Abstract
As researchers attempt to study quorum sensing in relevant clinical or environmental settings, it is apparent that many factors have the potential to affect signaling. These factors span a range of physical, chemical, and biological variables that can impact signal production, stability and distribution. Optimizing experimental systems to natural or clinical environments may be crucial for defining when and where quorum sensing occurs. These points are illustrated in our case study of S. aureus signaling in biofilms, where signal stability may be affected by the host environment. The basic signaling schemes have been worked out at the molecular level for a few of the major quorum-sensing systems. As these studies continue to refine our understanding of these mechanisms, an emerging challenge is to identify if and when the local environment can affect signaling.
Keywords: Clinical/Biomedical analysis, Cell systems/Single cell analysis, Biosensors, Biofilm, Quorum sensing
Introduction
As illustrated by many articles in this special issue, the diversity of mechanisms bacteria use to communicate with one another is remarkable. Quorum sensing is widespread among different bacterial species, emphasizing the importance of coordinating behavior as a group.
When considering the relevant context of quorum sensing, it is clear that the environment can be profoundly important. Many bacteria exist in spatially structured, multi-species communities, such as biofilms on submerged surfaces or flocs in the water column of aquatic environments [1, 2]. Within these communities bacteria may achieve the high cell numbers capable of generating the local signal concentrations required to produce a quorum sensing response. Quorum sensing undoubtedly occurs in situations where bacteria are growing planktonically, for example Vibrio harveyi quorum sensing-controlled bioluminescence in the ocean, thought to produce the “milky seas” phenomenon [3, 4]. An argument could, however, be made that quorum sensing is more likely to occur in structured communities. If the reader accepts this point, further considerations lead to some interesting questions for example:
Are certain signals better suited for signaling in different types of environments?
What are the important parameters influencing signaling in different environments?
Take, as an example, the well-studied, environmentally ubiquitous Pseudomonas aeruginosa. This bacterium has been isolated from a range of soil and aquatic samples [5–7]. The nature of signaling in these environments could be quite different, because signal production and diffusion dynamics may change.
This review will discuss factors that may affect signaling in structured microbial communities. In addition, quorum sensing in Staphylococcus aureus, a peptide-based signaling system, will be briefly reviewed and its role in biofilm formation will be discussed.
Factors affecting signal diffusion, stability, and distribution
The traditional way of studying quorum sensing in the laboratory is in shaken liquid-batch cultures. The quorum sensing response occurs at a specific point in the growth curve, coinciding with a threshold concentration of signal. Volume is fixed in a culture flask and signal molecules produced by bacteria usually increase in concentration with time (unless they are degraded). Batch cultures represent a closed system in which only gaseous exchange occurs. Most structured communities are in open systems in which exchange occurs with the surrounding liquid [1]. In an open, flowing system, signals may be “washed” away by the overlying fluid. In open systems the concentration of signal molecules within structured communities is primarily a function of:
signal-production rate;
the degradation rate or half-life of the signal;
the diffusion properties of the signal; and
the external hydrodynamic or mass-transfer conditions.
The prevailing environmental conditions and resident biology are, to different degrees, important for each of these processes.
The effects of signal and environmental chemistry on quorum sensing
Some of the best-studied quorum-sensing signals include the acyl-homoserine lactones (AHL) used by many Gram-negative bacterial species, small peptides used by some Gram-positive species, and the furans of the phylogenetically widely spread AI-2 signaling (reviews are available elsewhere [8–10]). Signal chemistry varies widely in different quorum-sensing systems and is important for determining its stability and diffusion characteristics in different environments.
All AHL signals have a characteristic homoserine lactone moiety. The homoserine lactone is derived from S-adenosylmethionine, one of the substrates for AHL synthesis [11–13]. Although stable at neutral and slightly acidic pH, the lactone ring is subject to chemical hydrolysis under basic conditions [14, 15]. The resulting product (e.g. butyryl-homoserine lactone would become butyryl-homoserine) lacks biological signaling activity and is capable of spontaneously re-lactonizing when the pH is lowered. The formula describing the relationship between pH and the lactone ring stability is 1/(1×107×[OH−]) [14], which gives the half life in days. For example, at pH 7 the homoserine lactone ring would be stable for hours whereas at pH 8.5 the stability would be of the order of minutes (Fig. 1). In many environments this might be insignificant. In some alkaline environments, however, like the photosynthetic mats at Octopus Springs in Yellowstone National Park, the pH is high enough to potentially affect signal gradients. Kaufmann et al. recently demonstrated that in addition to lactone hydrolysis, the 3-oxo-C12 AHL of P. aeruginosa is capable of spontaneously undergoing a Claisen-like condensation reaction in aqueous environments, forming a tetramic acid product, 3-hydroxydecylidene 5-(2-hydroxyethyl) pyrrolidine-2,4-dione (Fig. 2) [16]. This compound was shown to have significant antimicrobial activity against other species and a high affinity for iron. It was proposed this iron-binding activity constituted an alternative means of acquiring iron by P. aeruginosa.
Fig. 1.
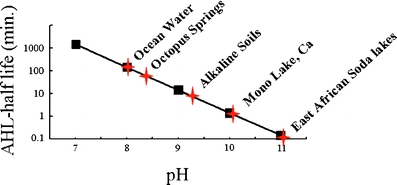
Predicted AHL half-lives in different alkaline environments
Fig. 2.
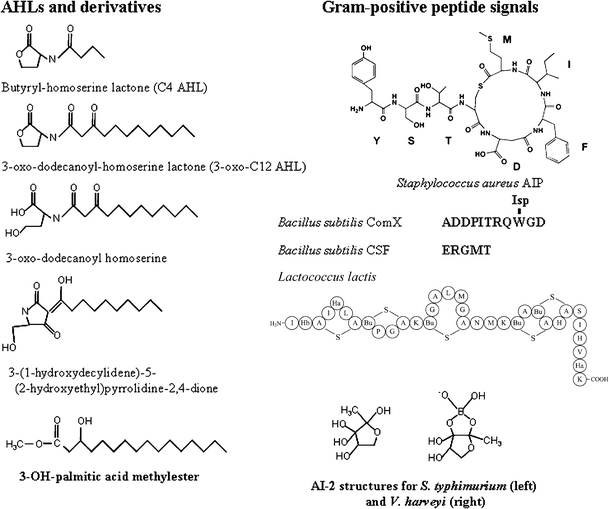
Structures of quorum-sensing signals and their derivatives. Letter designations for the Gram-positive peptide signals indicate amino acids. For the L. lactis signal nisin, the structural abbreviations were: Bu, dehydrobutirine with a lanthionine bridge; Ha, dehydroalanine; Hb, dehydrobutirine
Structured communities can create highly heterogeneous localized niches where chemistry can vary drastically over very small distances. For example, consumption of dissolved oxygen by laboratory biofilm cells can result in completely anaerobic regions within the biofilm only 100 μm from the fully oxygen saturated overlying liquid [17]. Similarly, pH can vary dramatically, as demonstrated by pH microelectrodes positioned in biofilms formed from the dental pathogen Streptococcus mutans. On addition of the growth substrate sucrose, the pH dropped from 7 outside the biofilm to 4.6 approximately 100 μm deep within the biofilm (Gieseke A, Nguyen D, von Ohle C, Stoodley P; unpublished results). Clearly, if the half-life of signaling molecules is a function of local chemistry it is reasonable to expect that degradation rates will vary in the community.
The acyl side-chains of AHLs range in length from 4 to 18 carbon atoms [18, 19]. The substitution chemistry of the acyl side-chain is also highly variable. Although most AHLs have a ketone group at the C1 position, some AHLs also have either a ketone or hydroxyl group at the C3 position. In addition, the acyl side-chain can vary in saturation. Although most AHLs have fully saturated acyl side-chains, some, for example the 3-hydroxy,7-cis-tetradecanoyl homoserine lactone of Rhizobium leguminosarum, are unsaturated [19, 20]. All these features of AHL acyl side-chain chemistry, in particular acyl chain length, affect the relative hydrophobicity of the signal. Pearson et al. observed this for P. aeruginosa, which produces two major AHL signals, a C4 (i.e. four-carbon side-chain) and 3-oxo-C12 AHL (Fig. 2) [21]. They found that the C4 signal diffused freely across the cell membrane whereas the 3-oxo-C12 signal partitioned to the cell membrane, where its extrusion was aided by the activity of the MexAB-OprM efflux pump.
The hydrophobicity of the local environment could have a large effect on AHL diffusion properties. For example, several AHL-utilizing bacterial species are also capable of producing highly hydrophobic, secreted polysaccharides (e.g. the pel/psl polysaccharides of P. aeruginosa) [22]. In a structured community, such as a biofilm, secreted polysaccharides can serve as the scaffolding holding the community together [23]. Under such conditions these polysaccharides might also act as an AHL-sequestering matrix. Charlton et al. provided evidence of this, indicating that the 3-oxo-C12 signal of P. aeruginosa partitioned into the extracellular biofilm matrix [24]. Perhaps the C4 signal has reduced reactivity with the biofilm matrix, making it a more effective signal in a biofilm and explaining why it is produced in greater amounts than 3-oxo-C12 by P. aeruginosa growing as a biofilm [25].
Another factor affecting AHL stability in the environment is biologically-driven signal degradation. Emphasizing this point, Wang et al. demonstrated that radiolabled AHLs fed to soil microbial communities were almost instantly mineralized [26]! Several bacterial species have the ability to use AHLs as carbon and nitrogen sources. These include species that produce their own AHLs and those known not to make them. Why some organisms both make and break down their own AHLs is a mystery. AHL degradation occurs as a result of two primary types of enzymatic activity, lactonases, which break the homoserine lactone ring, and acylases, which cleave the amide bond linking the acyl side-chain to the homoserine lactone ring [27–31]. Biological AHL degradation is a key consideration in multi-species environments. Leadbetter proposed the concept of “insulation” in which AHL-degrading organisms could prevent AHLs produced by bacteria spatially fixed in one location from reaching other bacteria of the same species [26].
Environmental chemistry also plays a key role in AI-2 signaling. The paradigm for this type of quorum sensing is the luxS system of Vibrio harveyi [8]. One of the key steps in AI-2 signal synthesis is the LuxS-catalyzed conversion of S-ribosyl homocysteine to homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) [32]. DPD is believed to be the key precursor to all AI-2 signals. DPD undergoes non-enzymatic cyclization and a number of spontaneous, reversible rearrangements to form the active signal-a furanosyl borate diester and other furans [33, 34]. The AI-2 signal for Salmonella typhimurium was recently solved and found to be, (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, a molecule related to, but structurally distinct from, the V. harveyi AI-2 (Fig. 2) [35]. Spontaneous extracellular chemical rearrangements of S. typhimurium AI-2 can produce the V. harveyi AI-2 signal; the reverse is also true [35]. Current thinking is that the local chemical environment can affect the ratio of the different furanosyl esters. In particular, the presence of borate may shift the equilibrium toward formation of the V. harveyi AI-2 [35].
The chemical structures of the Gram-positive quorum-sensing peptides vary greatly in the number of residues and the types of modification (Fig. 2). Because of the post-translational modifications on these peptides and their inability to diffuse across membranes, their biosynthetic pathways are more complex than those of the AHLs and dedicated signal-export systems are usually required. Many also induce their own biosynthesis, and are thus frequently termed “autoinducing peptides”. Several signals are simple linear peptides, for example the 17-residue competence-stimulating peptide (CSP) which regulates competence in Streptococcus pneumoniae [36]. The corresponding linear competence pheromone in Bacillus subtilis, called ComX, is more unusual in that an internal tryptophan residue has been cyclized and isoprenylated with a geranyl moiety [37], a modification that is catalyzed by ComQ [38].
Perhaps the largest and most complex peptide signals are the lantibiotics, which also have antimicrobial activity [39]. These molecules, for example nisin produced by Lactococcus lactis, are known to positively regulate their own biosynthesis in a density-dependent manner. They also have extensive post-translation modifications, including dehydrated residues, such as dehydroalanine and dehydrobutyrine, and thioether bridges called lanthionines (Fig. 2 depicts the base structure of the lantibiotic, nisin, of L. lactis). An emerging class of peptide signals are the cyclic lactones and thiolactones [40]. The first of this class to be discovered was the type I autoinducing peptide (AIP) of Staphylococcus aureus, an eight-residue thiolactone-containing peptide with the C-terminus constrained through linkage with a cysteine side-chain [41]. Since the discovery of AIPs, related signals have been identified in Enterococcus faecalis [42], Listeria monocytogenes [43], and other Staphylococci [44].
Environmental conditions can have a significant effect on peptide signal stability. The linear peptides are likely to have short half-lives as they are rapidly metabolized by the action of secreted proteases. Indeed, this issue has led some to question their role as a means of communication, raising the possibility they serve as a general strategy for a single cell to time regulatory events [45]. In contrast, the lantibiotics have multiple lanthionine bridges that block the activity of many proteases [46], greatly improving the longevity of these molecules. Like AHLs, lantibiotics are pH-sensitive, with solubility and stability dropping rapidly at higher pH; those with dehydrated residues are also reactive with thiols [47]. Because of these chemical constraints, the lantibiotic nisin has a half-life of only 0.9 h in mouse serum [48], the pH of which is approximately 7.3. For L. lactis in the environment, however, growth and production of nisin are optimum at lower pH, at which stability is greater, facilitating autoinduction. In a similar fashion, the thiolactone-containing peptides are resistant to proteolysis and sensitive to high pH and reactive thiols (chemistry of the S. aureus AIP is discussed in more detail below), and some have increased sensitivity to oxidative damage [49]. Although it remains to be demonstrated, protease resistance may be one of the most significant environmental constraints faced by bacteria using peptide communication signals and extensive post-translational modifications could have evolved to overcome this problem.
The effect of the hydrodynamic environment on quorum sensing
Mass transfer is the physical process by which molecules are transported in a system, and it has the potential to affect cell-to-cell signaling in many ways. Delivery of nutrients to the active biomass in the structured community is one example. This could affect the relative metabolic activity of the community, which in turn has the potential to affect signal-production rates. Mass transfer is affected by the hydrodynamics of the bulk fluid and the geometry of the structured community. These two factors affect each other, because a microbial community both shapes, and is shaped by, its external environment.
If liquid flow is a feature of the environment, it will wash signal away, diluting its concentration within the community. In these circumstances the system can be divided into three zones (Fig. 3). First, the biomass of the community which is producing signaling molecules, second, the viscous sublayer (δ) located at the biomass-bulk fluid interface, and, third, the well mixed turbulent core of the overlying fluid. In laminar flow there is no turbulent core and δ essentially extends to either the center of the channel if flow is in a closed conduit (e.g. pipe or catheter) or to the fluid surface if flow is in an open channel (e.g. river). Using this simplified system we have constructed a one-dimensional model based on a flat biofilm to illustrate how different values of δ may result in different concentration profiles while the thickness of the biofilm, the cell density in the biofilm, and the rate of signal-production in the biofilm remain constant. The model is based on coupling the diffusion of signal through the viscous sublayer with the simultaneous diffusion and production of signal that occurs within the biofilm itself. In the fluid outside the biofilm, the concentration of signal changes linearly:
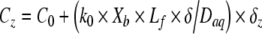 |
where Cz is the signal concentration (mg L−1) at depth z, C0 is the signal concentration outside the viscous sublayer (0 mg L−1), k0 is the signal production rate (2.96×10−17 mg cell−1 s−1), Xb is the active cell density in the biofilm (1×1010 CFU cm−3), Lf is the biofilm thickness (50 μm), δ is the thickness of the viscous sublayer (0, 25, or 50 μm), Daq is the diffusion coefficient of the signal in water (4.9×10−6 cm2 s−1), and δz is the proportional depth within δ (μm). Within the biofilm, the concentration of the signal changes quadratically with depth:
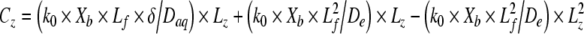 |
where De is the effective diffusion coefficient of signal in the biofilm (1.23×10−6 cm2 s−1) and Lz is the proportional depth within the biofilm (μm).
Fig. 3.
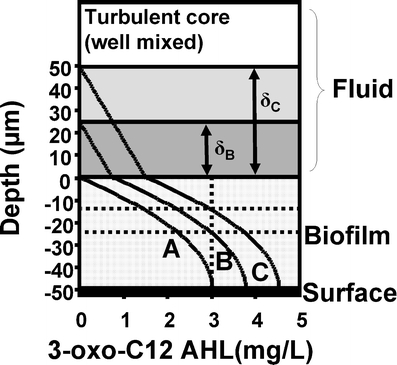
Schematic diagram illustrating how external flow may affect the concentration of a cell signal produced by biofilm cells. The biofilm is 50 μm thick (so there is no oxygen limitation and we can assume equal signal production throughout the depth of the biofilm) and the relative concentration profiles are for three flow conditions: (a) Very high shear, negligible diffusion boundary layer (δ). (b) Lower flow with a diffusion boundary layer of 25 μm (δB). (c) Lower flow, again, with a diffusion boundary layer of 50 μ (δC). The model is steady-state Fickian diffusion for flat-slab geometry with a uniform density of bacteria producing signals at a constant rate with no degradation. There is no convection within the biofilm and no flux at the substratum. The concentration of signal in the turbulent core of the flow is zero. The effective diffusion coefficient in the biofilm is assumed to be 0.25 times that of the bulk fluid based on the molecular weight of 3-oxo-C12 AHL [50]. For simplification the viscous sublayer is shown as the diffusion boundary layer. In reality the transition (the “buffer region” of the boundary layer) from the viscous sublayer to the turbulent core is not a sharp line, as depicted, but gradual. If we assume an inducing 3-oxo-C12 AHL concentration of 3 mg L−1 (10 μmol), depicted by the vertical dashed line, biofilm “A” is not induced at all, in biofilm “B” the bottom 50% is induced, and in biofilm “C” the bottom 75% is induced, as shown by the horizontal dashed lines where the inducing concentration intersects the concentration profile
Where known we used data as best we could, based on the signal 3-oxo-C12 AHL, unknown model data were hypothetical. All data were based on those used for illustrative purposes by Stewart [50].
These considerations led us to hypothesize that external flow conditions can affect quorum sensing in many ways. For example, in a somewhat closed, static system (e.g. a biofilm in a mud puddle or in the well of a microtitre plate), signals produced by a large, biofilm cell aggregate may induce a quorum-sensing response in neighboring bacteria that are not part of the aggregate. In contrast, in an open system subject to convective flow (e.g. a biofilm in a river, or the channel of a flow cell biofilm reactor), signals might be continuously removed from the system. Under these conditions mass transfer may prevent signals produced by a large biofilm cell aggregate from inducing cells in the vicinity of the aggregate. Experimental results on interspecies communications in dental plaque biofilms by Egland et al. [51] support this idea, and suggest that diffusible signals are adapted to function over short distances. At higher flows the higher flux of signal molecules from structured communities may reduce signal concentration in the interior or core of the community. A subsequent consequence is that more biomass would be necessary to produce an inducing concentration of signal.
Few studies have directly addressed the effect of hydrodynamics on quorum sensing in structured communities. Purevdorj et al. [52] found that at relatively high flow rates flow velocity was a stronger determinant of P. aeruginosa biofilm structure than quorum-sensing-required functions. Yarwood et al. [54] grew wild type (WT) and accessory gene regulator (agr) mutant biofilms of S. aureus by batch culture under static conditions, batch culture on spinning disks, and in flow cells. The QS mutation had the greatest effect under static conditions, resulting in an increase in biofilm formation, consistent with the study of Vuong et al. [55], who hypothesized that agr was a repressor of biofilm formation. Interestingly, there was no difference between WT and agr (quorum sensing) mutant biofilms when grown in flow cells. Care must, however, be taken in correlating growth system with phenotype, because media type and concentration were also varied in this study. Nevertheless, Fux et al. [56] obtained similar results for flow cell-grown biofilms. After 24 h the WT S. aureus biofilms had less biomass than a TRAP mutant but were significantly rougher. After 4 days, however, there was little difference between the biofilms as measured by COMSTAT confocal image-analysis software [57]. In the same study the opposite trend, as measured by viable plate counts, was observed for “filter colony biofilms” [58] grown under static conditions on nutrient agar. The WT biofilms contained an order of magnitude more cells than the TRAP mutant. Interestingly, a regulatory network mathematical model has predicted that at intermediate concentrations of 3-oxo-C12, bacteria can suddenly switch between induced and noninduced states [59]. This effect may help explain experimental observation of different biofilm phenotypes under what are assumed to be similar or even replicate growth conditions.
Quorum sensing in the gas phase with volatile signals
Quorum sensing is usually discussed and studied in the context of aqueous environments. Some environmental conditions, however, for example those found in soil, may not always be ideal for signaling in the liquid phase. Soil is subject to wetting and drying cycles, and even somewhat dry soils are teeming with metabolically active bacteria [60, 61]. Under these conditions it may be advantageous to some bacterial species to engage in quorum sensing. Local cell numbers may be quite high, but not linked to one another through the liquid phase. The production of signals able to act through the gas phase would circumvent these issues. There is precedence for volatile signaling in the microbial world. An interesting example is the signaling molecule, 3-OH palmitic acid methyl ester (3-OH PAME) of Ralstonia solanacearum (Fig. 2) [62]. The signal of this quorum-sensing system can act in the gas phase. This plant pathogen regulates the production of secreted polysaccharides (an important virulence factor) by use of 3-OH PAME [63, 64], the synthesis of which is catalyzed by the phcB gene product [63]. When 3-OH PAME concentrations reach ∼5 nmol L−1, 3-OH PAME-regulated genes are induced, presumably because of interaction of the signal with the membrane-associated PhcS sensor kinase. Although homologs of phcB are not found on many of the microbial genomes yet sequenced, signaling in the gas phase may be one of the next important frontiers in quorum sensing.
A case study of signaling in structured communities-Quorum-sensing in Staphylococcus aureus biofilms
Continuing with the themes of this review, a case study will be presented with a discussion on environmental, chemical, and biological factors discussed above that have the potential to affect quorum-sensing in S. aureus biofilms.
S. aureus is a human commensal that resides in a non-pathogenic state in the nasal airways. When there is a breach in the host defenses enabling access, S. aureus can convert to a pathogenic state and secrete an impressive array of toxins, hemolysins, and degratory enzymes [40], causing damage to host tissues. This lifestyle switch is mediated, in a cell-density-dependent manner, by the action of the AIP molecule. Early in the growth phase surface adhesins and antigens are produced, and when AIP reaches a critical concentration the quorum-sensing cascade is activated and these surface proteins are down-regulated and invasive factors are secreted. Microarray studies have revealed that 104 genes are up-regulated and 34 are down-regulated by the action of this quorum-sensing molecule [65]; this represents almost five percent of the genome. Intriguingly, there are four specific classes of AIP molecule that approximately correlate with the type of disease caused by the producing S. aureus strain. In a fascinating mechanism of bacterial cross-talk these different classes of AIP cross-inhibit quorum-sensing in other S. aureus groups and other staphylococcal species [41]; this may serve to isolate specific sub-populations for cooperative action.
The locus responsible for the quorum-sensing regulation in S. aureus is the accessory gene regulator or Agr locus and is known to contain two divergent transcripts, named RNAII and RNAIII [40]. The RNAII transcript is an operon of four genes, agrBDCA, that encode factors required to synthesize AIP and activate the regulatory cascade (Fig. 4). The biosynthetic pathway leading to functional AIP is not clear, but it is known that AgrD is the peptide precursor of AIP and AgrB is a membrane protease involved in its processing [66]. AgrC and AgrA form a typical two-component regulatory pair, and the binding of AIP to a surface receptor on AgrC activates this phosphoryl-transfer cascade [67]. When phosphorylated, AgrA is known to bind and induce expression of the RNAIII transcript [68], which encodes a regulatory RNA molecule that acts as the primary effector of S. aureus quorum-sensing [69]. AgrA also induces expression of the Agr proteins through the RNAII transcript, triggering the autoinduction phenomenon.
Fig. 4.
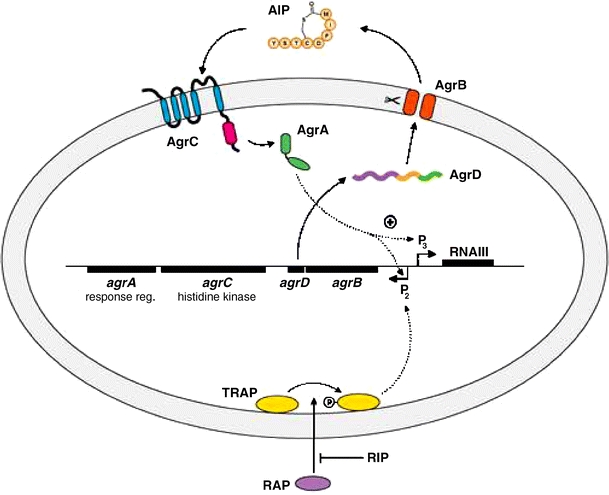
Schematic diagram of quorum-sensing systems of S. aureus. The gene locus for the agr system is shown in black and contains two divergent transcripts, RNAII and RNAIII, driven by the P2 and P3 promoters, respectively. The RNAII transcript encodes the agrBDCA operon, which encodes the signal, processing, and detection components for quorum-sensing in S. aureus (see text). The RNAIII transcript is a regulatory RNA that up-regulates and down-regulates all genes related to quorum-sensing. This transcript also encodes the amphipathic peptide δ-hemolysin. RAP and TRAP are part of a second quorum-sensing system thought to be a precursor of the agr system. RAP is secreted and induces TRAP phosphorylation, which in turn induces expression of the RNAII transcript. RIP is a heptapeptide known to inhibit the ability of RAP to induce TRAP phosphorylation
A second quorum-sensing cascade is thought to serve as a precursor to the Agr system [70], setting the stage for AIP regulation. As S. aureus cells multiply, the RNAIII-activating protein (RAP) is secreted and accumulates outside the cell, and at a threshold concentration, RAP triggers the phosphorylation of the cytoplasmic protein TRAP [71], which induces expression of the RNAII transcript (Fig. 3). Intriguingly, a linear heptapeptide called RIP is known to block the activity of RAP, enabling small-molecule control over S. aureus pathogenicity [72].
Chemistry of the S. aureus AIP signal
The reasons for evolving a cyclic thiolactone structure as a quorum-sensing signal are still not clear. The thioester bond and the presence of other labile amino acids reduce stability, because of the potential for oxidative damage, base-catalyzed hydrolysis, and thioester exchange. Indeed, the methionine residue in AIP is rapidly oxidized in vitro to a methionyl sulfoxide, converting the signal to an inactive byproduct [49]. Similarly, oxidants produced in vivo by the phagocyte NADPH oxidase are known to accelerate this inactivation [73]. Despite these issues, the AIP lifetime in host tissues is reported to be 3 h [74], which is more than adequate for regulation, bearing in mind that S. aureus doubles every 60 min in the host [75].
Structure-function studies have shown that the thiolactone cannot be replaced with a more stabile lactone, suggesting that the signal receptor might require acylation to activate the cascade [76], although a lactam substitution does activate at high concentrations [49]. As discussed above, constraining the peptide will improve metabolic stability to proteases [77], by protecting the C-terminus and impeding access to endoproteases, which probably increases the half-life compared with that of the linear AIP counterpart. Clearly, further study is necessary to determine the physiological benefit of this thiolactone structure.
The relationship between S. aureus quorum-sensing and biofilm communities
Biofilm formation is increasingly being recognized as an important virulence factor in S. aureus pathogenesis. Several biofilm-associated diseases, including osteomyelitis [78], endocarditis [79], medical device infections [80], and potentially even skin infections [81], have much clinical relevance. The formation of S. aureus biofilms progresses in a similar fashion to the Gram-negative biofilms, with attachment followed by development into a highly-structured cell community. Several attachment factors, for example the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) and the surface-attached Atl protein, have been implicated in this initial stage of biofilm formation (reviewed elsewhere [82, 83]). Secretion of a polysaccharide adhesin is thought to be critical for development of a structured biofilm [84], although some biofilm-forming S. aureus strains cannot produce this polymer [85]. Unlike some well-studied Gram-negative bacterial species, S. aureus are non-motile, which leads to significantly reduced biofilm architecture in flowing systems. The significance of this difference is not clear, because simple alterations of growth conditions for P. aeruginosa are known to eliminate higher-ordered biofilm structure [86].
Because the development of a robust P. aeruginosa biofilm under some conditions requires an active quorum-sensing system [53], one might assume the behavior of S. aureus is similar; the opposite seems to be true, however, because inactivation of the Agr system tends to enhance attachment [87]. Agr is known to down-regulate surface attachment factors, providing an explanation of this observation. The Agr cascade also up-regulates both the secretion of proteases that can degrade these attachment factors and the secretion of amphipathic peptides (phenol-soluble modulins) that facilitate detachment [83, 88], suggesting quorum-sensing may play a role in biofilm turnover. Despite these intriguing observations, deciphering the literature on this topic has been challenging, because of non-uniform biofilm culturing methods and strain-to-strain differences [89]. To emphasize this point, enhanced attachment by Agr mutants in static biofilm systems is not observed in flowing biofilm reactor systems [87, 90].
An elegant study by Yarwood et al. addressed some of the discrepancies in the literature and suggested an alterative role for S. aureus quorum-sensing in biofilms [87]. Under static conditions attachment of Agr mutants was markedly better than that of wild-type; as shear force increased with increased flow rate, however, this advantage was lost and under some experimental conditions became a disadvantage. As in P. aeruginosa biofilm studies, the method and type of experiment seemed to dictate the requirement for the Agr system. By following a quorum-sensing promoter in a flow cell biofilm, Yarwood et al. observed that only patches of surface cells activated the Agr cascade. Surprisingly, these cells detached from the biofilm in periodic waves, suggesting an alternative role for the Agr system in the detachment and recycling phase of biofilm development.
In a biofilm infection of the host it seems probable that S. aureus will be subjected to increased flow and/or shear force, in addition to many other factors, which will affect the quorum-sensing phenomenon. Increased flow will probably dictate the local AIP concentration, potentially deactivating quorum-sensing by washing away the signal, whereas increased shear force could also perform this function, while also accelerating cell detachment. We can only speculate on how this interplay will affect S. aureus in the context of an infection, because studies in this area are limited, but given the medical importance of these biofilms, it warrants further exploration.
Complicating these biofilm studies are the frequent occurrence of quorum-sensing negative isolates in clinical models. In one study, 36% of S. epidermidis isolates from joint prostheses infections were Agr mutants [80]; these mutants can also be isolated from chronic infections or through extended passage in vitro [91, 92]. It has been proposed that the metabolic burden of the Agr cascade leads to the mutations in this locus [40], with parallels to the in vitro conversion of mucoid P. aeruginosa strains to non-mucoid [93]. The frequent occurrence of Agr mutants suggests it may be advantageous for some portion of an S. aureus population to dispense with the quorum-sensing system. By inactivating Agr, a more heterogeneous, robust biofilm is likely to form, with attachment of the mutants improved by the higher level of surface factor expression. Although it remains to be investigated, this heterogeneity could mirror findings for P. aeruginosa biofilms, in which variants arise at a greater frequency than in planktonic cultures [94].
As studies on S. aureus progress, a plausible picture of the role of quorum-sensing in biofilm development is beginning to emerge. As S. aureus attaches and develops an initial biofilm, heterogeneity arising as a result of generation of Agr mutants may lead to a more robust structure with improved adherence properties. Communication between cells will be dictated by the secretion and sensing of AIP molecules, which will enable cross-activation or inhibition of quorum-sensing, depending on the staphylococcal subspecies present in close proximity. When a quorum is reached, a portion of the biofilm will slough off by down-regulation of adherence factors, which could potentially be dictated by the action of secreted proteases or amphipathic peptides. The detached S. aureus cells will have an activated quorum-sensing system that leads to the secretion of numerous invasive factors, enabling spread through the host tissues and the development of an infection. This model is in direct contrast to the well-studied P. aeruginosa paradigm, for which quorum sensing-regulated functions are important for maintaining the structural integrity of the biofilm. Clearly more studies are required to confirm or alter this hypothetical view of the S. aureus lifecycle, but as this model is refined, it may play a significant role in the development of treatment for staphylococcal diseases.
Acknowledgements
This work is supported by the (NSF), (NIH), and (CFF) and RO1 DC04173-04 (PI G.D. Ehrlich).
References
- 1.Hall-Stoodley L, Costerton JW, Stoodley P (2004) Nat Rev Microbiol 2(2):95–108 [DOI] [PubMed]
- 2.Davey ME, O’Toole GA (2000) Microbiol Mol Biol Rev 64(4):847–867 [DOI] [PMC free article] [PubMed]
- 3.Nealson KH, Hastings JW (2006) Appl Environ Microbiol 72(4):2295–2297 [DOI] [PMC free article] [PubMed]
- 4.Miller SD, Haddock SH, Elvidge CD, Lee TF (2005) Proc Natl Acad Sci USA 102(40):14181–14184, Epub 12005 Sep 14126 [DOI] [PMC free article] [PubMed]
- 5.Schmeisser C, Stockigt C, Raasch C, Wingender J, Timmis KN, Wenderoth DF, Flemming HC, Liesegang H, Schmitz RA, Jaeger KE, Streit WR (2003) Appl Environ Microbiol 69(12):7298–7309 [DOI] [PMC free article] [PubMed]
- 6.Troxler J, Azelvandre P, Zala M, Defago G, Haas D (1997) Appl Environ Microbiol 63(1):213–219 [DOI] [PMC free article] [PubMed]
- 7.Hu JY, Fan Y, Lin YH, Zhang HB, Ong SL, Dong N, Xu JL, Ng WJ, Zhang LH (2003) Res Microbiol 154(9):623–629 [DOI] [PubMed]
- 8.Waters CM, Bassler BL (2005) Annu Rev Cell Dev Biol 21:319–346 [DOI] [PubMed]
- 9.Fuqua C, Greenberg EP (2002) Nat Rev Mol Cell Biol 3(9):685–695 [DOI] [PubMed]
- 10.Lyon GJ, Novick RP (2004) Peptides 25(9):1389–1403 [DOI] [PubMed]
- 11.Parsek MR, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1999) Proc Natl Acad Sci USA 96(8):4360–4365 [DOI] [PMC free article] [PubMed]
- 12.More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC (1996) Science 272(5268):1655–1658 [DOI] [PubMed]
- 13.Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Proc Natl Acad Sci USA 93(18):9505–9509 [DOI] [PMC free article] [PubMed]
- 14.Schaefer AL, Hanzelka BL, Parsek MR, Greenberg EP (2000) Methods Enzymol 305:288–301 [DOI] [PubMed]
- 15.Voelkert E, Grant DR (1970) Anal Biochem 34:131–137 [DOI] [PubMed]
- 16.Kaufmann GF, Sartorio R, Lee S-H, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD (2005) PNAS 102(2):309–314 [DOI] [PMC free article] [PubMed]
- 17.deBeer D, Stoodley P, Roe F, Lewandowski Z (1994) Biotechnol Bioeng 43:1131–1138 [DOI] [PubMed]
- 18.Schaefer AL, Taylor TA, Beatty JT, Greenberg EP (2002) J Bacteriol 184(23):6515–6521 [DOI] [PMC free article] [PubMed]
- 19.Wagner-Dobler I, Thiel V, Eberl L, Allgaier M, Bodor A, Meyer S, Ebner S, Hennig A, Pukall R, Schulz S (2005) Chembiochem 6(12):2195–2206 [DOI] [PubMed]
- 20.Blosser-Middleton RS, Gray KM (2001) J Bacteriol 183(23):6771–6777 [DOI] [PMC free article] [PubMed]
- 21.Pearson JP, Van Delden C, Iglewski BH (1999) J Bacteriol 181(4):1203–1210 [DOI] [PMC free article] [PubMed]
- 22.Kolter R, Greenberg EP (2006) Nature 441(7091):300–302 [DOI] [PubMed]
- 23.Branda SS, Vik S, Friedman L, Kolter R (2005) Trends Microbiol 13(1):20–26 [DOI] [PubMed]
- 24.Charlton TS, de Nys R, Netting A, Kumar N, Hentzer M, Givskov M, Kjelleberg S (2000) Environ Microbiol 2(5):530–541 [DOI] [PubMed]
- 25.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP (2000) Nature 407(6805):762–764 [DOI] [PubMed]
- 26.Wang Y-J, Leadbetter JR (2005) Appl Environ Microbiol 71(3):1291–1299 [DOI] [PMC free article] [PubMed]
- 27.Dong Y-H, Xu J-L, Li X-Z, Zhang L-H (2000) PNAS 97(7):3526–3531 [DOI] [PMC free article] [PubMed]
- 28.Lee SJ, Park S-Y, Lee J-J, Yum D-Y, Koo B-T, Lee J-K (2002) Appl Environ Microbiol 68(8):3919–3924 [DOI] [PMC free article] [PubMed]
- 29.Zhang H-B, Wang L-H, Zhang L-H (2002) PNAS 99(7):4638–4643 [DOI] [PMC free article] [PubMed]
- 30.Huang JJ, Han J-I, Zhang L-H, Leadbetter JR (2003) Appl Environ Microbiol 69(10):5941–5949 [DOI] [PMC free article] [PubMed]
- 31.Leadbetter JR, Greenberg EP (2000) J Bacteriol 182(24):6921–6926 [DOI] [PMC free article] [PubMed]
- 32.Schauder S, Shokat K, Surette MG, Bassler BL (2001) Mol Microbiol 41(2):463–476 [DOI] [PubMed]
- 33.Meijler MM, Hom LG, Kaufmann GF, McKenzie KM, Sun C, Moss JA, Matsushita M, Janda KD. Angew Chem Int Ed 43(16):2106–2108 [DOI] [PubMed]
- 34.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Nature 415(6871):545–549 [DOI] [PubMed]
- 35.Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM (2004) Molecular Cell 15(5): 677–687 [DOI] [PubMed]
- 36.Havarstein LS, Coomaraswamy G, Morrison DA (1995) Proc Natl Acad Sci USA 92(24):11140–11144 [DOI] [PMC free article] [PubMed]
- 37.Okada M, Sato I, Cho SJ, Iwata H, Nishio T, Dubnau D, Sakagami Y (2005) Nat Chem Biol 1(1):23–24 [DOI] [PubMed]
- 38.Ansaldi M, Dubnau D (2004) J Bacteriol 186(1):15–21 [DOI] [PMC free article] [PubMed]
- 39.Kleerebezem M (2004) Peptides 25(9):1405–1414 [DOI] [PubMed]
- 40.Novick RP (2003) Mol Microbiol 48(6):1429–1449 [DOI] [PubMed]
- 41.Ji G, Beavis R, Novick RP (1997) Science 276(5321):2027–2030 [DOI] [PubMed]
- 42.Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, Nagasawa H (2001) Mol Microbiol 41(1):145–154 [DOI] [PubMed]
- 43.Autret N, Raynaud C, Dubail I, Berche P, Charbit A (2003) Infect Immun 71(8):4463–4471 [DOI] [PMC free article] [PubMed]
- 44.Ji G, Pei W, Zhang L, Qiu R, Lin J, Benito Y, Lina G, Novick RP (2005) J Bacteriol 187(9):3139–3150 [DOI] [PMC free article] [PubMed]
- 45.Perego M, Brannigan JA (2001) Peptides 22(10):1541–1547 [DOI] [PubMed]
- 46.Chatterjee C, Paul M, Xie L, van der Donk WA (2005) Chem Rev 105(2):633–684 [DOI] [PubMed]
- 47.Liu W, Hansen JN (1990) Appl Environ Microbiol 56(8):2551–2558 [DOI] [PMC free article] [PubMed]
- 48.Edwards JR, Bradley JS, Klugman KP (1998) J Antimicrob Chemother 42(2):265–266 [DOI] [PubMed]
- 49.McDowell P, Affas Z, Reynolds C, Holden MT, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CE, Bycroft BW, Chan WC, Williams P (2001) Mol Microbiol 41(2):503–512 [DOI] [PubMed]
- 50.Stewart PS (2003) J Bacteriol 185(5):1485–1491 [DOI] [PMC free article] [PubMed]
- 51.Egland PG, Palmer RJJ, Kolenbrander PE (2004) Proc Natl Acad Sci USA 101(48):16917–16922 [DOI] [PMC free article] [PubMed]
- 52.Purevdorj B, Costerton JW, Stoodley P (2002) Appl Environ Microbiol 68:4457–4464 [DOI] [PMC free article] [PubMed]
- 53.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP (1998) Science 280(5361):295–298 [DOI] [PubMed]
- 54.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP (2004) J Bacteriol 186(6):1838–1850 [DOI] [PMC free article] [PubMed]
- 55.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M (2004) J Infect Dis 190:1498–1505 [DOI] [PubMed]
- 56.Fux CA, Wilson S, Kim S, Nguyen D, Nistico L, Buchinsky FJ, Ehrlich GD, Balaban N, Stoodley P (2006) Antimicrob Agents Chemother Submitted
- 57.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll B, Molin S (2000) Microbiology 146:2395–2407 [DOI] [PubMed]
- 58.Anderl JN, Franklin MJ, Stewart PS (2000) Antimicrob Agents Chemother 44:1818–1824 [DOI] [PMC free article] [PubMed]
- 59.Fagerlinda MG, Rice SA, Nilsson P, Harlén M, James S, Charlton T, Kjelleberg S (2003) J Mol Microbiol Biotechnol 6:88–100 [DOI] [PubMed]
- 60.Janssen PH (2006) Appl Environ Microbiol 72(3):1719–1728 [DOI] [PMC free article] [PubMed]
- 61.Fierer N, Schimel JP, Holden PA (2003) Microb Ecol 45(1):63–71, Epub 2002 Dec 2010 [DOI] [PubMed]
- 62.Flavier AB, Clough SJ, Schell MA, Denny TP (1997) Mol Microbiol 26(2):251–259 [DOI] [PubMed]
- 63.Clough S, Lee K, Schell M, Denny T (1997) J Bacteriol 179(11):3639–3648 [DOI] [PMC free article] [PubMed]
- 64.Clough SJ, Flavier AB, Schell MA, Denny TP (1997) Appl Environ Microbiol 63(3):844–850 [DOI] [PMC free article] [PubMed]
- 65.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ (2001) J Bacteriol 183(24):7341–7353 [DOI] [PMC free article] [PubMed]
- 66.Qiu R, Pei W, Zhang L, Lin J, Ji G (2005) J Biol Chem 280(17):16695–16704 [DOI] [PubMed]
- 67.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick RP, Vandenesch F (1998) Mol Microbiol 28(3):655–662 [DOI] [PubMed]
- 68.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK (2004) J Bacteriol 186(22):7549–7555 [DOI] [PMC free article] [PubMed]
- 69.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S (1993) Embo J 12(10):3967–3975 [DOI] [PMC free article] [PubMed]
- 70.Balaban N, Novick RP (1995) Proc Natl Acad Sci USA 92(5):1619–1623 [DOI] [PMC free article] [PubMed]
- 71.Gov Y, Borovok I, Korem M, Singh VK, Jayaswal RK, Wilkinson BJ, Rich SM, Balaban N (2004) J Biol Chem 279(15):14665–14672 [DOI] [PubMed]
- 72.Balaban N, Goldkorn T, Nhan RT, Dang LB, Scott S, Ridgley RM, Rasooly A, Wright SC, Larrick JW, Rasooly R, Carlson JR (1998) Science 280(5362):438–440 [DOI] [PubMed]
- 73.Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, Otto M, Cheung AL, Gresham HD (2004) Proc Natl Acad Sci USA 101(38):13867–13872 [DOI] [PMC free article] [PubMed]
- 74.Wright JS 3rd, Jin R, Novick RP (2005) Proc Natl Acad Sci USA 102(5):1691–1696 [DOI] [PMC free article] [PubMed]
- 75.Domingue G, Costerton JW, Brown MR (1996) FEMS Immunol Med Microbiol 16(3-4):223–228 [DOI] [PubMed]
- 76.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW (1999) Proc Natl Acad Sci USA 96(4):1218–1223 [DOI] [PMC free article] [PubMed]
- 77.Chan WC, Coyle BJ, Williams P (2004) J Med Chem 47(19):4633–4641 [DOI] [PubMed]
- 78.Smeltzer MS, Thomas JR, Hickmon SG, Skinner RA, Nelson CL, Griffith D, Parr TR Jr, Evans RP (1997) J Orthop Res 15(3):414–421 [DOI] [PubMed]
- 79.Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer AS (1994) J Clin Invest 94(5):1815–1822 [DOI] [PMC free article] [PubMed]
- 80.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M (2004) J Infect Dis 190(8):1498–1505 [DOI] [PubMed]
- 81.Akiyama H, Kanzaki H, Tada J, Arata J (1996) J Dermatol Sci 11(3):234–238 [DOI] [PubMed]
- 82.Gotz F (2002) Mol Microbiol 43(6):1367–1378 [DOI] [PubMed]
- 83.Kong KF, Vuong C, Otto M (2006) Int J Med Microbiol 296(2-3):133–139 [DOI] [PubMed]
- 84.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F (1999) Infect Immun 67(10):5427–5433 [DOI] [PMC free article] [PubMed]
- 85.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS (2004) J Bacteriol 186(14):4665–4684 [DOI] [PMC free article] [PubMed]
- 86.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR (2006) Submitted [DOI] [PubMed]
- 87.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP (2004) J Bacteriol 186(6):1838–1850 [DOI] [PMC free article] [PubMed]
- 88.Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S (2001) Infect Immun 69(8):4742–4748 [DOI] [PMC free article] [PubMed]
- 89.Yarwood JM, Schlievert PM (2003) J Clin Invest 112(11):1620–1625 [DOI] [PMC free article] [PubMed]
- 90.Pratten J, Foster SJ, Chan PF, Wilson M, Nair SP (2001) Microbes Infect 3(8):633–637 [DOI] [PubMed]
- 91.Schwan WR, Langhorne MH, Ritchie HD, Stover CK (2003) FEMS Immunol Med Microbiol 38(1):23–28 [DOI] [PubMed]
- 92.Somerville GA, Beres SB, Fitzgerald JR, DeLeo FR, Cole RL, Hoff JS, Musser JM (2002) J Bacteriol 184(5):1430–1437 [DOI] [PMC free article] [PubMed]
- 93.Schurr MJ, Martin DW, Mudd MH, Deretic V (1994) J Bacteriol 176(11):3375–3382 [DOI] [PMC free article] [PubMed]
- 94.Boles BR, Thoendel M, Singh PK (2004) Proc Natl Acad Sci USA 101(47):16630–16635 [DOI] [PMC free article] [PubMed]


