe the effects of P and K on AOB community shifts in addition to the decisive factor N.
Only Nitrosospira and not Nitrosomonas species were detected in our soils, which is in agreement with many other studies suggesting the dominance of Nitrosospira species in soils (22). The DGGE bands derived from N-fertilized treatments belonged to Nitrosospira cluster 3, indicating that N fertilization resulted in the dominance of Nitrosospira cluster 3 in soil. The observed stimulation of Nitrosospira cluster 3 in N fertilizer treatments is in agreement with the results of Kowalchuk et al. (24, 25), who reported that members of Nitrosospira cluster 3 are dominant in early successional soils with relatively high ammonium concentrations. Furthermore, it is known that cultured strains from Nitrosospira cluster 3 grow well in high-ammonium culture media (8). In general, cluster 3 apparently becomes the dominant group in fertilized versus nonfertilized soils (10). In our study, Nitrosospira cluster 3 was also observed in the control and the PK treatment without N application. Our result is in agreement with the observation of Avrahami et al. (3), who indicated that cluster 3 is not necessarily dominant at high ammonium concentrations. Webster et al. (42) also observed that Nitrosospira cluster 3 is dominant in both improved (addition of N fertilizer) and unimproved (no addition of N fertilizer) grassland soils.
Most of the DGGE bands grouped in Nitrosospira cluster 3a, while bands 1 and 2, which were observed only in N-fertilized soils (band 1 was unique to mineral N), grouped in Nitrosospira cluster 3b. This result indicated that Nitrosospira cluster 3b had a higher N (perhaps NH4+-N) demand than Nitrosospira cluster 3a. Laboratory experiments have also shown that Nitrosospira clusters 3a and 3b exhibited different trends with respect to ammonium in two agricultural soils, reflecting the high versatility of AOB within Nitrosospira cluster 3 (3, 4). Band 3, which was observed only in nonfertilized soil, grouped in Nitrosospira cluster 9, which was consistent with observations that Nitrosospira amoA cluster 9 was found only at low fertilizer concentrations (3, 4) and also with the results of Oved et al. (34), who found this cluster in irrigated agricultural soils that had been treated with low ammonium concentrations.
We could not detect Nitrosospira clusters 2 and 4 in our soils. Nitrosospira cluster 2 has frequently been observed in acidic agricultural soils (25, 26, 40, 41). These soils had pH values between 3.3 and 5.4, confirming the preference of Nitrosospira cluster 2 for soils with a low pH. The soils in our study had high pH values, between 7.9 and 8.4, which might result in the absence of Nitrosospira cluster 2. Nitrosospira cluster 4 has so far been observed only in temperate and not in subtropical and tropical soils, and Avrahami and Conrad (5) speculated that Nitrosospira cluster 4 may be restricted to cold temperatures.
In conclusion, long-term application of N fertilizers leads to soil AOB community shifts and increased soil nitrification potential. Nitrosospira cluster 3 became dominant after long-term N fertilization. Community shifts could occur not only in N-fertilized soils with high nitrification potentials but also in soils that receive N-deficient (PK) treatment with a low nitrification potential. The different forms of N in mineral fertilizer N and organic manure N could affect the AOB community in different ways. Our results also indicate the effects of P and K, in addition to the decisive factor N, in AOB communities and the importance of balanced fertilization with N, P, and K in promoting soil nitrification functions in arable soils.
Acknowledgments
We thank Shengwu Qian of the Institute of Soil Science (ISS), Chinese Academy of Sciences (CAS), for his excellent field management, and Huayong Zhang and Rui Yin, also of ISS, CAS, for their support on soil sample collection. We also thank Naoto Ogawa, Yoriko Sakai, and Ganghua Lang of the National Institute for Agro-Environmental Sciences (NIAES) for their technical support and Sharon Avrahami, Stanford University, and Weixin Ding, ISS, CAS, for their useful discussions.
This work was supported by the National Basic Research Program of China (project 2005CB121108), a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Japan Society for the Promotion of Science in the form of a postdoctoral fellowship.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Aakra, A., J. B. Utaker, and I. F. Nes. 1999. RFLP of rRNA genes and sequencing of the 16S-23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int. J. Syst. Bacteriol. 49:123-130. [DOI] [PubMed] [Google Scholar]
- 2.Aakra, A., J. B. Utaker, and I. F. Nes. 2001. Comparative phylogeny of the ammonia monooxygenase subunit A and 16S rRNA genes of ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 205:237-242. [DOI] [PubMed] [Google Scholar]
- 3.Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. [DOI] [PubMed] [Google Scholar]
- 4.Avrahami, S., and R. Conrad. 2003. Patterns of community change among ammonia oxidizers in meadow soils upon long-term incubation at different temperatures. Appl. Environ. Microbiol. 69:6152-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avrahami, S., and R. Conrad. 2005. Cold-temperature climate: a factor for selection of ammonia oxidizers in upland soils? Can. J. Microbiol. 51:709-714. [DOI] [PubMed] [Google Scholar]
- 6.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barraclough, D., and G. Puri. 1995. The use of 15N pool dilution and enrichment to separate the heterotrophic and autotrophic pathways of nitrification. Soil Biol. Biochem. 27:17-22. [Google Scholar]
- 8.Belser, L. W., and E. L. Schmidt. 1978. Diversity in the ammonia-oxidizing nitrifier population of a soil. Appl. Environ. Microbiol. 36:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bremner, J. M. 1965. Total nitrogen, p. 1149-1178. In C. A. Black, D. D. Evans, L. E. Ensminger, J. L. White, F. E. Clark, and R. C. Dinauer (ed.), Methods of soil analysis, part 2. Chemical and microbiological properties. Agronomy series 9. American Society of Agronomy, Inc., Madison, WI.
- 10.Bruns, M. A., J. R. Stephen, G. A. Kowalchuk, J. I. Prosser, and E. A. Paul. 1999. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl. Environ. Microbiol. 65:2994-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carney, K. M., P. A. Matson, and B. J. M. Bohannan. 2004. Diversity and composition of tropical soil nitrifiers across a plant diversity gradient and among land-use types. Ecol. Lett. 7:684-694. [Google Scholar]
- 12.Carson, P. L. 1980. Recommended potassium test, p. 17-18. In W. C. Dahnke (ed.), Recommended chemical soil test procedures for the North Central Region. Bulletin 499. North Dakota Agricultural Experiment Station, Fargo, ND.
- 13.Chu, H. Y., Y. Hosen, K. Yagi, K. Okada, and O. Ito. 2005. Soil microbial biomass and activities in a Japanese Andisol as affected by controlled release and application depth of urea. Biol. Fertil. Soils 42:89-96. [Google Scholar]
- 14.Ebie, Y., N. Noda, H. Miura, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Comparative analysis of genetic diversity and expression of amoA in wastewater treatment processes. Appl. Microbiol. Biotechnol. 64:740-744. [DOI] [PubMed] [Google Scholar]
- 15.Hastings, R. C., C. Butler, I. Singleton, J. R. Saunders, and A. J. McCarthy. 2000. Analysis of ammonia-oxidizing bacteria populations in acid forest soil during conditions of moisture limitation. Lett. Appl. Microbiol. 30:14-18. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, R. C., M. T. Ceccherini, N. Miclaus, J. R. Saunders, M. Bazzicalupo, and A. J. McCarthy. 1997. Direct molecular biological analysis of ammonia oxidizing bacteria population in cultivated soil plots treated with swine manure. FEMS Microbiol. Ecol. 23:45-54. [Google Scholar]
- 17.Hayatsu, M., and N. Kosuge. 1993. Effects of difference in fertilization treatments on nitrification activity in tea soils. Soil Sci. Plant Nutr. 39:373-378. [Google Scholar]
- 18.Horz, H. P., A. Barbrook, C. B. Field, and J. M. Bohannan. 2004. Ammonia-oxidizing bacteria respond to multifactorial global change. Proc. Natl. Acad. Sci. USA 101:15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horz, H. P., J. H. Rotthauwe, T. Lukow, and W. Liesack. 2000. Identification of major subgroups of ammonia oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J. Microbiol. Methods 39:197-204. [DOI] [PubMed] [Google Scholar]
- 20.Kowalchuk, G. A., P. L. E. Bodelier, G. H. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidizing bacteria, in relation to oxygen availability in soils and root-oxygenated sediments using PCR, DGGE and oligonucleotide probe hybridization. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 21.Kowalchuk, G. A., Z. S. Naoumenko, P. J. Derikx, A. Felske, J. R. Stephen, and I. A. Arkhipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 23.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalchuk, G. A., A. W. Stienstra, G. H. Heilig, J. R. Stephen, and J. W. Woldendorp. 2000. Changes in the community structure of ammonia-oxidizing bacteria during secondary succession of calcareous grasslands. Environ. Microbiol. 2:99-110. [DOI] [PubMed] [Google Scholar]
- 25.Kowalchuk, G. A., A. W. Stienstra, G. H. Heilig, J. R. Stephen, and J. W. Woldendorp. 2000. Molecular analysis of ammonia-oxidizing bacteria in soil of successional grasslands of the Drentsche A (The Netherlands). FEMS Microbiol. Ecol. 31:207-215. [DOI] [PubMed] [Google Scholar]
- 26.Laverman, A. M., A. G. C. L. Speksnijder, M. Braster, G. A. Kowalchuk, H. A. Verhoef, and H. W. Van Verseveld. 2001. Spatiotemporal stability of an ammonia-oxidizing community in a nitrogen saturated forest soil. Microb. Ecol. 42:35-45. [DOI] [PubMed] [Google Scholar]
- 27.Mebius, L. J. 1960. A rapid method for determination of organic carbon in soil. Anal. Chim. Acta 22:120-124. [Google Scholar]
- 28.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1999. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl. Environ. Microbiol. 65:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng, L., W. Ding, and Z. Cai. 2005. Long-term application of organic manure and nitrogen fertilizer on N2O emissions, soil quality and crop production in a sandy loam soil. Soil Biol. Biochem. 37:2037-2045. [Google Scholar]
- 30.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schafer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 32.Nicolaisen, M. H., N. Risgaard-Petersen, N. P. Revsbech, W. Reichardt, and N. B. Ramsing. 2004. Nitrification-denitrification dynamics and community structure of ammonia oxidizing bacteria in a high yield irrigated Philippine rice field. FEMS Microbiol. Ecol. 49:359-369. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, S. R., C. V. Cole, F. S. Watanabe, and L. A. Dean. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate, p. 19. In USDA circular no. 939. U.S. Department of Agriculture, Washington, DC.
- 34.Oved, T., A. Shaviv, T. Goldrath, R. T. Mandelbaum, and D. Minz. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 37.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, W., and J. M. Norton. 2000. Microbial control of nitrate concentrations in an agricultural soil treated with dairy waste compost or ammonium fertilizer. Soil Biol. Biochem. 32:1453-1457. [Google Scholar]
- 40.Stephen, J. R., G. A. Kowalchuk, M.-A. V. Bruns, A. E. McCaig, C. J. Phillips, T. M. Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimens reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaman, M., H. J. Di, K. C. Cameron, and C. M. Frampton. 1999. Gross nitrogen mineralization and nitrification rates and their relationships to enzyme activities and the soil microbial biomass in soils treated with dairy shed effluent and ammonium fertilizer at different water potentials. Biol. Fertil. Soils 29:178-186. [Google Scholar]