Abstract
β-Catenins are conserved regulators of metazoan development that function with TCF DNA-binding proteins to activate transcription. In Caenorhabditis elegans, SYS-1/β-catenin and POP-1/TCF regulate several asymmetric divisions, including that of the somatic gonadal precursor cell (SGP). In the distal but not the proximal SGP daughter, SYS-1/β-catenin and POP-1/TCF transcriptionally activate ceh-22 to specify the distal fate. Here, we investigate the distribution of SYS-1/β-catenin and its regulation. Using a rescuing transgene, VNS::SYS-1, which fuses VENUS fluorescent protein to SYS-1, we find more VNS::SYS-1 in distal than proximal SGP daughters, a phenomenon we call “SYS-1 asymmetry.” In addition, SYS-1 asymmetry is seen in many other tissues, consistent with the idea that SYS-1 regulates asymmetric divisions broadly during C. elegans development. In particular, SYS-1 is more abundant in E than MS, and SYS-1 is critical for the endodermal fate. In all cases, SYS-1 is reciprocal to POP-1 asymmetry: cells with higher SYS-1 have lower POP-1, and vice versa. SYS-1 asymmetry is controlled posttranslationally and relies on frizzled and dishevelled homologs, which also control POP-1 asymmetry. Therefore, upstream regulators modulate the SYS-1 to POP-1 ratio by increasing SYS-1 and decreasing POP-1 within the same cell. By contrast, SYS-1 asymmetry does not rely on WRM-1, which appears specialized for POP-1 asymmetry. We suggest a two-pronged pathway for control of SYS-1:POP-1, which can robustly accomplish differential gene expression in daughters of an asymmetric cell division.
Keywords: dishevelled, frizzled, Wnt/MAPK signaling, asymmetric cell division
Beta-catenins are key regulators of metazoan development (1). When the canonical Wnt signaling pathway is inactive, β-catenin is destroyed, but when it is activated, β-catenin is stabilized, enters the nucleus, and functions as a transcriptional coactivator to transform the TCF/LEF DNA-binding protein from a repressor to an activator of target genes (ref. 2; reviewed in ref. 3). A variant Wnt signaling pathway controls asymmetric cell divisions in Caenorhabditis elegans (4, 5). A new player in that pathway is SYS-1/β-catenin (6).
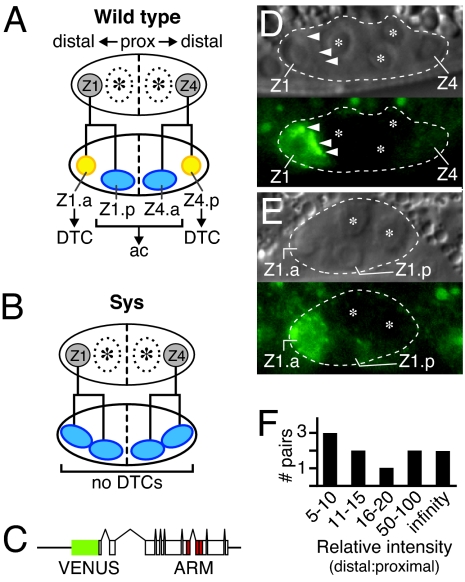
The sys-1 gene was discovered as a regulator of the asymmetric cell division of somatic gonadal precursor cells (SGPs) (7). Each SGP produces a distal daughter that generates a “distal tip cell” and a proximal daughter with potential to make an “anchor cell” (Fig. 1A). Each distal tip cell controls elongation into a gonadal “arm,” while the anchor cell induces the vulva. In sys-1 loss-of-function mutants, SGPs divide symmetrically and generate two daughters, both with proximal fates: no distal tip cells (and hence no gonadal arms) are produced (Fig. 1B); instead, extra anchor cells are made. This lack of distal tip cells coupled to extra anchor cells is a hallmark of the Sys phenotype.
Fig. 1.
Asymmetric expression of SYS-1/β-catenin in SGP daughters. (A) Schematic of wild-type SGP asymmetric division. The top shows gonadal primordium (large oval) with two SGPs (gray) and two germline precursors (asterisks). The anterior and posterior SGPs are called Z1 and Z4, respectively. Proximal (prox) and distal gonadal axes are indicated above primordium, and the dashed line indicates the plane of symmetry. The bottom shows that each SGP produces one distal (yellow) and one proximal (blue) daughter. The distal daughter makes a distal tip cell (DTC) and hence a gonadal arm; the proximal daughter has anchor cell (ac) potential. (B) Schematic of Sys SGP symmetrical division. Conventions are as in A. (C) Psys-1::VNS::SYS-1 reporter transgene. Boxes, exons; lines, noncoding regions. VENUS, yellow fluorescent protein; ARM, armadillo repeats (6). VNS::SYS-1 was integrated into the genome at two distinct sites (qIs94 and qIs95). (D) VNS::SYS-1 in SGP within 1 h of its division. Nomarski (Upper) and fluorescent (Lower) images of same L1 larva. Dashed line, outline of gonadal primordium; Z1 and Z4, SGPs; asterisks, germ cells; arrowheads, positions of VNS::SYS-1 granules. (E) VNS::SYS-1 in SGP daughters. Conventions as in D. VNS::SYS-1 is present in the distal daughter (Z1.a) but not the proximal daughter (Z1.p). (F) Relative intensities of VNS::SYS-1 in SGP daughter pairs ranged from 5-fold to infinity. x axis, relative intensity of VNS::SYS-1 in distal daughter compared with proximal daughter; numbers refer to fold difference. y axis, number of SGP pairs.
The sys-1 gene encodes a highly divergent β-catenin (6, 8). Several lines of evidence support the idea that SYS-1 is a functional β-catenin. First, transgenic SYS-1 can rescue a null mutant of bar-1, which encodes a typical β-catenin. Second, SYS-1 binds the β-catenin binding domain of POP-1/TCF. Third, SYS-1 acts as a transcriptional coactivator for POP-1 in a TOPFLASH reporter. Fourth, ceh-22 expression in distal SGP daughters depends on POP-1 binding sites in the ceh-22b promoter and also on SYS-1 and POP-1. Therefore, SYS-1 acts in many ways like a typical β-catenin.
Both SYS-1/β-catenin and POP-1/TCF control the SGP asymmetric cell division (7, 9). In pop-1 mutants, as in sys-1 mutants, the SGP daughters both adopt a proximal fate (Fig. 1B). Importantly, POP-1 is asymmetrically distributed to SGP daughters (10), a phenomenon that has been seen in many asymmetric cell divisions and has been dubbed “POP-1 asymmetry” (11). Although counterintuitive, nuclear POP-1 is reduced in distal SGP daughters relative to proximal daughters, a reduction that is critical for distal tip cell specification. One explanation is that abundant POP-1 represses transcription, but reduced POP-1/TCF activates transcription, as observed in the early embryo (2). Consistent with that model, a direct target gene of POP-1 and SYS-1 is activated in distal SGP daughters (8).
The ratio of SYS-1/β-catenin to POP-1/TCF is critical for transcriptional activation (6, 10). This idea is based on several findings. First, the generation of distal tip cells is sensitive to SYS-1 abundance: sys-1/+ heterozygotes have too few distal tip cells, and SYS-1 overexpression produces too many distal tip cells. Second, a decrease in POP-1 is critical for distal tip cell specification, which relies on transcriptional activation of the ceh-22b promoter. Third, the ratio of SYS-1/β-catenin to POP-1/TCF is critical for transcriptional activity in TOPFLASH reporter assays. Based on these data, the model was proposed that SYS-1 is made at a uniformly low level and that POP-1 must be decreased to accommodate that limiting level.
In this article, we examine SYS-1/β-catenin expression, using a rescuing reporter called VNS::SYS-1. VNS::SYS-1 is distributed asymmetrically to SGP daughters, a phenomenon we dub “SYS-1 asymmetry.” Importantly, SYS-1 asymmetry is reciprocal to POP-1 asymmetry. In the early embryo, SYS-1 is critical for the endodermal fate. We focus on the SGP division to investigate the control of SYS-1 asymmetry. That control is posttranslational and depends on both frizzled and dishevelled, two conserved components of the Wnt pathway. Taking our results together with those of previous studies, we suggest that the SYS-1:POP-1 ratio is controlled by a forked pathway that drives both high SYS-1 and low POP-1 in the responding cell. The reciprocal asymmetry of SYS-1 and POP-1 provides an elegant way to adjust the SYS-1:POP-1 ratio both rapidly and robustly in daughters of an asymmetric cell division.
Results
VNS::SYS-1 Reporter.
To visualize SYS-1/β-catenin, we made a transgene, called Psys-1::VNS::SYS-1, that encodes wild-type SYS-1 tagged with VENUS yellow fluorescent protein (Fig. 1C). In our attempts to make a SYS-1 reporter, most transgenic lines were lethal, sick, or produced no detectable protein (A.R.K., unpublished data). The lethal and sick lines were likely the result of transgene overexpression, because they were frequent when DNA was injected at high concentrations; when the concentration was dropped, most lines did not express the reporter. VNS::SYS-1 was the exception, in that it was both detectable and viable. The level of VNS::SYS-1 expression is in fact low, and its signal is easily bleached. Therefore, our analyses rely on still images rather than continuous observation. Importantly, VNS::SYS-1 rescued gonadogenesis defects of sys-1(q544) mutants and rendered them fertile. Furthermore, VNS::SYS-1 did not induce extra gonadal arms, a hallmark of SYS-1 overexpression (6). We conclude that VNS::SYS-1 is functional and that its distribution is likely to be representative of endogenous protein.
SYS-1 Asymmetry in the SGP Lineage.
VNS::SYS-1 was not found in SGPs of newly hatched L1 larvae (n = 23) but was detectable ≈1 h before their division (n = 48) (Fig. 1D). VNS::SYS-1 was present in both nucleus and cytoplasm but was enriched in granules often localized to the proximal edge of the SGP (Fig. 1D). During the SGP division, VNS::SYS-1 localized to large puncta in each of the two daughters, at a position suggesting localization to the centrosome; no SYS-1 asymmetry was seen at this point. After the division, VNS::SYS-1 was easily seen in distal daughters but either was not seen or was barely detectable in proximal daughters (Fig. 1E). In the next division, VNS::SYS-1 was in the distal-most SGP granddaughters (Z1aa, Z4pp) but not their proximal sisters (Z1ap, Z4pa); by contrast, VNS::SYS-1 was not in SGP proximal granddaughters (Z1pa, Z1pp, Z4aa, Z4ap).
The relative abundance of VNS::SYS-1 was estimated by using ImageJ software (http://rsb.info.nih.gov/ij). For this quantitation, signal in each SGP daughter was normalized to background fluorescence in germ cells. Signal in distal SGP daughters was, on average, 25-fold higher than that in the proximal sister (n = 8). The broad range of relative abundance (Fig. 1F) may reflect changes during the cell cycle or variability between experiments. We conclude that VNS::SYS-1 is distributed asymmetrically to SGP daughters. Importantly, this “SYS-1 asymmetry” is reciprocal to POP-1 asymmetry (see Discussion).
Reciprocal SYS-1 and POP-1 Asymmetries in Other Tissues.
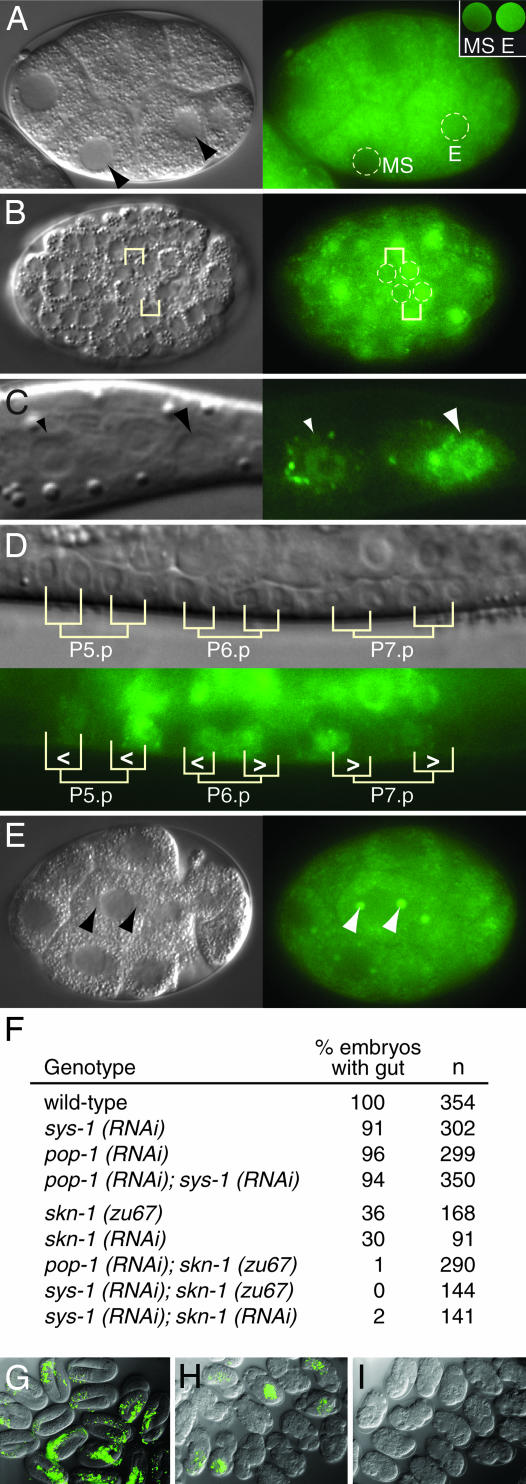
The sys-1 locus affects the development of several tissues: the sys-1 null phenotype is embryonic lethal (6), and weak sys-1 mutants are defective in both SGP and T cell asymmetric divisions (10). Consistent with the idea that SYS-1 functions broadly, VNS::SYS-1 was expressed in many cells during embryogenesis and larval development (Fig. 2). Furthermore, SYS-1 was asymmetrically distributed in daughters of many asymmetric divisions, and SYS-1 asymmetry was the reciprocal of POP-1 asymmetry in all cases. For example, the E blastomere has lower POP-1 than the MS blastomere (11), but E has higher SYS-1 than MS (Fig. 2A). In addition, posterior daughters in the embryonic hypodermis and larval T cell have lower POP-1 than anterior daughters (11), but those posterior daughters have higher SYS-1 than anterior daughters (Fig. 2 B and C). A similar situation holds true for distal daughters and granddaughters in the vulval lineage (12) (Fig. 2D). As seen in the SGPs, SYS-1 localized to large puncta in each daughter at a predicted centrosomal position, and in all cases, SYS-1 levels were similar at the two putative centrosomes (Fig. 2E). Therefore, SYS-1 asymmetry is likely to be induced, once the daughter has formed.
Fig. 2.
SYS-1 asymmetry during embryonic and larval development. (A–E) Nomarski (Left) and fluorescent (Right) images of same embryo or animal. (A) Early embryo. Dotted lines outline nuclei. For clarity, nuclear fluorescence is shown separately in the Inset. Images were treated identically for comparison. (B) Mid-stage embryo. SYS-1 asymmetry was observed in several daughter pairs (two are indicated). Anterior is to the left. (C) L1 larva. VNS::SYS-1 was more abundant in the posterior T cell daughter (large arrowhead) than its anterior sister (small arrowhead). (D) Developing vulva. Vulval precursors and their divisions are marked. Carats show SYS-1 asymmetry, with open end toward daughter with more SYS-1. (E) VNS::SYS-1 is enriched at a position typical of centrosomes during cell division. (F) SYS-1 controls formation of the gut, which is derived from the E blastomere. n, number of embryos scored. (G) Wild-type embryos showing gut autofluorescence (green). (H) skn-1 embryos. Only some embryos make gut (green). (I) skn-1; sys-1(RNAi) embryos. No embryos make gut (green).
SYS-1/β-Catenin Controls Gut Development but Not Spindle Orientation.
In early embryos, the EMS blastomere divides asymmetrically to generate E (endoderm) and MS (mesoderm) daughters. The E daughter is responsible for generating the gut, which is a simple tube composed entirely of endodermal cells; by contrast, MS makes primarily mesodermal derivatives (13). To test whether sys-1 affects E specification, we scored embryos for production of gut after depletion of sys-1 gene activity by RNA-mediated interference (RNAi) (see Methods). Guts were present in all control embryos (Fig. 2 F and G), but they were missing from nearly 10% of sys-1(RNAi) embryos (Fig. 2F). That relatively low penetrance of gutless embryos was comparable with the effect seen in pop-1(RNAi) and pop-1(zu189) mutant embryos (Fig. 2F) (14, 15). In animals depleted for both sys-1 and pop-1 by RNAi, the penetrance remained low (Fig. 2F), which suggests that POP-1 and SYS-1 are part of the same pathway.
The pop-1(RNAi) effect on gut formation is dramatically enhanced by depletion of the SKN-1 transcription factor by using either skn-1 RNAi or a skn-1 mutant (15). We therefore asked whether sys-1(RNAi) could be similarly enhanced. Indeed, intestines were absent from virtually all sys-1(RNAi); skn-1(RNAi) or sys-1(RNAi); skn-1(zu67) embryos (Fig. 2 F and I), an effect comparable with that seen for pop-1(RNAi); skn-1(RNAi) (Fig. 2F) (15). We conclude that sys-1 is critical for the endodermal fate.
The VNS::SYS-1 localization to centrosomes is intriguing, because other Wnt signaling components have been implicated in control of spindle orientation in early embryos. For example, rotation of the EMS spindle from its initial left/right orientation to its final anterior/posterior orientation is delayed in mutants deficient for mom-2 (Wnt ligand), mom-5 (Fz), dsh-1/2 and mig-5 (dishevelled), or gsk-3 (GSK-3β) (16–20). Similarly, the ABar spindle does not align properly in pop-1 or wrm-1 mutants, an effect that appears to rely on transcriptional activation, because similar effects have been observed in mutants deficient for ama-1 (large subunit of RNA polymerase II) (19). To learn whether SYS-1 is required for spindle orientation, we examined EMS and ABar spindles in sys-1(RNAi) embryos. However, no defects were observed (n = 29), indicating that SYS-1 does not affect Wnt-dependent spindle orientations in the early embryo. We suggest that POP-1 may control spindle orientation by working with a different transcriptional coactivator.
SYS-1 Asymmetry Is Controlled Posttranslationally.
To test whether control of SYS-1 asymmetry was transcriptional, translational, or posttranslational, we generated a transgene that is comparable with VNS::SYS-1 but does not produce SYS-1 protein. This transgene, called Psys-1::VNS::SYS-1(stops), includes nonsense codons engineered into exons 1 and 3 (Fig. 3A). In a smg-1 mutant, VNS::SYS-1(stops) is not subject to nonsense-mediated mRNA decay (21). Transgenic lines carrying VNS::SYS-1(stops) did not affect viability and was highly expressed, consistent with a lack of SYS-1 protein. Expression from VNS::SYS-1(stops) shows where vns::sys-1 mRNA is synthesized and translated but expression from the VNS::SYS-1(stops) transgene is impervious to SYS-1 posttranslational regulation.
Fig. 3.
Regulation of SYS-1 asymmetry. (A) Psys-1::VNS::SYS-1(stops) transgene. Conventions as in Fig. 1C. Asterisks, positions of nonsense codons. (B) VNS::SYS-1(stops) is expressed equivalently in both SGP daughters (arrowheads). (C) Regulators assayed for control of SYS-1 asymmetry, including method of depletion. (D) VNS::SYS-1 partially rescues Sys phenotype. (E–H) Nomarski (Upper) and fluorescent (Lower) images of mutants carrying VNS::SYS-1. (I) SYS-1 asymmetry is controlled by frizzled receptors and dishevelled homologs but not by WRM-1 or POP-1. Circle pairs represent SGP daughter pairs. The dark green/white pair represents VNS::SYS-1 asymmetry; the light green pair represents symmetrical VNS::SYS-1 but does not indicate its absolute level. For frizzled, the level was low or undetectable; but for dishevelled, levels were variable (see text for further explanation).
Unlike VNS::SYS-1, the VNS::SYS-1(stops) reporter was seen at equivalent levels in both SGP daughters (Fig. 3B). Because VNS::SYS-1(stops) expresses at a higher level than VNS::SYS-1, we placed VNS::SYS-1(stops) into a wild-type background, which reintroduced nonsense-mediated decay. VNS::SYS-1 abundance was reduced as expected, but the pattern was unchanged: VNS::SYS-1(stops) was still expressed at equivalent levels in both SGP daughters. In addition, it was broadly expressed in both embryos and larvae. In mid-stage embryos, expression was widespread in head, tail, and body hypodermis but low or absent in the developing gut. Postembryonically, VNS::SYS-1(stops) was expressed in the nervous system, hypodermis (including the T cell), developing vulva, and somatic gonad. In all tissues, its expression appeared uniform, in stark contrast to expression of VNS::SYS-1. We conclude that SYS-1 asymmetry is likely to be regulated at the level of SYS-1 protein, perhaps at the level of protein stability.
WRM-1 and POP-1 Do Not Affect SYS-1 Asymmetry.
A divergent Wnt pathway controls the SGP asymmetric division (6, 9, 10, 22). This pathway, called Wnt/MAPK (23), controls diverse asymmetric divisions (reviewed in ref. 24). Its name can be confusing, because no Wnt ligand has been identified for several affected divisions (e.g., SGP), and because the relevant kinase is more closely related to Nemo-like kinase (NLK) than MAPK. Nonetheless, the Wnt part of the pathway does include frizzled receptors, dishevelled homologs, POP-1/TCF, and SYS-1/β-catenin (Fig. 3C), and the MAPK part includes a divergent β-catenin called WRM-1 and the LIT-1/NLK homolog, which together control POP-1 asymmetry (23, 25).
To explore the control of SYS-1 asymmetry, we introduced VNS::SYS-1 into strains defective for relevant constituents of the Wnt/MAPK pathway (Fig. 3C). We first examined VNS::SYS-1 in five mutants, monitoring Sys penetrance by counting gonadal arms as a simple read-out for distal tip cell generation. Previous studies demonstrated that the Sys phenotype is sensitive to both sys-1 dosage and relative abundance of SYS-1 and POP-1 proteins (6, 10). Therefore, even a modest increase in SYS-1 abundance might be expected to have an effect. Indeed, VNS::SYS-1 decreased the Sys penetrance of all five mutants (Fig. 3D). For example, Sys penetrance was 67% in dsh-2 mutants on their own, but only 10% in dsh-2 mutants carrying VNS::SYS-1. Therefore, strains with VNS::SYS-1 appear to have more SYS-1 than normal.
We scored SYS-1 asymmetry in wrm-1; VNS::SYS-1 and pop-1; VNS::SYS-1 larvae, which retained a significant Sys phenotype (Fig. 3D). For both of them, VNS::SYS-1 was dramatically higher in the distal SGP daughter than its proximal sister (Fig. 3 E, F, and I). Thus, SYS-1 asymmetry appeared unaffected by either WRM-1 or POP-1. We also scored SYS-1 asymmetry in lit-1/NLK mutants and again saw no effect (data not shown); however, the near absence of Sys defects in these strains makes this result less compelling (Fig. 3D). The failure of wrm-1 to affect the normal VNS::SYS-1 pattern suggests that the MAPK branch does not control SYS-1 asymmetry. We conclude that SYS-1 asymmetry does not rely on either WRM-1/β-catenin or POP-1/TCF.
Frizzled and Dishevelled Regulate SYS-1 Asymmetry.
The frizzled receptor, LIN-17, causes a partially penetrant Sys phenotype (9, 26), and its paralog, MOM-5, is also implicated in gonadogenesis (27). To enhance Sys penetrance in the presence of VNS::SYS-1, we depleted mom-5 by RNAi in lin-17; VNS::SYS-1 animals. The phenotype changed from a penetrance of 1% Sys in lin-17; VNS::SYS-1 single mutants to 50% Sys in lin-17; mom-5(RNAi); VNS::SYS-1 animals (Fig. 3I). Therefore, LIN-17 and MOM-5 have overlapping functions in control of the SGP division. We next assessed SYS-1 asymmetry. In the single mutant (lin-17; VNS::SYS-1), SYS-1 asymmetry appeared normal (n = 13), but in lin-17; mom-5(RNAi); VNS::SYS-1 animals, SYS-1 asymmetry was often lost (Fig. 3I). Specifically, both SGP daughters expressed low or undetectable VNS::SYS-1 in animals depleted for both frizzled receptors (Fig. 3G). We conclude that the two frizzled receptors are critical for maintenance of SYS-1/β-catenin in distal SGP daughters.
Two dishevelled homologs, DSH-2 and MIG-5, cause partially penetrant Sys phenotypes when removed individually (22, 28). We attempted to deplete both dishevelleds to enhance the Sys phenotype. However, most dsh-2; mig-5(RNAi); VNS::SYS-1 embryos died, and attempts to dilute mig-5(RNAi) met with limited success. In one experiment, penetrance of the Sys phenotype was 37% in the few escapers found (n = 38) with a corresponding effect on SYS-1 asymmetry (Fig. 3I). We therefore examined SYS-1 asymmetry in dsh-2 single mutants despite their low Sys penetrance and found to our surprise that SYS-1 asymmetry was defective in about half of the SGP daughter pairs (Fig. 3 H and I). In dsh-2 mutants, SYS-1 was present at detectable levels in both SGP daughters, suggesting that DSH-2 affects SYS-1 protein levels in both daughters. In one final attempt to increase Sys penetrance, we examined lin-17; dsh-2 double mutants and found an effect similar to that of dsh-2 single mutants: SYS-1 asymmetry was defective with low levels of SYS-1 in both SGP daughters. We conclude that both frizzled and dishevelled activities are critical regulators of SYS-1 asymmetry.
Frizzled and Dishevelled Regulate ceh-22 Expression.
The ceh-22 gene is a direct target of transcriptional activation by SYS-1/β-catenin and POP-1/TCF in SGP distal descendants (8). We therefore asked how ceh-22b expression is affected in frizzled and dishevelled mutants, which alter both SYS-1 asymmetry (this work) and POP-1 asymmetry (10, 22). Normally, Pceh-22b::VENUS is bright in the distal SGP daughter and remains high in the distal granddaughters (Z1.aa, Z1.ap, Z4.pa, Z4.pp) (8). We therefore scored ceh-22-expressing cells in early L2 somatic gonads depleted for frizzled or dishevelled. The reporter was expressed in the distal granddaughters of both SGPs in all wild-type animals (100%, n = 23). By contrast, in lin-17; Pceh-22b::VENUS and lin-17; mom-5(RNAi) Pceh-22b::VENUS larvae, ceh-22-expressing cells were seen in 90% (n = 10) and 42% (n = 24) of larvae, respectively; for dsh-2; Pceh-22b::VENUS, they were found in only 36% (n = 25) of larvae. These numbers correspond well with Sys penetrance in animals expressing Pceh-22b::VENUS (lin-17: 5% Sys penetrance; lin-17; mom-5(RNAi), 74% Sys penetrance; dsh-2, 50% Sys penetrance). We conclude that frizzled and dishevelled regulators affect ceh-22 expression. The simplest interpretation is that the regulation of SYS-1 and POP-1 asymmetry by frizzled and dishevelled homologs affects the transcriptional activation of the target genes of SYS-1/β-catenin and POP-1/TCF.
Discussion
SYS-1/β-Catenin Broadly Controls Asymmetric Cell Divisions.
Asymmetric cell divisions occur throughout C. elegans development (13, 29–31). Many of these asymmetric divisions rely on Wnt/MAPK signaling: the EMS division in early embryos (16, 17), hypodermal divisions in later embryogenesis (11, 32, 33), the SGP division in the newly hatched larva (9), and various other postembryonic hypodermal divisions, including V and T cells (34–37), the vulval lineage (27, 38, 39), and B cells (40).
SYS-1/β-catenin affects both SGP and T cell divisions (7, 10), and sys-1 null mutants die as mid-stage embryos (6). Here we show that SYS-1 asymmetry is typical of many asymmetric divisions throughout development, including daughters of the EMS division. In addition, we find that SYS-1/β-catenin controls formation of the gut, which derives exclusively from the E blastomere. Therefore, SYS-1/β-catenin is likely to function together with POP-1 in many and perhaps most asymmetric divisions during development.
The relevance of a Wnt/MAPK-like pathway to vertebrate asymmetric divisions is not known. Canonical Wnt signaling has been implicated in the control of vertebrate stem cells, which may divide asymmetrically (41–45), and a variant called the planar cell polarity pathway certainly affects asymmetric divisions (see ref. 46). In Xenopus and sea urchins, NLK homologs antagonize TCF activity, whereas zebrafish NLK affects cell fates (47). Therefore, a broader role for a Wnt/MAPK-like pathway in controlling asymmetric divisions may be emerging.
Reciprocal Asymmetry of SYS-1/β-Catenin and POP-1/TCF.
Asymmetric divisions generate daughters with distinct fates (Fig. 4A). In C. elegans, the most common pathway-controlling asymmetric division is Wnt/MAPK signaling. The terminal-pathway-specific regulators that effect differential transcription in the two daughters are SYS-1/β-catenin and POP-1/TCF (reviewed in refs. 4, 6, 8, and 10). A major conclusion of our findings is that SYS-1/β-catenin and POP-1/TCF have a reciprocal distribution in the daughters of asymmetric divisions and that daughters with higher SYS-1 and lower POP-1 activate expression of a target gene (Fig. 4A).
Fig. 4.
Reciprocal asymmetry of both POP-1 and SYS-1 and its control. (A) Reciprocal asymmetry of POP-1 and SYS-1. Nuclear POP-1 is higher in cell B, and cellular SYS-1 is higher in cell A. The lower POP-1 and higher SYS-1 transcriptionally activates target genes in cell A. PC, precursor cell. (B) Limiting coactivator model (Upper) and reciprocal asymmetry model (Lower). (C) A forked Wnt/MAPK pathway elevates SYS-1 and lowers POP-1 to dramatically change the SYS-1:POP-1 ratio and control transcriptional activation of target genes (see text for further explanation).
Previous studies showed that POP-1/TCF levels are lower in the nucleus of daughters that require POP-1/TCF activity for fate specification (10, 14, 16, 48, 49). This enigma was explained by discovery of SYS-1/β-catenin, a dose-sensitive regulator that, together with POP-1/TCF, acts to transcriptionally activate target genes (6, 8). In vitro, the ratio of SYS-1/β-catenin to POP-1 controls their transcriptional activity, and in vivo, the levels of SYS-1/β-catenin dictate its activity (6). These findings led to a model in which the abundance of POP-1/TCF was lowered to accommodate a limiting concentration of SYS-1/β-catenin (6). In its simplest guise, this model suggests that SYS-1/β-catenin would be maintained at a uniformly low level in both daughters of the asymmetric cell division and that the ratio of SYS-1:POP-1 would be controlled solely by changing POP-1 abundance (Fig. 4B Upper).
In this article, we report that SYS-1 levels are higher in the daughter that requires SYS-1 and POP-1 for fate specification and lower in cells that do not require them (Figs. 1 and 4A). Therefore, SYS-1 is distributed asymmetrically, and that asymmetry is the reciprocal of POP-1 asymmetry. The discovery of SYS-1 asymmetry eliminates the simplest form of the limiting transcriptional coactivator model (Fig. 4B Upper). We therefore propose a different model, based on reciprocal SYS-1 and POP-1 asymmetries (Fig. 4B Lower). Although we suggest that SYS-1 and POP-1 levels may be similar in the activated cell, which is the simplest version of this model, the relative abundance of SYS-1 and POP-1 is not known.
Frizzled and Dishevelled Homologs Regulate the SYS-1:POP-1 Ratio.
Previous studies showed that frizzled and dishevelled homologs control POP-1 asymmetry in many asymmetric divisions, including EMS, T cells, and SGPs (refs. 10, 16, 19, 22, and 24, and this work). Here, we demonstrate that SYS-1 asymmetry is also controlled by frizzled and dishevelled homologs (Figs. 3 and 4C). Indeed, the same homologs (fz: LIN-17, MOM-5; dsh: DSH-2, MIG-5) control both SYS-1 and POP-1 asymmetries.
In animals that lack either frizzled or dishevelled homologs, SYS-1 asymmetry is often abolished in SGP daughters. In the distal daughter, which requires SYS-1/β-catenin for its fate, both frizzled and dishevelled homologs control SYS-1 accumulation; but intriguingly, in the proximal daughter, dishevelled may also play a role in SYS-1 disappearance, which suggests that dishevelled has a more complex role in SYS-1 regulation than expected. Whatever that role may be, both frizzled and dishevelled homologs influence SYS-1 asymmetry, and their effects are opposite on SYS-1 and POP-1 (Fig. 4C). Frizzled and dishevelled homologs therefore profoundly affect the SYS-1:POP-1 ratio, which is critical for transcriptional activation (6). To test whether transcriptional activation was affected by frizzled and dishevelled, we examined expression from the ceh-22b promoter, which is a direct target of SYS-1 and POP-1 (8). Indeed, ceh-22b expression was dramatically reduced in frizzled- and dishevelled-depleted animals. Therefore, both frizzled and dishevelled homologs affect the ability of their terminal regulators (SYS-1/β-catenin and POP-1/TCF) to activate transcription.
Distinct Branches of Control for SYS-1 and POP-1 Asymmetry.
POP-1 asymmetry is controlled by the MAPK branch, which is composed of WRM-1/β-catenin and LIT-1/NLK (23, 25, 50). By contrast, SYS-1 asymmetry is not controlled by WRM-1 (this work). Because WRM-1 and LIT-1 work together (25), the simplest interpretation is that SYS-1 asymmetry is not controlled by the MAPK branch.
The regulators of SYS-1 asymmetry that act downstream of dishevelled remain unknown, but two lines of evidence suggest that they control SYS-1 stability. First, a reporter transgene that does not make SYS-1 protein [VNS::SYS-1(stops)] was expressed uniformly in SGP daughters, which suggests that SYS-1 asymmetry is controlled at the level of SYS-1 protein rather than the gene or mRNA. Second, SYS-1 protein is present at equivalent levels in centrosomal positions of both daughters during division, but it virtually disappears from the proximal SGP daughter after division. The idea that SYS-1 is degraded in proximal SGP daughters is attractive, in part because canonical Wnt signaling stabilizes β-catenin in vertebrates by inhibiting the β-catenin elimination of the destruction complex (axin, APC, GSK-3β). The destruction complex itself does not appear to affect SYS-1 asymmetry, at least in the SGP, because no effect on SGPs was observed in mutants or RNAi of destruction complex components (ref. 51; B.T.P., unpublished work). Therefore, SYS-1 stability may be regulated by a divergent destruction complex or a distinct mechanism. The elucidation of the mechanism controlling SYS-1 asymmetry is an important challenge for future work.
Methods
Strains.
C. elegans Bristol strain N2 was used as wild type. Fig. 3C lists most mutations used in this study. SU307 [skn-1(zu67) IV/nT1[qIs51] IV;V] was used to generate skn-1(zu67) homozygotes; NG3124 [dsh-2(or302)/mIn1[dpy-10(e128) mIs14] II] was used for dsh-2 homozygotes.
Transgenes.
Psys-1::VENUS::SYS-1 (pJK1105) includes genomic sys-1 (3.6 kb upstream, all exons and introns, 1.1 kb downstream) with VENUS ORF inserted between codons one and three of first exon; qEx554 was integrated to generate qIs94 and qIs95(III). Psys-1::VENUS::SYS-1(stops) (pJK1186) was made by site-directed mutagenesis of pJK1105, changing codons 3 and 92 into stop codons. qEx654 was integrated to generate qIs130.
SYS-1 and ceh-22 Expression.
Synchronized L1 larvae were plated on bacteria for 18–36 h at 15°C and scored for reporter expression soon thereafter. For wrm-1(ne1982) and lit-1(or131), L1s were raised at 25°C for 12 h. Feeding RNAi was done by means of standard methods.
Embryo Analyses.
Nomarski images were collected as described in ref. 19 and analyzed by using modified versions of NIH Image and ImageJ (available on request from J.H.). To assess endoderm induction, RNAi was performed by injection of L4 or young adults with 2 μg/μl double-stranded RNA synthesized by standard methods. For SYS-1 expression, live embryos were digitally recorded, and fluorescent images were captured in nascent sister cells after division.
Image Quantitation.
Images were captured on a Hamamatsu (Hamamatsu City, Japan) C4742-95 camera, using OpenLab 5.0.1 (Improvision, Lexington, MA). ImageJ software was used to outline the area to be quantitated in a Nomarski image, and the level of fluorescence was measured in the corresponding fluorescence image. The entire cell was included for VNS::SYS-1, but only the nucleus was included for GFP::POP-1. The fold difference between distal and proximal SGP daughters was calculated as the ratio of net distal daughter fluorescence to net proximal daughter fluorescence, in which net fluorescence was calculated by subtracting background in nearby germ cells from total signal in the cell of interest. For nuclear GFP::POP-1 quantitation, the inverse calculation was used to find the fold difference between the proximal and distal nuclei (net proximal/net distal). ImageJ quantitation of SGP daughter pairs in VNS::SYS-1(stops) varied from 0.89 to 0.99 (n = 4 cell pairs).
Acknowledgments
We thank members of the J.K. laboratory for comments on the manuscript and A. Helsley-Marchbanks and L. Vanderploeg for manuscript and figure preparation. This work was supported by National Institutes of Health Postdoctoral Fellowship F32 GM075598 (to B.T.P.), National Institutes of Health Predoctoral Training Grant in Molecular Biosciences T32 GM07215 (A.R.K. and R.K.), and National Science Foundation Grant IOB 0518081 (to J.H.). J.K. is an Investigator of The Howard Hughes Medical Institute.
Abbreviations
- NLK
Nemo-like kinase
- SGP
somatic gonadal precursor cell.
Footnotes
The authors declare no conflict of interest.
References
- 1.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Shetty P, Lo M-C, Robertson SM, Lin R. Dev Biol. 2005;285:584–592. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 3.van Noort M, Clevers H. Dev Biol. 2002;244:1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- 4.Herman MA, Wu M. Front Biosci. 2004;9:1530–1539. doi: 10.2741/1306. [DOI] [PubMed] [Google Scholar]
- 5.Korswagen HC, Coudreuse DYM, Betist MC, van de Water S, Zivkovic D, Clevers HC. Genes Dev. 2002;16:1291–1302. doi: 10.1101/gad.981802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd AR, III, Miskowski JA, Siegfried KR, Sawa H, Kimble J. Cell. 2005;121:761–772. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Miskowski J, Li Y, Kimble J. Dev Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- 8.Lam N, Chesney MA, Kimble J. Curr Biol. 2006;16:287–295. doi: 10.1016/j.cub.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegfried K, Kimble J. Development (Cambridge, UK) 2002;129:443–453. doi: 10.1242/dev.129.2.443. [DOI] [PubMed] [Google Scholar]
- 10.Siegfried KR, Kidd AR, III, Chesney MA, Kimble J. Genetics. 2004;166:171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin R, Hill RJ, Priess JR. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- 12.Deshpande R, Inoue T, Priess JR, Hill RJ. Dev Biol. 2005;278:118–129. doi: 10.1016/j.ydbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Sulston JE, Schierenberg E, White JG, Thomson JN. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 14.Lin R, Thompson S, Priess JR. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 15.Maduro MF, Kasmir JJ, Zhu J, Rothman JH. Dev Biol. 2005;285:510–523. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Priess JR, Mello CC. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 17.Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 18.Schlesinger A, Shelton CA, Maloof JN, Meneghini M, Bowerman B. Genes Dev. 1999;13:2028–2038. doi: 10.1101/gad.13.15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, Wood W, Hardin J. Dev Cell. 2004;7:831–841. doi: 10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Walston TD, Hardin J. Semin Cell Dev Biol. 2006;17:204–213. doi: 10.1016/j.semcdb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Grimson A, O'Connor S, Newman CL, Anderson P. Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang W, Lloyd CE, Zarkower D. Mech Dev. 2005;122:781–789. doi: 10.1016/j.mod.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 24.Herman MA. Semin Cell Dev Biol. 2002;13:233–241. doi: 10.1016/s1084-9521(02)00051-4. [DOI] [PubMed] [Google Scholar]
- 25.Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 26.Sternberg PW, Horvitz HR. Dev Biol. 1988;130:67–73. doi: 10.1016/0012-1606(88)90414-9. [DOI] [PubMed] [Google Scholar]
- 27.Gleason JE, Szyleyko EA, Eisenmann DM. Dev Biol. 2006;298:442–457. doi: 10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 28.Walston T, Guo C, Proenca R, Wu M, Herman M, Hardin J, Hedgecock E. Dev Biol. 2006;298:485–497. doi: 10.1016/j.ydbio.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 29.Sulston JE, Albertson DG, Thomson JN. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- 30.Kimble J, Hirsh D. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 31.Sulston JE, Horvitz HR. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 32.Park CS, Kim SI, Lee MS, Youn C-y, Kim DJ, Jho E-h, Song WK. J Biol Chem. 2004;279:19592–19599. doi: 10.1074/jbc.M314208200. [DOI] [PubMed] [Google Scholar]
- 33.Bischoff M, Schnabel R. PLoS Biol. 2006;4:e396. doi: 10.1371/journal.pbio.0040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MA, Horvitz HR. Development (Cambridge, UK) 1994;120:1035–1047. doi: 10.1242/dev.120.5.1035. [DOI] [PubMed] [Google Scholar]
- 35.Whangbo J, Harris J, Kenyon C. Development (Cambridge, UK) 2000;127:4587–4598. doi: 10.1242/dev.127.21.4587. [DOI] [PubMed] [Google Scholar]
- 36.Takeshita H, Sawa H. Genes Dev. 2005;19:1743–1748. doi: 10.1101/gad.1322805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstein B, Takeshita H, Mizumoto K, Sawa H. Dev Cell. 2006;10:391–396. doi: 10.1016/j.devcel.2005.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenmann DM, Kim SK. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. Development (Cambridge, UK) 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- 40.Wu M, Herman MA. Dev Biol. 2006;293:316–329. doi: 10.1016/j.ydbio.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 41.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Joh K, Hedgecock EM, Hori K. DNA Seq. 1999;10:207–217. doi: 10.3109/10425179909033950. [DOI] [PubMed] [Google Scholar]
- 43.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 44.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 46.Barrow JR. Semin Cell Dev Biol. 2006;17:185–193. doi: 10.1016/j.semcdb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Thorpe CJ, Moon RT. Development (Cambridge, UK) 2004;131:2899–2909. doi: 10.1242/dev.01171. [DOI] [PubMed] [Google Scholar]
- 48.Herman MA. Development (Cambridge, UK) 2001;128:581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- 49.Maduro MF, Lin R, Rothman JH. Dev Biol. 2002;248:128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- 50.Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Bei Y, Davis RJ, Mello CC. Mol Cell. 1999;4:275–280. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- 51.Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. Development (Cambridge, UK) 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]