Abstract
Mutations in the human methyl-CpG-binding protein gene MECP2 cause the neurological disorder Rett syndrome and some cases of X-linked mental retardation (XLMR). We report that MeCP2 interacts with ATRX, a SWI2/SNF2 DNA helicase/ATPase that is mutated in ATRX syndrome (α-thalassemia/mental retardation, X-linked). MeCP2 can recruit the helicase domain of ATRX to heterochromatic foci in living mouse cells in a DNA methylation-dependent manner. Also, ATRX localization is disrupted in neurons of Mecp2-null mice. Point mutations within the methylated DNA-binding domain of MeCP2 that cause Rett syndrome or X-linked mental retardation inhibit its interaction with ATRX in vitro and its localization in vivo without affecting methyl-CpG binding. We propose that disruption of the MeCP2–ATRX interaction leads to pathological changes that contribute to mental retardation.
Keywords: DNA methylation, Rett syndrome, X-linked mental retardation
Epigenetic phenomena have recently been implicated in development of the brain (1). The protein MeCP2, for example, which binds to certain methylated CpG sites in the genome and can recruit transcriptional corepressors (2–4), is essential for normal brain function. Females heterozygous for mutations in the X-linked MECP2 gene develop Rett syndrome (RTT), a severe neurological disorder that becomes apparent within the first 2 years of life (5–7). Mouse models of this disorder in which the Mecp2 gene is deleted also show a delayed-onset neurological phenotype (8, 9). Conditional deletion of Mecp2 in the brain alone causes indistinguishable symptoms, confirming that this organ is most affected by absence of MeCP2 (8, 9). More specifically, expression of MeCP2 in neurons alone is sufficient to prevent the onset of the neurological phenotype (10). The data suggest that MeCP2 is required to interpret the DNA methylation signal in neurons, but little is known about the molecular details of this essential role.
The function of a protein can be illuminated by identification of its interacting partners. For example, the association of MeCP2 with the corepressor molecule mSin3a established a link between DNA methylation and deacetylation of chromatin (2, 11). Subsequently, MeCP2 has been reported to interact with several other protein partners, including histone H3 lysine 9 methyltransferase activity (12), c-Ski (13), DNMT1 (14), CoRest (15), LANA (16), PU1 (17), splicing factors (18), Y box-binding protein 1 (19), Brm (20), and RNA (21). The relative importance of these interactions, in particular their contributions to the function of MeCP2 in neurons, remains to be clarified. In this study, we performed a further search for proteins that interact with MeCP2. We identified ATRX (22, 23), a SWI2/SNF2 DNA helicase/ATPase that is mutated in a separate neurological disorder, ATRX syndrome (α-thalassemia/mental retardation, X-linked). We mapped the interaction sites on each protein and demonstrated that MeCP2 could target the C-terminal domain of ATRX to heterochromatic foci in cultured mouse cells. Strikingly, the heterochromatic localization of ATRX (24) was disturbed in neurons of the Mecp2-null brain. Moreover, mutation in the human MECP2 gene that cause mental retardation were found to inhibit the MeCP2–ATRX interaction. These genetic findings raise the possibility that MeCP2 and ATRX collaborate during brain development, and that mutations that disturb their interaction interfere with neuronal function.
Results
ATRX Associates with MeCP2.
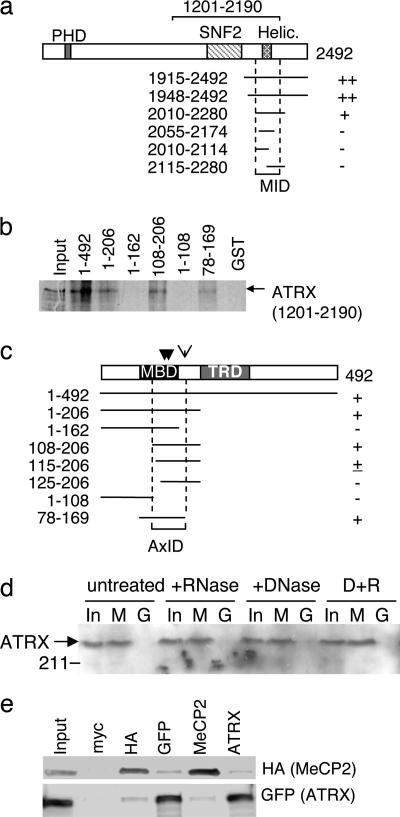
To identify MeCP2-interacting proteins, we performed a yeast two-hybrid screen. Intact MeCP2 caused transcriptional inhibition in yeast because of sequences in the C-terminal half of the protein. We therefore used the N-terminal half of the e2 isoform of rat MeCP2 (amino acids 1–206) as bait. Human and rat MeCP2 are 98% identical in sequence from amino acid 16 to 206 (human). Screening of a human fetal brain library yielded 67 clones that activated the HIS reporter gene, of which 12 verified clones matched the human ATRX cDNA. We defined the region of ATRX that interacted with MeCP2 by deletion analysis. An ATRX polypeptide that contained the C-terminal helicase motif (2010–2280) interacted with MeCP2 as assayed with ADE and HIS reporters in the yeast PJ69 strain, but shorter polypeptides failed to activate either reporter (Fig. 1a). The interaction strength was quantified in the MaV203 strain with a lacZ reporter (data not shown). We next asked whether MeCP2 interacted with ATRX in vitro using GST-pulldown assays with recombinant proteins. In vitro translated ATRX (1201–2190) bound to the immobilized GST-MeCP2 fusion protein but not to GST alone (Fig. 1b). The related protein Lsh/HELLS (25), which contains a SWI2/SNF2 DNA helicase domain that shows 32% amino acid identity to ATRX, did not bind to GST-MeCP2 [supporting information (SI) Fig. 6]. By testing a panel of GST fusion proteins containing truncated MeCP2 fragments (Fig. 1b and SI Fig. 7), we established that the ATRX-binding domain lies between amino acids 108 and 169 and therefore overlaps the methyl-CpG-binding domain of MeCP2 (Fig. 1c) (26). To test for interactions between the full-length proteins, we exposed a recombinant MeCP2-GST fusion protein to a nuclear extract from mouse brain that had been solubilized by extensive sonication. Western blotting established that endogenous ATRX was bound by full-length MeCP2 (Fig. 1d). The interaction was insensitive to DNase I or RNase A treatments, indicating that it is not mediated by nucleic acid (Fig. 1d).
Fig. 1.
MeCP2 interacts with ATRX. (a) Yeast two-hybrid assays identified a region of ATRX that interacts with MeCP2. The diagram depicts ATRX and its domains. Fragments 1915–2492 and 1948–2492 (amino acids numbered from the N terminus) were original clones from the library screen. Symbols ++, +, or − indicate the strength of the MeCP2 interaction based on expression of yeast HIS, ADE, and LacZ reporters (data not shown). The MeCP2 interaction domain (MID) is marked. (b) The ATRX-interaction region overlaps the MBD of MeCP2. The MeCP2 region that binds ATRX was identified by GST pulldown by using a deletion series of MeCP2-GST fusion proteins (for polyacrylamide gel of proteins, see SI Fig. 7) incubated with in vitro-translated [35S]-labeled ATRX fragment 1201–2190 (bracket; a). The input lane (Top) contained 25% of labeled protein used for the pull-down assay. (c) Summary of GST-pulldown showing presence (+) or absence (−) of an ATRX interaction leading to identification of an ATRX-interacting domain (AxID). Point mutations that inhibit ATRX binding (R133C, A140V, and R168X; see Fig. 5) are marked by arrows. (d) Native ATRX in nuclear extracts from brain is “pulled down” by immobilized full-length MeCP2. A MeCP2-GST fusion protein (M) or GST alone (G) were immobilized on glutathione beads and mixed with extract. Bound proteins and input (In; 10%) were separated and probed with anti-ATRX antibody on a Western blot. (e) CoIP of HA-tagged MeCP2 and GFP-fusion ATRX(NLS1201–2492) expressed transiently in mouse L cells. “Input” lane shows 10% of input amount. Antibodies used for coIP and blot visualization are labeled above and beside the figure, respectively.
We asked whether MeCP2 and ATRX interact in vivo by cotransfecting expression constructs that encoded HA-tagged full-length MeCP2 and a GFP-ATRX (1201–2492) fusion protein including a nuclear localization signal into mouse L cells. Both anti-HA and anti-MeCP2 antibodies coprecipitated GFP-ATRX, whereas anti-GFP and anti-ATRX antibodies both reproducibly coimmunoprecipitated HA-MeCP2 (Fig. 1e). An irrelevant antibody, anti-myc, precipitated neither MeCP2 nor ATRX (Fig. 1e). The results demonstrate that ATRX and MeCP2 can interact in vivo. We attempted to perform coimmunoprecipitation of endogenous proteins from mouse brain. ATRX was insoluble even after extraction of nuclei with 2 M salt (see SI Fig. 8) and has been classified as a “matrix-associated” protein (27). Under conditions where ATRX could be solubilized (1 M urea/300 mM NaCl/1% Nonidet P-40), we were unable to detect coimmunoprecipitation with MeCP2 (P. Skene, unpublished observations).
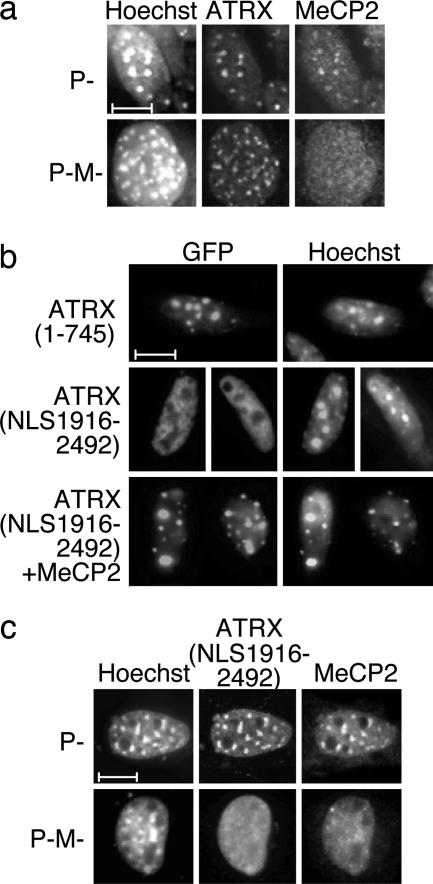
Studies have established that, in mouse cells, both MeCP2 and ATRX localize to pericentromeric heterochromatin (24, 28, 29). MeCP2 localization depends on binding to abundant methyl-CpG sites in these nuclear foci, whereas ATRX localization may require interaction with heterochromatin protein 1 (HP1) via its N-terminal domain (24). To test whether MeCP2 affects ATRX localization, we analyzed the localization of each endogenous protein in mouse fibroblasts lacking detectable DNA methylation [P-M-cells (30)]. As expected (29), MeCP2 was not efficiently targeted to the heterochromatic nuclear foci in nonmethylated cells, whereas correct targeting was seen in control P-cells (Fig. 2a). ATRX was concentrated in heterochromatin of both P- and P-M-fibroblasts (Fig. 2a), demonstrating that heterochromatic targeting of ATRX in these cells does not depend on MeCP2 localization. We confirmed that the N terminus of ATRX was sufficient for correct localization in wt cultured fibroblasts by showing that the N terminus of ATRX, which lacks the MeCP2 interaction domain, was targeted to heterochromatic foci when fused to GFP ]GFP-ATRX (1–745); Fig. 2b Top). As the dominance of the N-terminal domain would mask a role of MeCP2 in ATRX targeting, we expressed a GFP-ATRX (NLS1916–2492) fusion protein lacking the N-terminal domain but retaining the C-terminal DNA helicase motif that binds MeCP2. This protein showed a dispersed nuclear distribution (Fig. 2b Middle). Coexpression of the ATRX (NLS1916–2492) protein with HA-tagged MeCP2, however, led to efficient targeting of ATRX to heterochromatic foci (Fig. 2b Bottom). When the same cotransfection was performed by using DNA methylation-deficient P-M-cells, neither MeCP2 nor the C-terminal fragment of ATRX localized to the heterochromatic foci (Fig. 2c). These results raise the possibility that ATRX recruitment in vivo can involve both MeCP2 and DNA methylation.
Fig. 2.
ATRX localizes to heterochromatic foci in cultured mouse cells lacking DNA methylation, but exogenous MeCP2 can direct the ATRX C-terminal domain to heterochromatin in a DNA methylation-dependent manner. (a) Localization of endogenous ATRX in mouse fibroblasts is normally independent of MeCP2. Control P cells and the DNA methylation-deficient P-M cells were immunostained with anti-ATRX and anti-MeCP2 antibodies and counterstained with Hoechst 33258. (b) The C-terminal region of ATRX is targeted to heterochromatin by MeCP2. Constructs expressing GFP fused to the ATRX N terminus (1–745) or the ATRX C terminus (1916–2492), including an SV40 NLS, were transfected singly, or in combination with full-length MeCP2, into mouse L cells. (c) Targeting of the ATRX C terminus by MeCP2 depends on DNA methylation. ATRX (NLS1916–2492) and HA-MeCP2 were transiently coexpressed in control P or DNA methylation-deficient P-M cells. Localization was monitored by using anti-GFP and anti-HA antibodies. (Scale bars: 10 μm.)
ATRX Delocalization in the Mecp2-Null Brain.
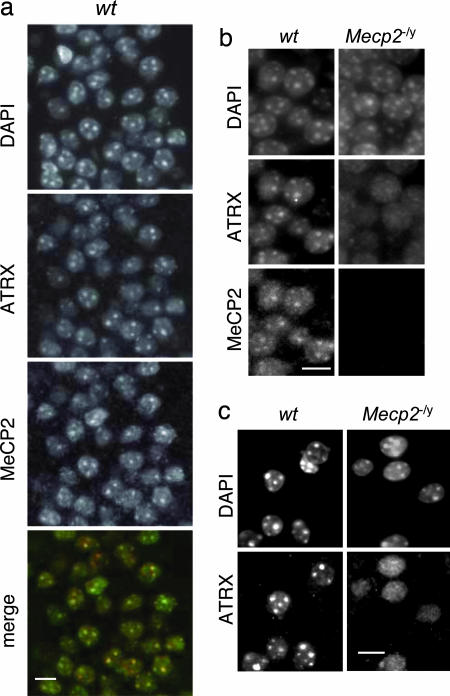
The ATRX N terminus is the dominant signal for nuclear sublocalization in cultured fibroblasts (Fig. 2b), but MeCP2 is not highly expressed in these cells. Knowing that MeCP2 is abundant in neurons (31, 32), we asked whether ATRX localization in the brains of Mecp2-null mice (9) was affected by absence of MeCP2. In the wild-type adult brain, ATRX and MeCP2 were both concentrated in heterochromatic foci (70–80% of nuclei; Fig. 3a). In each of four Mecp2-null brains from animals with overt neurological symptoms (8–11 weeks postnatal age), however, only a minority of nuclei displayed focal ATRX (≈15%; see Fig. 4a). Diffuse ATRX staining was seen reproducibly in mutant animals in the dentate gyrus of the hippocampus (Fig. 3b and SI Fig. 9), the cortex (Fig. 3c), and other regions of the Mecp2-null brain. In contrast to the vast majority of brain cells, tissues in which MeCP2 is less abundant (e.g., kidney and heart) showed no apparent difference in ATRX localization in the presence or absence of MeCP2 (SI Fig. 10).
Fig. 3.
Mislocalization of endogenous ATRX in the Mecp2-null mouse brain. (a) Immunostaining of ATRX, MeCP2, and DNA (DAPI) in wt hippocampus (CA1 region) confirms colocalization of MeCP2 and ATRX in neuronal nuclei (see yellow stain in merge). (b) Loss of ATRX localization in the Mecp2-null hippocampus. Speckled nuclear staining of ATRX coincides with MeCP2 foci in the wt dentate gyrus (for merged images, see SI Fig. 9), but dispersed nuclear ATRX staining is reproducibly seen in the Mecp2-null brain. (c) Immunostaining of ATRX in cortical neurons of wt and Mecp2−/y brains. (Scale bars: 10 μm.)
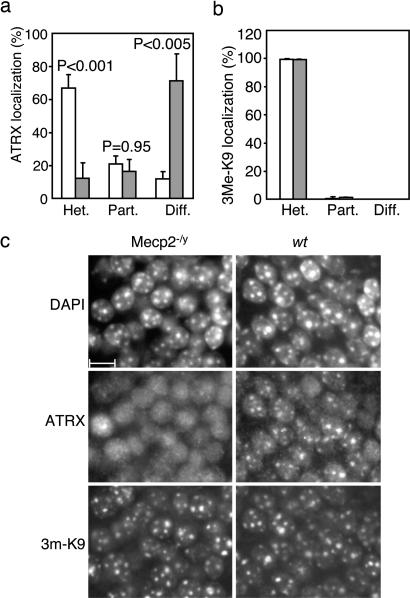
Fig. 4.
Heterochromatic foci persist in the Mecp2-null brain but do not sequester ATRX. (a) Dentate gyrus nuclei (≈100 per section, three brains per genotype) were graded for ATRX localization: predominantly heterochromatic staining (het.), heterochromatic plus diffuse staining (part.), and diffuse staining only (diff.). Significance of differences between wt (open bars) and mutant (shaded bars) are denoted by P values. (b) Dentate gyrus was also graded for staining with antibodies against trimethyl histone H3 lysine 9. (c) ATRX is delocalized in mutant nuclei that retain normal histone H3 lysine 9 trimethylation of heterochromatic foci. (Scale bar: 10 μm.)
Loss of ATRX-positive foci was not due to reduced heterochromatic integrity, because DAPI bright spots and anti-trimethyl histone H3 lysine 9 immunostaining were indistinguishable from wt in mutant brain sections that lacked focal ATRX staining (Fig. 4 b and c). The heterochromatin protein HP1β also showed a punctate localization that was indistinguishable between the wt and mutant dentate gyrus (SI Fig. 11). These findings suggest that heterochromatin structure is normal in Mecp2-null cells. The absence of a correlation between ATRX and HP1β staining also indicates that any influence of HP1β on ATRX localization (33) is minimal in these brain cells. Diffuse ATRX staining was not due to reduced ATRX expression, because wt and Mecp2-null brains expressed equivalent levels of ATRX protein by Western blot (SI Fig. 12). We also tested the possibility that the cellular composition of the hippocampus was radically altered in the mutants, rendering these regions nonequivalent in the wt and mutant brains. Morphologically, the hippocampus appeared identical in wt and Mecp2-null brains. Immunostaining confirmed that nuclei in the dentate gyrus with delocalized ATRX were NeuN-positive postmitotic neurons, as found in equivalent regions of the wt hippocampus (SI Fig. 13). Our results are compatible with the hypothesis that ATRX localization to heterochromatic foci in neurons is directed by the MeCP2–ATRX interaction demonstrated elsewhere in this study, although we are unable to exclude the possibility that the effect is an indirect consequence of MeCP2 deficiency.
MeCP2 abundance has been shown to increase dramatically in neurons as they mature (31), leading to an estimated average concentration of 6 × 106 molecules per brain cell (see ref. 29). We asked whether ATRX was incorrectly localized in newborn (postnatal day 1) Mecp2-null brain, where MeCP2 is usually present at relatively low concentrations. The results showed no obvious ATRX delocalization (SI Fig. 14), suggesting that MeCP2-dependent ATRX targeting is a feature of mature neurons only.
Certain MECP2 Mutations Affect the ATRX Interaction but Not DNA Binding.
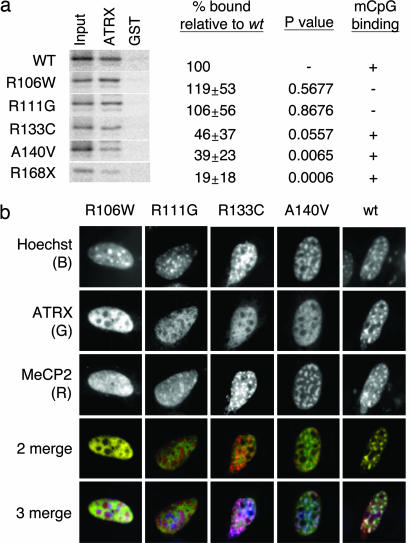
The disruption of ATRX localization in the Mecp2-null mouse brain suggested that human MECP2 mutations that cause neurological disorders might affect the interaction with ATRX. We tested three mutant forms of MeCP2 (R106W, R133C, and R168X) that occur in RTT, one mutant whose clinical significance is not well characterized (R111G) and one mutant (A140V) that occurs in male XLMR (34–36), for binding to ATRX by using the GST pull-down assay. The relative ATRX binding affinities of R111G and R106W were indistinguishable from wt, but A140V, R133C, and R168X bound very weakly (Fig. 5a). Analysis of the DNA-binding activity of the same mutant polypeptides by bandshift analysis showed that the methyl-CpG affinity was abolished by the R106W and R111G mutations and slightly reduced compared with wild type by the R168X mutation (summary in Fig. 5a; see SI Fig. 15). Neither the R133C nor the A140V mutations affected binding to methylated DNA, although weak binding of R133C to the nonmethylated probe was observed, suggesting reduced specificity for methylated DNA (SI Fig. 15). In summary, the A140V, R168X, and, to a lesser extent, R133C mutations reduce the affinity of MeCP2 for ATRX in vitro without major effects on its affinity for methylated DNA.
Fig. 5.
Mutations in human in the MBD of human MeCP2 that cause mental retardation disturb the MeCP2–ATRX interaction without affecting methyl-CpG binding. (a) A GST fusion of ATRX (1915–2492) was immobilized and exposed to in vitro-translated [S35]-labeled MeCP2 (1–206) with the mutations indicated at the left. Shown is 10% of input, the amounts retained by ATRX (1915–2492) and by GST. Densitometric results (average of three experiments) are expressed relative to wt (% bound) together with P values (Student t test). The “mCpG binding” column summarizes the methyl-CpG binding as assayed in SI Fig. 15. (b) MeCP2 mutants A140V and R133C target heterochromatic foci but cannot direct ATRX to heterochromatin. HA-tagged full-length mutant MeCP2 forms were coexpressed with GFP-ATRX (NLS1916–2492) fusion proteins in mouse fibroblasts. Localization was monitored by immunostaining using anti-HA (MeCP2) and anti-GFP (ATRX) antibodies. In the merge, Hoechst, ATRX, and MeCP2 are blue, green, and red, respectively. R133C and A140V mutants colocalize with Hoechst bright spots (pink spots, triple merge), but ATRX staining is diffuse (absence of yellow spots, double merge). Note that MeCP2 R111G partially localizes to nucleoli.
We next asked whether in vivo-expressed full length MeCP2 carrying these mutations could (i) localize to methyl-CpG-rich heterochromatic foci; (ii) recruit ATRX (NLS1916–2492) to these foci. The R168X mutant was not assayed because it lacks a nuclear localization signal. R106W and R111G polypeptides neither localized themselves nor recruited ATRX to pericentric heterochromatin (Fig. 5b), in agreement with Fig. 5a and previous findings (37). In contrast, R133C and A140V mutants targeted heterochromatin (Fig. 5b), as predicted by their affinity for methyl-CpG in vitro (Fig. 5a), but neither mutant protein could recruit ATRX (NLS1916–2492) to heterochromatin (Fig. 5b). We conclude that the clinically relevant R133C and A140V mutations, which have no discernable effect on targeting of MeCP2 to methylated sites in this assay, disrupt the interaction between MeCP2 and ATRX in vitro and in vivo.
Discussion
Our data show that MeCP2 and ATRX, two proteins that are required for normal brain function, interact in vitro and in living cells. The interaction is supported by five independent lines of evidence: (i) a positive yeast two-hybrid assay; (ii) in vitro GST pulldowns; (iii) coimmunoprecipitation of the in vivo coexpressed proteins; (iv) MeCP2-dependent and DNA methylation-dependent targeting of the ATRX C-terminal domain in vivo to nuclear heterochromatic foci; and (v) widespread delocalization of ATRX in neuronal nuclei of Mecp2-null mice.
Certain MECP2 mutations that cause XLMR interfere with the MeCP2–ATRX interaction. Specifically, the A140V and R133C mutant proteins localize efficiently to heterochromatic foci, but are unable to recruit the C terminus of ATRX to these sites. Despite the overlap of the MBD and ATRX-binding domains of MeCP2, only binding to ATRX is affected by the A140V mutation, which therefore uncouples the DNA- and ATRX-binding functions of MeCP2. The R133C and R168X mutations also reduce binding to ATRX, but both may affect additional functions of MeCP2, because the former enhances binding to nonmethylated DNA, and the latter eliminates the C-terminal two-thirds of MeCP2. Interestingly, the R133C mutation, although frequent in RTT patients, tends to cause a relatively mild phenotype, which may be related to incomplete loss of methylated DNA binding. The A140V mutation is therefore of particular interest, because its only measurably altered property is loss of the ability to interact with ATRX. A140V is not an RTT mutation but has been reported in hemizygous males of several independent XLMR families (34–36, 38). The A140V phenotype is evidently less severe than that of mutations causing RTT, because heterozygous females are either normal or occasionally suffer mild mental retardation. An intriguing possibility is that inappropriate targeting of ATRX contributes to the neurological phenotype of RTT patients and Mecp2-null mice.
In cultured cells, it has been demonstrated that the ATRX N-terminal region, which contains the conserved PHD domain, is sufficient to target the protein to heterochromatic foci (33). In mature MeCP2-deficient neurons, however, ATRX adopts a diffuse nuclear distribution, suggesting that the N terminus is not sufficient for localization in these cells. Dispersal of ATRX is not caused by a general loss of heterochromatin integrity, because both trimethylation of histone H3 lysine 9 and localization of HP1β remain normal in these neurons. A previous report found that overexpressed MeCP2 in muscle cells alters heterochromatin aggregation (39), but neither the number [Mecp2−/y mean = 5.33 (SD = 1.46); wt mean = 4.97; (SD = 1.81); n = 75 and 71] nor the distribution of nuclear foci differed noticeably between wt and Mecp2-null neurons in 8- to 11-week-old mice. In the absence of gross structural alterations in Mecp2-null neuronal heterochromatin, it seems that loss of ATRX targeting in neurons depends on MeCP2 rather than the N-terminal domain. The reasons for this altered dependence are currently unknown, but it is conceivable that either ATRX or its usual targeting partner (perhaps HP1) are posttranslationally modified in neurons, thereby preventing interaction. Alternatively, the normal ATRX N-terminal ligand protein may be inaccessible in neurons because of physical sequestration or saturation of binding sites by competing proteins. It is also possible that the exceptionally high abundance of MeCP2 in brain (29) gives it the dominant role in ATRX localization, particularly if the abundance of ATRX itself exceeds that of its conventional targeting proteins in mature neurons.
Patients with mutations in ATRX exhibit severe psychomotor retardation, characteristic facial features, α-thalassaemia, and genital abnormalities (40). Of relevance is the observation that conditional deletion of ATRX in the mouse forebrain causes a major decrease in forebrain size because of a reduction in the number of neuronal precursors surviving to reach the cortex (41). These findings demonstrate the importance of ATRX in early neuronal development and raise the possibility that its continued presence is important in mature neurons, where MeCP2 is also required (31). Little is known of the normal function of ATRX, but it belongs to a family of proteins that share the Snf2 motor domain. Several family members can modify the structure of nucleosomal chromatin (42), and ATRX has itself been shown to modify chromatin in an ATP-dependent manner in vitro (43). In addition, ATRX interacts with the chromatin-associated proteins HP1α (24, 44), the polycomb group protein EZH2 (45) and the transcription cofactor Daxx (43, 46). Heterochromatic foci provide a convenient visual assay for correct localization of ATRX, but major genomic sites of ATRX function may be euchromatic. A speculative hypothesis is that ATRX contributes to epigenetic regulation of certain MeCP2-bound genes. Several other mammalian candidates are known, including the rodent brain-derived neurotrophic factor (Bdnf) gene (47, 48) and others that are reportedly misregulated in the absence of MeCP2 (49–51). The next step is to determine whether these and other emerging MeCP2 targets are affected directly or indirectly by ATRX.
Materials and Methods
Construction of Plasmids, Cell Lines, Protein Immunostaining, Extraction of Nuclear Proteins, GST Pulldowns of Endogenous ATRX, and Western Blotting.
For additional information on these topics, see SI Methods.
Yeast Two-Hybrid Screening.
A human fetal brain yeast two-hybrid library constructed in prey vector pACT2 (Clontech, Palo Alto, CA) was transformed into yeast PJ69α. The bait constructs were transformed to PJ69a and subsequently mated with the pretransformed library in yeast PJ69α. Approximately 107 independent clones were screened. Initial screening was performed by using histidine-minus SD plates, on which only HIS-expressing yeast can grow, according to User Manual PT3183–1 (Clontech). Interaction was confirmed in yeast PJ69 on adenine-minus SD plates by using the GAL2-ADE2 reporter. To quantitatively measure the strength of interaction by using a LacZ reporter, prey and bait plasmids were cotransformed into yeast MaV203(MATα) (GIBCO BRL, Carlsbad, CA). The activity of β-galactosidase was measured in liquid culture by using the ONPG substrate as described in ProQuest Two-Hybrid System Instruction Manual (GIBCO BRL). Neither the MeCP2 bait nor the ATRX prey constructs could alone activate the yeast reporters.
GST Pull-Down Assay.
Expression and purification of GST–MeCP2 and GST-ATRX(1915–2492) fusion proteins were described (4, 26). [35S]methionine-labeled ATRX fragment (amino acid 1201–2190) and MeCP2 fragment (amino acid 1–206) were generated by using a coupled transcription–translation T7 RNA polymerase TNT system (Promega, Madison, WI). GST fusion proteins (3 μg) were first bound to glutathione–Sepharose beads. For pulldown with GST-MeCP2 fusion protein, [35S]-labeled ATRX was incubated with immobilized GST fusion protein in 100 μl of binding buffer (20 mM Hepes, pH 7.9/0.1 M NaCl/1 mM EDTA/0.5 mM DTT/10% glycerol/0.1% Triton X-100/1 mg/ml BSA/protease inhibitor mixture) at 4°C for 2 h, and beads were subsequently washed with RIPA buffer (50 mM Tris, pH 8.0/150 mM NaCl/1% Nonidet P-40/0.1% SDS/protease inhibitor mixture). For pulldown with the GST-ATRX fusion, [35S]-labeled MeCP2 or its mutant derivatives were incubated with immobilized GST fusion protein in 400 μl of binding buffer at 4°C for 2 h, and beads were subsequently washed with 0.2 M NaCl washing buffer (20 mM Hepes, pH7.9/0.2 M NaCl/1 mM EDTA/0.5 mM DTT/0.1% Triton X-100/10% glycerol). Bound proteins were eluted in Laemmli loading buffer, fractionated by SDS/PAGE, and detected by autoradiography or by PhosphorImager. For GST pulldown of endogenous ATRX, see SI Methods.
DNA Binding Assay.
Affinity purified MeCP2-GST fusion protein was incubated on ice with a 73-bp 32P-labeled DNA probe 5′ GTTTTCCCAGTCACTACCGCAGCACGGTGGGGGGCCGGAGTTAAGGACTCGTTGTCGTCATAGCTGTTTCCTG that was either methylated at a single central HpaII site (bold) or nonmethylated. MeCP2 bound probe was separated from free probe on 5% acrylamide gel by using a standard protocol (26).
Coimmunoprecipitation (coIP).
L cells were transfected with HA-MeCP2 expressing plasmid pCMV/HA-MeCP2 and GFP-NLS-ATRX expressing plasmid pcDNA3/GFP-ATRX(NLS1201–2190) by using the transfection reagent Lipofectamine2000 (Invitrogen, Carlsbad, CA). After 2 d, nuclei were isolated, and nuclear extract was prepared (4). Nuclear extract was precleared by incubation with protein A-Sepharose (Amersham Pharmacia, Piscataway, NJ) before coIP. Aliquots (100 μg of protein) of precleared nuclear extract were incubated for 2 h at 4°C, with 20 μl of affinity-purified HA-Tag polyclonal antibody (Clontech), 5 μl of GFP antiserum (Living colors full-length A.v. polyclonal antibody; Clontech), 5 μl of MeCP2 antiserum (674) (4), 20 μl of affinity-purified anti-ATRX antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or with 20 μl of affinity-purified anti-myc antibody (Invitrogen). The final volume was 500 μl in CSK buffer (10 mM Pipes, pH6.8/100 mM NaCl/3 mM MgCl2/1 mM EDTA/0.5% Triton X-100/EDTA-free protease inhibitors). Antibody-bound material was pelleted with protein A-Sepharose (Amersham Pharmacia), washed three times with RIPA buffer [150 mM NaCl/1% Nonidet P-40/50 mM Tris, pH 8.0/0.1% SDS/1× Complete Protease inhibitors (EDTA-free; Roche, Indianapolis, IN)]. Immunoprecipitated material was detected by Western blot using rat anti-HA high-affinity antibody (Roche) and mouse anti-GFP antibody (Roche).
Supplementary Material
Acknowledgments
We thank Patricia Yeyati, Rob Klose, Lars Schmiedeberg, Richard Meehan, and Wendy Bickmore for advice and Peter Skene and Irina Stancheva for comments on the manuscript. This work was supported by the Medical Research Council (J.N.), the Wellcome Trust, Rett Syndrome U.K., and a Postdoctoral Research Fellowship from the Rett Syndrome Research Foundation (to S.K.).
Abbreviations
- CoIP
coimmunoprecipitation
- RTT
Rett syndrome.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608056104/DC1.
References
- 1.Hong EJ, West AE, Greenberg ME. Curr Opin Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 3.Nan X, Campoy FJ, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 4.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 5.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 6.Hagberg B, Aicardi J, Dias K, Ramos O. Ann Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 7.Rett VA. Weiner Medizinische Wochenschrift. 1966;37:723–726. [PubMed] [Google Scholar]
- 8.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 9.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 10.Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Proc Natl Acad Sci USA. 2004;101:6033–6038. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nan X, Cross S, Bird A. Novartis Found Symp. 1998;214:6–21. doi: 10.1002/9780470515501.ch2. [DOI] [PubMed] [Google Scholar]
- 12.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 13.Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. J Biol Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 14.Kimura H, Shiota K. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 15.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Science. 2002;298:1747–1751. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 16.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. J Virol. 2002;76:11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki M, Yamada T, Kihara-Negishi F, Sakurai T, Oikawa T. Oncogene. 2003;22:8688–8698. doi: 10.1038/sj.onc.1207182. [DOI] [PubMed] [Google Scholar]
- 18.Buschdorf JP, Stratling WH. J Mol Med. 2004;82:135–143. doi: 10.1007/s00109-003-0497-9. [DOI] [PubMed] [Google Scholar]
- 19.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Nat Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 21.Jeffery L, Nakielny S. J Biol Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 22.Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 23.Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 24.McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nan X, Meehan RR, Bird A. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berube NG, Smeenk CA, Picketts DJ. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 28.Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 29.Nan X, Tate P, Li E, Bird A. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen HF, Ben-Porath I, Bird AP. Mol Cell Biol. 2004;24:3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishi N, Macklis JD. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY. Hum Mol Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 33.McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre AC, Souville I, et al. Hum Mol Genet. 2001;10:941–946. doi: 10.1093/hmg/10.9.941. [DOI] [PubMed] [Google Scholar]
- 35.Orrico A, Lam C, Galli L, Dotti MT, Hayek G, Tong SF, Poon PM, Zappella M, Federico A, Sorrentino V. FEBS Lett. 2000;481:285–288. doi: 10.1016/s0014-5793(00)01994-3. [DOI] [PubMed] [Google Scholar]
- 36.Klauck SM, Lindsay S, Beyer KS, Splitt M, Burn J, Poustka A. Am J Hum Genet. 2002;70:1034–1037. doi: 10.1086/339553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo S, Nomura Y, Segawa M, Fujita N, Nakao M, Schanen C, Tamura M. J Med Genet. 2003;40:487–493. doi: 10.1136/jmg.40.7.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen D, Lazar G, Couvert P, Desportes V, Lippe D, Mazet P, Heron D. Am J Psychiatry. 2002;159:148–149. doi: 10.1176/appi.ajp.159.1.148-a. [DOI] [PubMed] [Google Scholar]
- 39.Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. J Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 41.Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. Proc Natl Acad Sci USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 45.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontes M, Colleaux L. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 46.Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- 47.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 48.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 49.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 50.Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, Bird A. Hum Mol Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- 51.Samaco RC, Hogart A, LaSalle JM. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.