Abstract
Periodontal infections have a microbial etiology. Association of species with early disease would be useful in determining which microbes initiate periodontitis. We hypothesized that the microbiota of subgingival and tongue samples would differ between early periodontitis and health. A cross-sectional evaluation of 141 healthy and early periodontitis adults was performed with the use of oligonucleotide probes and PCR. Most species differed in associations with sample sites; most subgingival species were associated with subgingival samples. Few species were detected more frequently in early periodontitis by DNA probes. Porphyromonas gingivalis and Tannerella forsythia (Tannerella forsythensis) were associated with early periodontitis by direct PCR. In conclusion, the microbiota of tongue samples was less sensitive than that of subgingival samples in detecting periodontal species, and there was overlap in species detected in health and early periodontitis. Detection of periodontal pathogens in early periodontitis suggests an etiology similar to that of more advanced disease.
Keywords: microbiology, tongue, subgingival, health, early periodontitis
INTRODUCTION
While periodontal diseases affect over half of the adults in the United States (Albandar et al., 1999), with appropriate interceptive treatment, periodontitis can be prevented. It has been difficult to identify those young adults who could develop periodontitis from those who remain periodontally healthy. Rather than universal prophylactic treatment, it would be preferable to identify and selectively treat individuals at high risk for disease before significant periodontal attachment loss occurs. Early periodontitis is detected clinically by loss of periodontal attachment, which, in younger adults, is strongly associated with gingivitis (Tanner et al., 2005). Gingivitis, with or without early attachment loss, however, cannot indicate which individuals will develop periodontitis. The primary etiology of periodontitis is the presence of specific bacteria in the subgingival plaque biofilm. Thus, it seems likely that identification of certain species could aid in the risk assessment for early periodontitis.
The composition of the subgingival microbiota of chronic periodontitis in adults has been described by culture (Moore and Moore, 1994; Tanner et al., 1998; van Winkelhoff et al., 2002), immunological (Riviere et al., 1996; Clerehugh et al., 1997; Machtei et al., 1999), and molecular methods (Socransky et al., 1998; Paster et al., 2001; Kumar et al., 2003; de Lillo et al., 2004). Culture analysis of subgingival plaque samples of early periodontitis detected Tannerella forsythia (Tannerella forsythensis), Campylobacter rectus, and Selenomonas noxia associated with progressing, compared with non-progressing, subjects, whereas Porphyromonas gingivalis was associated, by whole genomic DNA probes, with progressing periodontitis (Tanner et al., 1998). These species have also been associated with moderate and advanced periodontitis (Moore and Moore, 1994; Socransky et al., 1998). Molecular PCR cloning and sequencing methods have identified several species that are rarely or not detected by culture methods (Paster et al., 2001), some of which show strong associations with adult periodontitis (Kumar et al., 2003). Several periodontal species, including P. gingivalis and Prevotella melaninogenica, have been identified from tongue samples of adults (Van der Velden et al., 1986; Lee et al., 1999; Mager et al., 2003). The aims of this study were to compare the microbiota detected in subgingival and tongue samples and to evaluate associations between recognized species and uncultivated phylotypes with early periodontitis. Bacterial species that showed positive associations with early attachment loss could be of value in assessing periodontal risk, particularly if from easily obtained tongue samples.
MATERIALS & METHODS
We conducted a cross-sectional cohort study comparing the microbiota of subgingival and tongue samples and the microbiota of periodontally healthy and early periodontitis individuals.
Study Population and Clinical Measurements
Periodontally healthy and early periodontitis individuals (Table) underwent clinical monitoring as previously described (Tanner et al., 2005). Individuals were medically healthy, aged between 20 and 40 yrs, had not taken antibiotics in the previous 6 mos or had any previous periodontal therapy. Study participants had at least 24 teeth, and minimal periodontal attachment loss. Participants' rights were protected by approval of the study protocol from the IRBs of all participating institutions, and participants who signed an informed consent were accepted into the study. Participants completed medical and dental histories and a questionnaire that included race/ethnicity and history of cigarette smoking. Comprehensive clinical measurements were taken as previously described (Tanner et al., 2005). Healthy participants had a mean periodontal attachment level of ≤1.5 mm and no sites with > 2 mm attachment loss. Early periodontitis 1 individuals also had a mean periodontal attachment level of ≤1.5 mm but at least 1 site with 2 mm attachment loss. Early periodontitis 2 individuals had > 1.5 mm to ≤2.0 mm mean periodontal attachment loss.
Table.
Clinical and Demographic Characteristics of the Study Population
| Healtha (n = 28) | Early Periodontitis 1b (n = 71) | Early Periodontitis 2c (n = 42) | Significance (H, EP1, EP2) | |
|---|---|---|---|---|
| Mean age (in yrs) ± SEMd | 26.9 ± 1.03 | 30.5 ± 0.72 | 31.7 ± 1.04 | p = 0.0051e |
| Gender, n (% male) | 8 (29%) | 33 (46%) | 23 (55%) | p = 0.0367f |
| Never-smoker, n (%) | 24 (86%) | 46 (65)% | 26 (62%) | |
| Smoking history: | ||||
| Former smoker, n (%) | 2 (7%) | 13 (18)% | 7 (17%) | p = 0.0522f |
| Current smokers, n (%) | 2 (7%) | 12 (17)% | 9 (21%) | (all groups) |
| Race/Ethnicity: | ||||
| Asian, n (%) | 3 (23%) | 7 (54%) | 3 (23%) | |
| African-American, n (%) | 0 | 11 (52%) | 10 (48%) | |
| Hispanic, n (%) | 2 (20%) | 2 (20%) | 6 (60%) | |
| Caucasian, n (%) | 21 (23%) | 48 (53%) | 22 (24%) | p = 0.0409g |
| Other, n (%) | 2 (33%) | 3 (50%) | 1 (17%) | (4 major groups) |
| Gingival Index (mean ± SEM) | 0.50 ± 0.07 | 0.75 ± 0.05 | 1.30 ± 0.08 | p < 0.0001e |
| Plaque Index (mean ± SEM) | 0.51 ± 0.07 | 0.72 ± 0.06 | 0.93 ± 0.10 | p < 0.0222e |
| Percent sites BOP* (mean ± SEM) | 6 ± 1 | 13 ± 2 | 41 ± 4 | p < 0.0001e |
| Pocket depth (mm, mean ± SEM) | 1.98 ± 0.03 | 2.04 ± 0.02 | 2.53 ± 0.04 | p < 0.0001e |
| Attachment level (AL) (mm, mean ± SEM) | 1.14 ± 0.02 | 1.28 ± 0.02 | 1.70 ± 0.02 | |
Health: mean AL ≤1.5 mm, no sites > 2 mm AL loss.
Early periodontitis 1: mean AL ≤1.5 mm, ≥1 site more than 2 mm AL loss.
Early periodontitis 2: mean AL > 1.5 mm.
SEM = Standard Error of the Mean.
Kruskal-Wallis test.
Mantel-Haenszel Chi-square test.
Chi-square.
Bleeding on probing
Microbial Analyses
Sample-taking
Supragingival plaque was removed with a Gracey curette, after which samples were taken with a fresh sterile curette from 4 subgingival sites, usually distolingual, on 1st or 2nd molars, and the samples were pooled. Tongue samples were also taken with a Gracey curette by stroking the posterior 1/3 of the dorsum of the tongue. Samples were placed in 100 μL of Tris-EDTA buffer and stored frozen at −70°C.
DNA Probe Analysis
Bacterial samples were analyzed by the PCR-based, reverse-capture oligonucleotide checkerboard assay (Socransky et al., 1994; Paster et al., 1998), as described in the modified procedure (Becker et al., 2002). Probe species (Fig. 1) were selected to include frequently encountered periodontally related species or clones of novel unidentified, including not-yet-cultivated, phylotypes from PCR-cloning studies of human dental plaque (Paster et al., 2001) and the tongue (Kazor et al., 2003). The detection limit for the assay was about 103 cells.
Figure 1.

Species detected by oligonucleotide DNA probes in reverse-capture checkerboard assay from paired tongue and subgingival samples of 121 healthy and early periodontitis individuals. Species detected more frequently from tongue samples are at the top of the Fig., and those detected more frequently subgingivally are at the bottom of the Fig. #Species prevalence differs, tongue vs. subgingival, McNemar’s Chi-square p < 0.05. *Species-associated tongue and subgingival Chi-square for associations, p < 0.05.
Multiplex PCR Analysis
To improve detection sensitivity, particularly of P. gingivalis, we performed a direct PCR assay (Tran and Rudney, 1999) using 2 μL of sample. Amplicons were visualized by agarose gel electrophoresis. The detection limit for P. gingivalis was about 102 cells, and for T. forsythia, 103 cells.
Reactions from both assays were scored as species detected or not detected.
Statistical Analyses
Species Association with Subgingival and Tongue Samples
We analyzed data from paired subgingival and tongue samples taken at the same visit from each individual. Contingency tables evaluated species detection in subgingival and tongue samples. Differences in species prevalence in tongue and subgingival samples were evaluated by McNemar's Chi-square test, and similarities of species detection in tongue and subgingival samples were evaluated by the Chi-square test for association.
Species Associations with Early Periodontitis
Individuals were grouped into healthy and two early periodontitis disease categories (Table). Comparisons of species prevalence across clinical groups were done by the Mantel-Haenszel chi-square for trend.
Significance levels were not adjusted for the multiple comparisons over species.
RESULTS
Clinical Characteristics of Study Population
The clinical and demographic characteristics of 141 healthy and early periodontitis individuals (Table) are from the previously described study population of 221 individuals (Tanner et al., 2005). Although samples were taken from all study participants, there was insufficient DNA from some participants to get an amplicon from PCR, resulting in differences in numbers of individuals with microbial data from the clinical study population. Most participants were never-smokers and Caucasian (65%); 21 were African-American. There was a positive association between early periodontitis and gingival index, percent sites bleeding on probing, pocket depth, participant age, gender, and cigarette smoking (Table).
Microbiology
Reverse-capture Oligonucleotide Probe Data
Subgingival and tongue samples
There were differences in species detection in 121 paired subgingival and tongue samples (Fig. 1). Many Streptococcus and Streptococcus-like species (Gemella and Granulicatella) were detected more frequently from, and were associated with, tongue samples (p < 0.05, McNemar's Chi-square test). Eubacterium sulci, Actinomyces odontolyticus, Campylobacter concisus, and Prevotella melaninogenica were also associated with tongue samples.
Most of the Gram-negative taxa and not-yet-cultivated phylotypes were detected more frequently from subgingival samples (p < 0.05, McNemar's Chi-square test), including Treponema species, T. forsythia, Porphyromonas endodontalis, A. actinomycetemcomitans, Prevotella oris, TM7 phylum, Dialister invisus, and Fusobacterium nucleatum subsp. animalis. Subgingivally associated Gram-positive species included Streptococcus mutans, Streptococcus sanguinis, Streptococcus anginosus, Actinomyces naeslundii II, Actinomyces gerencseriae, and Filifactor alocis (p < 0.05, McNemar's Chi-square test). Detection of F. nucleatum subsp. polymorphum and Prevotella denticola subgingivally was associated with detection on the tongue (p < 0.05, Chi-square test for associations). P. gingivalis was detected in less than 5% of samples when DNA probes were used.
Health and early periodontitis samples
Subgingival samples from 141 individuals were compared between clinical categories (Fig. 2). Species detected more frequently in early periodontitis than in health included Treponema denticola, F. alocis, P. endodontalis, Bacteroidetes oral clone AU126, F. nucleatum subsp. animalis, and Fusobacterium nucleatum subsp. polymorphum, although none significantly, except Actinomyces odontolyticus.
Figure 2.

Species detected by oligonucleotide DNA probes from pooled molar subgingival plaque samples from 28 healthy, 71 early periodontitis 1, and 42 early periodontitis 2 individuals. Health-associated species are at the top of the Fig., whereas species detected more frequently in early periodontitis 2 are at the bottom of the Fig. Mantel-Haenszel Chi-square for Trend: * p ≤0.05, ** p ≤0.01.
Multiplex PCR Analysis of P. gingivalis and T. forsythia
The multiplex PCR assay detected more P. gingivalis-positive samples than did the DNA probe assay, a similar proportion of T. forsythia samples in 124 paired subgingival and tongue samples (Fig. 3). The detection frequency for P. gingivalis was similar from subgingival and tongue samples. T. forsythia was detected more frequently from subgingival as compared with tongue samples (McNemar Chi-square, p = 0.001). Detection of T. forsythia from the tongue was not associated with subgingival detection within individuals.
Figure 3.
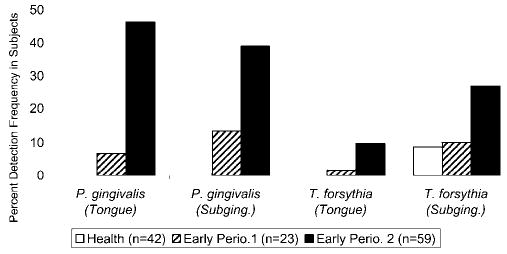
Detection of P. gingivalis and T. forsythia in 124 paired tongue and subgingival samples of 23 healthy, 59 early periodontitis 1, and 42 early periodontitis 2 individuals, by means of the multiple PCR assay. There was no difference in P. gingivalis detection between tongue and subgingival samples, but T. forsythia was detected more frequently subgingivally (p = 0.001). P. gingivalis in tongue and subgingival samples (p < 0.001) and T. forsythia in subgingival samples (p < 0.03) were associated with early periodontitis.
P. gingivalis from tongue and subgingival sample sites was significantly associated with early periodontitis (Mantel-Haenszel Chi-square, p < 0.001). T. forsythia detection subgingivally was also associated with early periodontitis (p = 0.03). There were insufficient T. forsythia-positive tongue samples to test their association with early periodontitis.
DISCUSSION
We compared the microbiota of tongue and subgingival samples of adults showing early stages of periodontitis. Clinically, there was a strong association of early periodontitis with gingival inflammation, as has been observed in other populations of young adults with periodontitis and in the source population of the current study (Tanner et al., 2005). The microbiota of early periodontitis included several species associated with moderate to advanced periodontitis. The association of P. gingivalis with early periodontitis was of particular interest, because, using culture techniques, we had not associated this species with early periodontitis (Tanner et al., 1998). By immunoassays, however, P. gingivalis was detected in 3- to 5-mm pockets more than in shallower sulci (Hamlet et al., 2001), and in early periodontitis more frequently than in health (Riviere et al., 1996). In the latter study, however, higher levels of P. gingivalis were detected in gingivitis than in early periodontitis, which is consistent with the elevated gingival inflammation in early periodontitis observed in the current study. P. gingivalis was also detected more frequently and in higher levels in adolescent individuals with early periodontitis as compared with health (Clerehugh et al., 1997), and with deeper pocket and bleeding sites in a population of 11- to 13-year-old children (Ellwood et al., 1997). In all these studies, P. gingivalis was detected in only a proportion of early periodontitis individuals, which may reflect the detection sensitivities of assays, or the involvement of additional species in the initiation of periodontitis.
In contrast to P. gingivalis, T. forsythia was associated with early periodontitis by culture (Tanner et al., 1998). Furthermore, T. forsythia, but not P. gingivalis, was associated with, and predictive of, progressing attachment loss in older individuals with minimal periodontitis (Tran et al., 2001), by the PCR assay used in the current study. T. forsythia was detected more frequently from adolescents with progressing attachment loss than from non-progressing individuals (Hamlet et al., 2004). The current study also detected T. forsythia in health, consistent with its detection in periodontally healthy children, adolescents (Sirinian et al., 2002), and adults (Gmür et al., 1999). While a different health-associated T. forsythia-like phylotype has been reported (Leys et al., 2002; Kumar et al., 2003), T. forsythia and P. gingivalis were infrequently detected from individuals without periodontal bone loss, when culture methods were used (van Winkelhoff et al., 2002). Analysis of these data suggests that while T. forsythia colonizes periodontally healthy individuals, it colonizes more frequently and at higher levels in periodontitis.
The oligonucleotide probes facilitated assay of uncultivated phylotypes and fastidious species likely underrepresented by culture. Few differences were detected between health and early periodontitis, probably reflecting the small differences between clinical categories inherent in our design to characterize the earliest stages of periodontal loss. Species or phylotypes detected more frequently from early periodontitis, previously associated with moderate to advanced periodontitis by molecular methods (Paster et al., 2001; Kumar et al., 2003), included F. alocis, T. denticola, Peptostreptococcus micros, P. endodontalis, and Bacteroidetes clone AU126. T. denticola was also detected, by immunofluorescence, more frequently from early periodontitis than health (Riviere et al., 1996). F. alocis and P. endodontalis, while both previously identified from periodontal sites by culture (Petit et al., 1993; Tanner et al., 1998), are fastidious species that likely require detection by molecular methods for association with periodontitis (Kumar et al., 2003). The Obsidian Pool and TM7 phylotypes belong in bacterial groups with no known cultivated representatives. Their detection in health and early periodontitis indicates the complexity of the oral microbiota not previously recognized from culture studies.
Differences in species colonization patterns between subgingival sites and mucous membranes have been recognized for many decades, and have been attributed to different adherence patterns to host tissues. With the recognition that individual species can have distinct colonization preferences, detection of the periodontal (subgingival) pathogen P. gingivalis, on the tongue surface and other mucous membranes (Van der Velden et al., 1986), was a notable observation. Our finding, however, that the majority of subgingival species were detected more frequently from subgingival sites than on the tongue suggests that subgingival samples would be more reliable than tongue samples for the detection of periodontal pathogens.
While other studies have not examined the species set of this report from paired samples, associations were consistent with literature reports, particularly a recent report comparing the microbiota of several intra-oral sites by DNA probes (Mager et al., 2003). Subgingivally associated species from both reports included A. actinomycetemcomitans, A. naeslundii II, A. gerencseriae, and T. forsythia. While S. mutans is most frequently associated with dental caries, it was detected subgingivally in health (Tanner et al., 1998) and after periodontal therapy (van der Reijden et al., 2001). Several "subgingival" species, in addition to P. gingivalis, were detected at similar or higher frequencies from tongue than from subgingival samples. Tongue-associated species included C. concisus, a pathogen of gastric enteritis (Vandamme et al., 1989). Other tongue-associated species—S. salivarius, S. parasanguinis, Megasphera phylotype, and Rothia mucilaginosa—were included as "positive controls" of the tongue samples (Kazor et al., 2003).
We used two PCR-based methods to analyze the microbiota of samples. Both assays avoided problems associated with the anaerobic sample-handling needed for culture analysis and could detect uncultivated taxa with only sequence data for identification. The oligonucleotide probe assay allowed for the simultaneous detection of multiple species, but direct PCR of samples was more sensitive for P. gingivalis. The detection frequency was similar for T. forsythia, but there was only reasonable agreement between methods (data not shown), likely resulting from the differences in primers between the assays. The strongest disease associations were from the direct PCR assay. Whether species other than P. gingivalis and T. forsythia would show stronger associations with early periodontitis if assayed by direct PCR would require further analysis.
In conclusion, this cross-sectional analysis indicated that P. gingivalis from tongue and subgingival samples and T. forsythia from subgingival samples were associated with adults with early periodontitis. Several other taxa previously associated with moderate to advanced periodontitis were detected more frequently from early periodontitis than from health, although associations were not significant. This suggests similarities in the microbial etiology in early and advanced periodontitis. The microbiota of tongue and subgingival samples differed, reflecting site-specificity in oral species, although all species were detected in both sites.
Acknowledgments
We acknowledge the assistance of Tahani Boumenna in microbial analyses, and that of Lora Murray, Melissa Martins, and Lisa Newell in subject recruitment and clinical measurements. This project was supported by NIH grants DE-09513 and DE-11443 from the National Institute of Dental and Craniofacial Research, and by RR-00533 and RR-01032 from the National Center for Research Resources.
References
- Albandar JM, Brunelle JA, Kingman A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J Periodontol. 1999;70:13–29. doi: 10.1902/jop.1999.70.1.13. [DOI] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerehugh V, Seymour GJ, Bird PS, Cullinan M, Drucker DB, Worthington HV. The detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Prevotella intermedia using an ELISA in an adolescent population with early periodontitis. J Clin Periodontol. 1997;24:57–64. doi: 10.1111/j.1600-051x.1997.tb01185.x. [DOI] [PubMed] [Google Scholar]
- de Lillo A, Booth V, Kyriacou L, Weightman AJ, Wade WG. Culture-independent identification of periodontitis-associated Porphyromonas and Tannerella populations by targeted molecular analysis. J Clin Microbiol. 2004;42:5523–5527. doi: 10.1128/JCM.42.12.5523-5527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood R, Worthington HV, Cullinan MP, Hamlet S, Clerehugh V, Davies R. Prevalence of suspected periodontal pathogens identified using ELISA in adolescents of differing ethnic origins. J Clin Periodontol. 1997;24:141–145. doi: 10.1111/j.1600-051x.1997.tb00482.x. [DOI] [PubMed] [Google Scholar]
- Gmür R, Marinello CP, Guggenheim B. Periodontitis associated bacteria in supragingival plaque of dental hygienists: stability of carrier state and clinical development. Eur J Oral Sci. 1999;107:225–228. doi: 10.1046/j.0909-8836...x. [DOI] [PubMed] [Google Scholar]
- Hamlet SM, Cullinan MP, Westerman B, Lindeman M, Bird PS, Palmer J, et al. Distribution of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Prevotella intermedia in an Australian population. J Clin Periodontol. 2001;28:1163–1171. doi: 10.1034/j.1600-051x.2001.281212.x. [DOI] [PubMed] [Google Scholar]
- Hamlet S, Ellwood R, Cullinan M, Worthington H, Palmer J, Bird P, et al. Persistent colonization with Tannerella forsythensis and loss of attachment in adolescents. J Dent Res. 2004;83:232–235. doi: 10.1177/154405910408300309. [DOI] [PubMed] [Google Scholar]
- Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- Lee KH, Tanner AC, Maiden MF, Weber HP. Pre- and post-implantation microbiota of the tongue, teeth, and newly placed implants. J Clin Periodontol. 1999;26:822–832. doi: 10.1111/j.1600-051x.1999.tb02527.x. [DOI] [PubMed] [Google Scholar]
- Leys EJ, Lyons SR, Moeschberger ML, Rumpf RW, Griffen AL. Association of Bacteroides forsythus and a novel Bacteroides phylotype with periodontitis. J Clin Microbiol. 2002;40:821–825. doi: 10.1128/JCM.40.3.821-825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtei EE, Hausmann E, Dunford R, Grossi S, Ho A, Davis G, et al. Longitudinal study of predictive factors for periodontal disease and tooth loss. J Clin Periodontol. 1999;26:374–380. doi: 10.1034/j.1600-051x.1999.260607.x. [DOI] [PubMed] [Google Scholar]
- Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Bartoszyk IM, Dewhirst FE. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 1998;20:223–231. [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MD, van Winkelhoff AJ, van Steenbergen TJ, de Graaff J. Porphyromonas endodontalis: prevalence and distribution of restriction enzyme patterns in families. Oral Microbiol Immunol. 1993;8:219–224. doi: 10.1111/j.1399-302x.1993.tb00563.x. [DOI] [PubMed] [Google Scholar]
- Riviere GR, Smith KS, Tzagaroulaki E, Kay SL, Zhu X, DeRouen TA, et al. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J Periodontol. 1996;67:109–115. doi: 10.1902/jop.1996.67.2.109. [DOI] [PubMed] [Google Scholar]
- Sirinian G, Shimizu T, Sugar C, Slots J, Chen C. Periodontopathic bacteria in young healthy subjects of different ethnic backgrounds in Los Angeles. J Periodontol. 2002;73:283–288. doi: 10.1902/jop.2002.73.3.283. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Tanner A, Maiden MF, Macuch PJ, Murray LL, Kent RL., Jr Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol. 1998;25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x. [DOI] [PubMed] [Google Scholar]
- Tanner AC, Kent R, Jr, Van Dyke T, Sonis ST, Murray LA. Clinical and other risk indicators for early periodontitis in adults. J Periodontol. 2005;76:573–581. doi: 10.1902/jop.2005.76.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran SD, Rudney JD. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J Clin Microbiol. 1999;37:3504–3508. doi: 10.1128/jcm.37.11.3504-3508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran SD, Rudney JD, Sparks BS, Hodges JS. Persistent presence of Bacteroides forsythus as a risk factor for attachment loss in a population with low prevalence and severity of adult periodontitis. J Periodontol. 2001;72:1–10. doi: 10.1902/jop.2001.72.1.1. [DOI] [PubMed] [Google Scholar]
- van der Reijden WA, Dellemijn-Kippuw N, Stijne-van Nes AM, de Soet JJ, van Winkelhoff AJ. Mutans streptococci in subgingival plaque of treated and untreated patients with periodontitis. J Clin Periodontol. 2001;28:686–691. doi: 10.1034/j.1600-051x.2001.028007686.x. [DOI] [PubMed] [Google Scholar]
- Van der Velden U, van Winkelhoff AJ, Abbas F, de Graaff J. The habitat of periodontopathic micro-organisms. J Clin Periodontol. 1986;13:243–248. doi: 10.1111/j.1600-051x.1986.tb01467.x. [DOI] [PubMed] [Google Scholar]
- van Winkelhoff AJ, Loos BG, van der Reijden WA, Van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29:1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Falsen E, Pot B, Hoste B, Kersters K, De Ley J. Identification of EF group 22 campylobacters from gastroenteritis cases as Campylobacter concisus. J Clin Microbiol. 1989;27:1775–1781. doi: 10.1128/jcm.27.8.1775-1781.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]


