Abstract
The objective of this study was to model a typical dairy waste stream, monitor the chemical and bacterial population dynamics that occur during aerobic or anaerobic treatment and subsequent storage in a simulated lagoon, and compare them to those of waste held without treatment in a simulated lagoon. Both aerobic and anaerobic treatment methods followed by storage effectively reduced the levels of total solids (59 to 68%), biological oxygen demand (85 to 90%), and sulfate (56 to 65%), as well as aerobic (83 to 95%), anaerobic (80 to 90%), and coliform (>99%) bacteria. However, only aerobic treatment reduced the levels of ammonia, and anaerobic treatment was more effective at reducing total sulfur and sulfate. The bacterial population structure of waste before and after treatment was monitored using 16S rRNA gene sequence libraries. Both treatments had unique effects on the bacterial population structure of waste. Aerobic treatment resulted in the greatest change in the type of bacteria present, with the levels of eight out of nine phyla being significantly altered. The most notable differences were the >16-fold increase in the phylum Proteobacteria and the approximately 8-fold decrease in the phylum Firmicutes. Anaerobic treatment resulted in fewer alterations, but significant decreases in the phyla Actinobacteria and Bacteroidetes, and increases in the phyla Planctomycetes, Spirochetes, and TM7 were observed.
California is the largest dairy-producing state in the United States, housing over 2.5 million dairy cows on approximately 2,300 dairies, with the average farm maintaining 1,000 cows (35). The average 450-kg dairy cow produces approximately 37 kg of waste (manure and urine) per day (27); thus a 1,000 cow dairy produces approximately 37,000 kg of waste per day or 13.5 million kg of waste per year. The waste is usually held in storage lagoons until it can be applied to agricultural fields as a soil amendment/fertilizer for crops destined for animal or human consumption. The average herd size in California has increased by approximately 8% a year for the last 10 years (35), and new challenges associated with the waste stream have emerged. For example, many of the larger dairies produce more waste than they can apply to nearby fields due to excessive nutrient levels (e.g., nitrogen, phosphate, potassium, etc.) and transporting waste to distant agricultural fields is an economic liability. Cow manure has also been associated with pathogenic bacteria such as Escherichia coli O157:H7 (14), Salmonella sp. (37), Campylobacter sp. (38), and Mycobacterium avium subsp. paratuberculosis (8), and crops fertilized with this material may transmit these pathogens to the consumer. Furthermore, waste lagoons can impair air quality via the release of odorous compounds, leading to nuisance complaints from surrounding residential communities (17). One possible solution to these problems is to treat the waste before it enters the storage lagoons. The most commonly used treatment methodologies for both municipal and agricultural wastes are aerobic and anaerobic digestion (11, 29, 34, 36). Previous studies have shown these techniques to be effective for organic matter, nutrient (19, 33), and pathogen (13) reduction, but little is known about the microbial population dynamics associated with these processes.
Because cultivation methods are estimated to support the growth of only a small fraction of the naturally occurring biodiversity (1), the use of small-subunit rRNA gene (16S) analysis has proven to be a powerful tool to describe the microbial population structure of the human gut (10) and soil (9) and to compare the populations associated with different types of dairy waste storage lagoons (25). Techniques such as terminal fragment length polymorphism (24), denaturing gradient gel electrophoresis (30), and length heterogeneity PCR (32) are popular because they are relatively rapid and inexpensive, but do not provide the detailed information that 16S rRNA gene sequencing does. However, all of these techniques are subject to caveats, including PCR amplification and cloning bias, uneven bacterial cell lysis, and copy number variations of 16S rRNA genes within different species. In this study, 16S rRNA gene sequence analysis was used to determine the bacterial population dynamics of dairy waste treated in aerobic or anaerobic reactors followed by storage in simulated waste storage lagoons and to compare it to the dynamics of untreated waste stored in simulated lagoons. This was accomplished by pumping fresh dairy waste through lab-scale aerobic and anaerobic reactors and holding the effluent in stagnant storage tanks that simulated dairy waste storage lagoons (Fig. 1) or simply holding the waste in simulated storage lagoons. Samples were collected from the fresh waste material, the reactors, and the storage tanks for a period of 6 months and monitored for their chemical composition and bacterial population structure. Our results confirm that both aerobic and anaerobic treatment are more effective at reducing nutrient levels than storage alone and that each treatment method has a unique effect on the bacterial population structure of dairy waste.
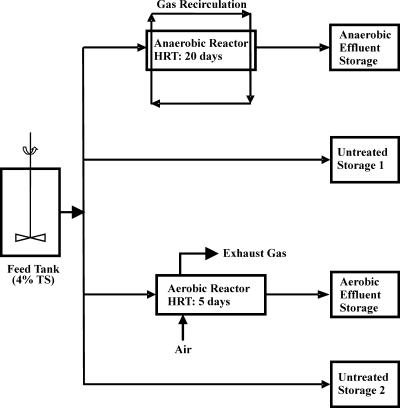
FIG. 1.
Diagram of the aerobic and anaerobic reactors. Fresh waste was placed into the feed tank and was pumped into aerobic or anaerobic reactors at an HRT of 5 or 20 days, respectively. Waste was also pumped into untreated storage tanks at the same rates. Treated waste exiting the digesters was collected in holding tanks for the duration of the experiment.
MATERIALS AND METHODS
Sample collection and preparation.
Fresh dairy cow waste (manure and urine <12 h postexcretion) was collected from the research dairy located on the campus of the University of California at Davis weekly from 15 November 2004 until 9 May 2005. The waste was passed through a screen with 2-mm openings to remove large particles that would clog the lines of the reactors. The screened waste was diluted with tap water to yield a slurry of approximately 4% total solids (TS), loaded into a feed tank maintained at 4°C, and used to feed the aerobic and anaerobic reactors. Fresh waste material was added into the feed tank weekly, and any material remaining in the feed tank was discarded when fresh material was added. The aerobic and anaerobic reactors were designed and operated to reduce total solids and biological oxygen demand (BOD5) by approximately 35 and 80%, respectively, and are described schematically in Fig. 1. Aerobic treatment was performed at room temperature (approximately 25°C) in a 3-liter reactor with a 2-liter working volume and 1-liter headspace with dimensions of 15 cm in diameter and 37 cm in depth, and with a hydraulic retention time (HRT) of 5 days. Atmospheric air was pumped continuously through the reactor to maintain a dissolved oxygen concentration of approximately 2 mg liter−1. Effluent from the aerobic reactor was collected and held in a 100-liter storage tank for the duration of the experiment. Anaerobic treatment was performed in a 5-liter reactor with a 4-liter working volume and a 1-liter headspace with dimensions of 15 cm in diameter and 74 cm in depth. The contents of the anaerobic reactor were maintained at 37°C and mixed for 2 min every hour by recirculating the headspace gas through the liquid. The anaerobic reactor had an HRT of 20 days, and its effluent was collected and stored in a 100-liter tank for the duration of the experiment. Both aerobic and anaerobic reactors were fed once a day from the same feed tank. In addition to the reactors described above, feed material was pumped directly into two storage tanks (100 liters each) to simulate the storage of untreated waste. One tank was fed at the same rate as the aerobic reactor, and the other tank was fed at the same rate as the anaerobic reactor. The material in these tanks, as in the effluent storage tanks for the aerobic and anaerobic reactors, received no mixing except when samples were taken and was maintained at room temperature for the duration of the experiment. Previous studies in our laboratory have shown that both aerobic and anaerobic treatments of manure reduce microbial diversity and chemical variability (26); thus, a sampling scheme was developed on the hypothesis that the feed material would have the most microbial and chemical variability. Therefore, the feed material was assayed weekly as described below when fresh material was added. It was hypothesized the contents of the aerobic and anaerobic reactors contained the next greatest variability, and they were sampled biweekly. Finally, the aerobic and anaerobic reactor effluent storage tanks and the untreated manure storage tanks were sampled every 4 weeks.
Viable counts of bacteria and chemical analysis of wastewater.
Samples were quantified for viable bacteria by performing serial dilutions in phosphate-buffered saline that were vortex agitated for 2 min prior to being plated onto brain heart infusion agar plates (BHI) and incubated at 25°C for 2 days under normal atmospheric conditions or in an anaerobic chamber. To quantify the number of coliform bacteria, samples were diluted as described above, plated onto MacConkey agar plates, and incubated at 37°C for 18 h. All media were purchased from Difco (Detroit, MI) as dehydrated powders. Chemical analysis was performed at A&L Western Agricultural Labs (Modesto, CA), a State of California accredited agricultural and environmental testing laboratory, using standard protocols (2).
DNA extraction from waste samples.
Two-milliliter samples of wastewater were centrifuged at 10,000 × g for 10 min, and the resultant pellets, or 0.5-g manure samples, were used for DNA extraction. DNA was extracted from the samples using a modification of the MoBio UltraClean fecal DNA isolation kit (MoBio, Solano Beach, CA) as described previously (26).
PCR amplification of 16S rRNA gene sequences and library construction.
PCR amplification of 16S rRNA gene sequences was carried out using the primers 27f (5′ AGAGTTTGATCCTGGCTCAG 3′) and 1392r (5′ GACGGGCGGTGTGTAC 3′) (21). PCRs were performed as recommended by Polz and Cavanaugh (31) to reduce bias in amplification. Briefly, 50-μl reaction volumes contained 200 μM deoxynucleoside triphosphates, 100 ng genomic DNA, and 2 U Expand high-fidelity enzyme mix (Roche, Nutley, NJ) in Expand high-fidelity buffer with 1.5 mM MgCl2 and 1 μM of each primer. PCRs were performed in a Tetrad Thermocycler (Bio-Rad, Hercules, CA) under the following conditions: one cycle of 95°C for 5 min; 15 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min; and one cycle of 5 min at 72°C. PCR products were purified by ethanol precipitation, cloned using the QIAGEN PCR cloning kit (QIAGEN, Valencia, CA) as per the manufacturer's instructions, and transformed into E. coli TOP10F′ cells (Invitrogen, Carlsbad, CA). Clones were plated on LB agar plates containing kanamycin (50 μg ml−1), isopropyl-β-d-thiogalactopyranoside (IPTG) (20 mM), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg ml−1). White colonies were selected and grown in 96-well plates in LB broth supplemented with kanamycin. Two PCRs and cloning experiments were performed for each sample, and 96 clones were picked from each PCR to minimize potential PCR bias.
DNA template preparation and sequencing.
DNA templates were prepared using the TempliPhi 100 amplification kit (Amersham Biosciences, Sunnyvale, CA) as per the manufacturer's instructions. Sequencing reactions were performed in one direction using the primer 1392r and the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Sequencing reactions were purified using the DyeEx 96 kit (QIAGEN, Valencia, CA); electrophoresis and readout were performed using an Applied Biosystems 3730XL genetic analyzer (Applied Biosystems, Foster City, CA). Two 96-well plates of 16S rRNA gene sequences were analyzed for each sample: a total of 13,824 sequences were analyzed.
DNA sequence analysis and dendrogram construction.
DNA sequences were edited manually to correct falsely called bases and trimmed at both the 5′ and 3′ ends using Chromas (version 2.31; Technelysium Pty. Ltd., Helensvale, Australia). Only sequences with unambiguous reads of >500 bp were used; each read used averaged approximately 600 bp. The predicted 16S rRNA sequences from this study were compared to 16S rRNA sequences in a BLASTable database constructed from sequences downloaded from the Ribosomal Database Project (release 8.1; http://rdp8.cme.msu.edu). Comparisons were made using the program BLASTALL (ftp://ftp.ncbi.nih.gov/BLAST/executables/LATEST/) and a FASTA-formatted file containing the predicted 16S rRNA sequences. Operational taxonomic units (OTUs) were defined as clones with >97% sequence identity. For dendrogram construction, partial 16S rRNA gene sequences representing the 10 most prevalent OTUs from each environment (feed material, aerobic and anaerobic reactor effluents, effluents held in storage tanks, and control untreated material in storage) and the most similar 16S rRNA gene sequences to each OTU from the NCBI nonredundant (nr) database were aligned using CLUSTALX. The 16S rRNA gene sequences from the nr database were first reverse complemented and trimmed to approximate the start point and length of the OTU sequences. Phylogenetic and molecular evolutionary analyses were performed using MEGA version 2.1 (20); the dendrogram was constructed using the neighbor-joining algorithm and the Kimura two-parameter distance estimation method.
Rarefaction analysis and statistical methods.
Rarefaction analysis was performed using the approximation algorithm of Hurlbert (18) with 95% confidence intervals estimated as described by Heck (16) using the freeware program aRarefactWin by S. Holland (University of Georgia, Athens; http://www.uga.edu/∼strata/AnRareReadme.html). The percent coverage of the total OTUs identified in each sample was calculated using the equation C = [1 − (n/N)] × 100, where C is the percent coverage, n is the number of OTUs, and N is the number of clones examined. Student's t test, available in the SAS STAT package, was employed to test for differences between 16S rRNA gene libraries as well as the cultural and chemical parameters measured. Each OTU was assigned to a phylum using the Classifier software (7), which assigns an OTU sequence to a phylum using a naïve Bayesian rRNA classifier trained on the known type strain 16S sequences. Once the OTUs of each library were assigned to a phylum, pairwise comparisons of the phyla within the libraries were performed using Student's t test. In addition, comparisons of the 16S rRNA libraries were analyzed using the Library Compare software (7), which estimates the likelihood that the frequency of membership in a given taxon is the same for the two libraries using the equation:
 |
where N1 and N2 are the total number of sequences for libraries 1 and 2, respectively, and x and y are the number of sequences assigned to an OTU from libraries 1 and 2, respectively. The percentage of a phylum in one library was considered significantly different from that in another library if both statistical methods (Student's t test and Compare) were in agreement. The diversity within the libraries was measured with the Shannon-Wiener index (H), species richness (S), and evenness (E) using the equations H = −Σpi ln (pi), where pi is the proportion of the total number of OTUs made up to the ith OTUs; and E = H/log(S), where S = total number of OTUs in the community.
Nucleotide sequence accession numbers.
DNA sequences representative of the 10 most prevalent OTUs from each library were deposited into GenBank under accession no. DQ673153 to DQ673212.
RESULTS
Cultural and chemical analyses of waste, reactor effluents, and stored material.
Changes in aerobic, anaerobic, and coliform plate counts, TS, BOD5, total Kjedahl N (TKN), and NH4, S, SO4, K2O, and Na concentrations were measured before and after treatment and are presented in Table 1. After aerobic treatment, significant reductions in BOD5 (77%), NH4 (87%), anaerobic (92%), and coliform (95%) plate counts were observed as compared to those in the feed material at the P < 0.05 level. Analysis of the aerobic reactor effluent stored in stagnant holding tanks showed significant reductions in TS (68%), BOD5 (83%), TKN (48%), NH4 (69%), SO4 (56%), and aerobic (90%) and coliform (99%) plate counts as compared to those in the feed material (P < 0.05). After anaerobic digestion, significant reductions in TS (43%), BOD5 (87%), S (58%), SO4 (43%), and anaerobic (99.9%) and coliform (99.7%) plate counts were observed as compared to those in the feed material. Anaerobic reactor effluent stored in stagnant holding tanks had significant reductions in TS (59%), BOD5 (85%), S (61%), SO4 (65%), and anaerobic (81%) and coliform (99.9%) plate counts compared to the feed material. The feed material held in an untreated control tank showed significant reductions in BOD5 (29%), SO4 (70%), and anaerobic (91%) and coliform (96%) plate counts compared to the feed material at the P < 0.05 level.
TABLE 1.
Chemical and cultural analyses of aerobic and anaerobic waste treatment systems
| Treatment and parametera | Avg (range) result for treatment by parameter for:
|
|||
|---|---|---|---|---|
| Feed material | Reactor effluent | Effluent storage | Untreated storage | |
| Aerobic | ||||
| Total solids (g liter−1) | 44.0 (29.0-58.0) | 30.0 (16.0-45.0) | 14.0 (7.0-29.0) | 28.0 (17.0-54.0) |
| BOD5 (g liter−1) | 14.9 (13.1-18.0) | 3.5 (2.4-4.5) | 1.5 (0.9-2.3) | 10.6 (8.0-15.3) |
| Total Kjedahl N (g liter−1) | 2.9 (1.5-6.1) | 2.2 (1.3-4.7) | 1.5 (0.6-4.3) | 3.0 (1.2-6.0) |
| NH4 concn (g liter−1) | 0.8 (0.4-1.6) | 0.1 (0.06-0.2) | 0.2 (0.1-0.5) | 1.1 (0.9-1.5) |
| S concn (g liter−1) | 0.3 (0.1-0.6) | 0.4 (0.2-0.6) | 0.2 (0.1-0.5) | 0.2 (0.2-0.4) |
| SO4 concn (g liter−1) | 0.5 (0.2-1.0) | 0.5 (0.3-0.9) | 0.2 (0.1-0.3) | 0.1 (0.1-0.2) |
| K2O concn (g liter−1) | 2.2 (1.3-3.0) | 2.2 (1.2-3.2) | 2.0 (1.0-3.2) | 1.9 (1.2-3.0) |
| Na concn (g liter−1) | 0.6 (0.3-1.0) | 0.6 (0.4-0.9) | 0.6 (0.5-0.8) | 0.5 (0.5-0.6) |
| APC (CFU ml−1) | 5.4 × 107 (0.04 × 107-7.2 × 107) | 6.1 × 107 (0.02 × 108-2.8 × 108) | 9.1 × 106 (0.08 × 107-2.7 × 107) | 3.0 × 106 (0.5 × 106-5.0 × 106) |
| AnPC (CFU ml−1) | 1.7 × 107 (0.1 × 107-5.1 × 107) | 1.4 × 106 (0.01 × 106-4.4 × 106) | 1.7 × 106 (0.02 × 106-8.6 × 106) | 1.5 × 106 (0.04 × 106-5.1 × 106) |
| CPC (CFU ml−1) | 1.8 × 105 (0.02 × 106-1.0 × 106) | 9.9 × 103 (0.0 × 104-1.7 × 104) | 1.4 × 103 (0.0 × 103-6.1 × 103) | 7.2 × 103 (0.03 × 104-2.9 × 104) |
| Anaerobic | ||||
| Total solids (g liter−1) | 44.0 (29.0-58.0) | 25.0 (16.0-36.0) | 18.0 (7.8-22.0) | 28.0 (17.0-54.0) |
| BOD5 (g liter−1) | 14.9 (13.1-18.1) | 1.9 (1.4-3.0) | 2.2 (0.9-4.7) | 10.6 (8.0-15.3) |
| Total Kjedahl N (g liter−1) | 2.9 (1.5-6.1) | 2.6 (1.5-4.8) | 2.4 (0.7-5.6) | 3.0 (1.2-6.0) |
| NH4 concn (g liter−1) | 0.8 (0.4-1.6) | 1.0 (0.7-1.6) | 1.0 (1.0-1.4) | 1.1 (0.9-1.5) |
| S concn (g liter−1) | 0.3 (0.1-0.6) | 0.1 (0.1-0.3) | 0.1 (0.1-0.2) | 0.2 (0.2-0.4) |
| SO4 concn (g liter−1) | 0.5 (0.2-1.0) | 0.3 (0.1-0.7) | 0.2 (0.1-0.3) | 0.1 (0.1-0.2) |
| K2O concn (g liter−1) | 2.2 (1.3-3.0) | 1.8 (1.2-2.6) | 1.5 (0.8-1.9) | 1.9 (1.2-3.0) |
| Na concn (g liter−1) | 0.6 (0.3-1.0) | 0.5 (0.2-0.8) | 0.5 (0.4-0.6) | 0.5 (0.5-0.6) |
| APC (CFU ml−1) | 5.4 × 107 (0.04 × 107-7.2 × 107) | 2.2 × 107 (0.04 × 107-7.3 × 107) | 2.7 × 106 (0.08 × 106-3.9 × 106) | 3.0 × 106 (0.5 × 106-5.0 × 106) |
| AnPC (CFU ml−1) | 1.7 × 107 (0.1 × 107-5.1 × 107) | 1.9 × 105 (0.3 × 105-2.2 × 105) | 3.3 × 106 (0.04 × 107-1.8 × 107) | 1.5 × 106 (0.04 × 106-5.1 × 106) |
| CPC (CFU ml−1) | 1.8 × 105 (0.02 × 106-1.0 × 106) | 5.4 × 102 (0.1 × 103-1.7 × 103) | 1.2 × 102 (0.1 × 102-2.2 × 102) | 7.2 × 103 (0.03 × 104-2.9 × 104) |
Abbreviations: APC, aerobic plate count; AnPC, anaerobic plate count; CPC, coliform plate count.
Analysis of 16S rRNA libraries derived from dairy waste, reactor effluents, and stored material.
To determine the effect of aerobic and anaerobic digestion on the bacterial population structure of dairy waste, we constructed 16S rRNA libraries from DNA extracted from waste, the effluent of the aerobic and anaerobic reactors, the effluent held in storage tanks, as well as control untreated material held in storage tanks over a 6-month period (Table 2). These sequences were analyzed using the Classifier software to determine the type of bacteria from which the sequences were most likely derived. At the phylum level, the majority of the 16S rRNA sequences derived from the feed material were assigned to the Firmicutes, followed by the Bacteroidetes, the Actinobacteria, the Proteobacteria, and the Spirochetes (Table 2). The library derived from the aerobic reactor effluent showed the greatest difference from the feed material, with the levels of eight out of nine phyla being significantly different. The most notable differences were the >16-fold increase in the phylum Proteobacteria and the approximately 8-fold decrease in the phylum Firmicutes. Other significant differences included the phyla Actinobacteria, Deinococcus-Thermus, Planctomycetes, Spirochetes, TM7, and Verrucomicrobia. The sequences derived from the aerobic reactor effluent held in storage vessels showed significant increases in the phyla Firmicutes, Planctomycetes, and Spirochetes and a decrease in the phylum TM7 as compared to the aerobic reactor-derived library. After anaerobic digestion, the bacterial population structure showed statistically significant decreases in the phyla Actinobacteria and Bacteroidetes and a statistically significant increase in the phyla Planctomycetes, Spirochetes, and TM7. The sequences derived from the anaerobic reactor effluent held in a storage tank showed a significant increase in the phylum Deinococcus-Thermus, while all other phyla in the library showed no significant change from those of the anaerobic reactor. Comparisons between the libraries derived from the feed material and the control untreated material held in storage tanks revealed a significant increase in the level of Spirochetes, while all other phyla levels remained unchanged. Comparisons of libraries derived from the two untreated control storage tanks, which differed from each other only in the volume of material that was pumped into them each day, showed no significant differences in any of the phyla. Comparisons of the aerobic and anaerobic reactor effluent libraries showed significant differences in all phyla except the Planctomycetes and the Bacteroidetes.
TABLE 2.
Percentage of rRNA gene clones assigned to phyla before and after treatment and storage
| Treatment and phylum assignment | Avg (range) % of clones assigned to phylum
|
|||
|---|---|---|---|---|
| Feed material | Reactor effluent | Effluent storage | Untreated storage | |
| Aerobic | ||||
| Actinobacteria | 5.3 (2.1-10.1) | 10.9 (6.1-34.9) | 5.1 (1.9-9.3) | 3.0 (1.7-4.7) |
| Bacteroidetes | 16.1 (9.0-24.3) | 15.4 (4.7-22.0) | 12.8 (9.8-17.8) | 8.9 (6.1-10.5) |
| Deinococcus-Thermus | NDa | 1.2 (0.0-1.7) | 0.8 (0.7-1.3) | ND |
| Firmicutes | 74.8 (61.8-88.0) | 9.5 (4.0-17.4) | 21.7 (13.4-27.4) | 78.0 (72.2-80.1) |
| Planctomycetes | ND | 1.1 (0.6-2.3) | 5.7 (1.3-7.5) | ND |
| Proteobacteria | 3.4 (0.0-5.1) | 55.1 (46.0-59.9) | 48.8 (37.5-58.2) | 3.3 (2.1-3.8) |
| Spirochetes | 0.2 (0.0-1.6) | ND | 0.4 (0.0-1.2) | 1.5 (0.6-3.2) |
| TM7 | ND | 4.5 (0.6-11.0) | 1.1 (0.6-1.6) | ND |
| Verrucomicrobia | ND | 1.1 (0.0-1.7) | 0.3 (0.0-0.7) | ND |
| Unknown | 0.7 (0.0-1.9) | 1.1 (0.6-1.8) | 3.4 (0.7-7.4) | 4.9 (2.3-6.1) |
| Anaerobic | ||||
| Actinobacteria | 5.3 (2.1-10.1) | 2.0 (1.2-3.7) | 1.8 (1.2-2.9) | 2.2 (1.3-3.9) |
| Bacteroidetes | 16.1 (9.0-24.3) | 9.9 (5.6-15.9) | 10.8 (8.4-14.5) | 11.5 (6.5-13.8) |
| Deinococcus-Thermus | ND | ND | 4.6 (0.0-13.4) | ND |
| Firmicutes | 74.8 (61.8-88.0) | 74.3 (64.0-79.3) | 64.2 (74.5-56.2) | 76.5 (73.2-81.7) |
| Planctomycetes | ND | 0.5 (0.0-0.7) | 0.5 (0.0-1.4) | ND |
| Proteobacteria | 3.4 (0.0-5.1) | 4.9 (2.2-11.2) | 9.3 (2.3-17.3) | 4.5 (0.8-7.8) |
| Spirochetes | 0.2 (0.0-1.6) | 0.8 (0.5-1.4) | 0.5 (0.0-1.9) | 0.8 (0.6-1.3) |
| TM7 | ND | 0.2 (0.0-0.7) | 0.2 (0.0-0.6) | ND |
| Verrucomicrobia | ND | ND | ND | ND |
| Unknown | 0.7 (0.0-1.9) | 7.2 (3.4-10.8) | 8.0 (4.2-9.8) | 4.1 (1.4-6.7) |
ND, not determined.
Identification of the 10 most prevalent OTUs from each library.
The 10 most numerous OTUs identified in each library are presented in Table 3 and Fig. 2. These results are consistent with the phylum assignment data. For example, 7 of the 10 most prominent OTUs from the feed material are members of the phylum Firmicutes (the most predominant phylum), 2 are from the phylum Bacteroidetes (the second largest phylum), and 1 is from the phylum Proteobacteria (the fourth most predominant phylum). This trend continues throughout the aerobic effluent-, anaerobic effluent-, and storage-derived libraries. Samples subjected to a similar treatment regimen yielded libraries displaying similar 16S rRNA gene sequence composition. For example, the feed material-derived library shares 50% of the 10 most prevalent OTUs with the untreated storage-derived library, but only 10% with the aerobic reactor effluent-derived library. Likewise, the anaerobic reactor effluent-derived library shares 60% of the OTUs with the anaerobic effluent storage-derived library but has none in common with the aerobic effluent storage-derived library.
TABLE 3.
The 10 most commonly isolated OTUs from each library
| OTUa | No. of clones | % of total | Phylum (% confidence threshold)b | Best match in GenBank | % Similarity |
|---|---|---|---|---|---|
| F1 | 204 | 6.0 | Firmicutes (100) | Trichococcus flocculiformis | 97-98 |
| F2 | 151 | 4.4 | Firmicutes (100) | AY438851 | 98-99 |
| F3 | 142 | 4.2 | Firmicutes (100) | AF371787 | 99 |
| F4 | 96 | 2.8 | Firmicutes (100) | AY100573 | 98-99 |
| F5 | 86 | 2.5 | Firmicutes (100) | AY438899 | 98 |
| F6 | 83 | 2.4 | Firmicutes (100) | AY438880 | 97-98 |
| F7 | 77 | 2.3 | Firmicutes (100) | Clostridium lituseburense | 98-100 |
| F8 | 60 | 1.8 | Bacteroidetes (100) | AY438832 | 98-99 |
| F9 | 58 | 1.7 | Bacteroidetes (100) | AB219992 | 92-94 |
| F10 | 58 | 1.7 | Proteobacteria (100) | Pseudomonas sp. strain SKU | 98-99 |
| AR1 | 154 | 9.6 | Proteobacteria (100) | Thauera terpenica | 99-100 |
| AR2 | 57 | 3.5 | Actinobacteria (100) | Aeromicrobium marinum | 97-98 |
| AR3 | 54 | 3.4 | Proteobacteria (100) | Pseudomonas sp. strain SKU | 98-100 |
| AR4 | 48 | 3.0 | Proteobacteria (100) | Dyella japonica | 96-98 |
| AR5 | 40 | 2.5 | Proteobacteria (100) | Roseobacter sp. strain YS-57 | 99 |
| AR6 | 35 | 2.2 | Bacteroidetes (100) | Sphingobacterium thalpophilum | 97-98 |
| AR7 | 34 | 2.1 | Proteobacteria (100) | Xanthomonas axonopodis | 95-97 |
| AR8 | 34 | 2.1 | Bacteroidetes (96) | UBA318142 | 95-96 |
| AR9 | 33 | 2.1 | Proteobacteria (100) | Dyella koreensis | 97-98 |
| AR10 | 26 | 1.6 | Bacteroidetes (99) | AF507866 | 96-97 |
| AnR1 | 144 | 8.6 | Bacteroidetes (100) | CR933150 | 97-99 |
| AnR2 | 139 | 8.3 | Firmicutes (100) | Sedimentibacter sp. B4 | 96-97 |
| AnR3 | 130 | 7.8 | Firmicutes (88) | DQ191708 | 96-97 |
| AnR4 | 125 | 7.5 | Firmicutes (100) | Clostridium lituseburense | 98-100 |
| AnR5 | 58 | 3.5 | Actinobacteria (62) | AB092855 | 99-100 |
| AnR6 | 43 | 2.7 | Firmicutes (100) | AY438851 | 98 |
| AnR7 | 32 | 1.9 | Proteobacteria (62) | AB232562 | 96-97 |
| AnR8 | 31 | 1.8 | Firmicutes (100) | Trichococcus flocculiformis | 97-98 |
| AnR9 | 31 | 1.8 | Firmicutes (100) | Eubacterium tenue | 99-100 |
| AnR10 | 30 | 1.8 | Firmicutes (100) | AY100573 | 98-99 |
| AS1 | 194 | 17.6 | Proteobacteria (100) | Thauera terpenica | 99-100 |
| AS2 | 58 | 5.3 | Planctomycetes (100) | Pirellula sp. | 98-99 |
| AS3 | 31 | 2.8 | Proteobacteria (100) | Roseobacter sp. YS-57 | 97-99 |
| AS4 | 20 | 1.8 | Proteobacteria (100) | Rhodobacter gluconicum | 99-100 |
| AS5 | 19 | 1.7 | Firmicutes (100) | AY570630 | 98-99 |
| AS6 | 18 | 1.6 | Proteobacteria (100) | Xanthomonas axonopodis | 95-96 |
| AS7 | 16 | 1.5 | Firmicutes (100) | Tissierella praeacuta | 93-95 |
| AS8 | 16 | 1.5 | Proteobacteria (78) | AY438740 | 99 |
| AS9 | 16 | 1.5 | Proteobacteria (100) | Thermomonas hydrothermalis | 95-97 |
| AS10 | 15 | 1.4 | Bacteroidetes (100) | Petrimonas sulfuriphila | 98-99 |
| AnS1 | 80 | 7.3 | Firmicutes (100) | Clostridium lituseburense | 98-100 |
| AnS2 | 64 | 5.9 | Firmicutes (100) | DQ191708 | 95-96 |
| AnS3 | 53 | 4.9 | Firmicutes (100) | Turicibacter sanguinis | 99-100 |
| AnS4 | 48 | 4.4 | Planctomycetes (100) | Pirellula sp. | 98-100 |
| AnS5 | 47 | 4.3 | Firmicutes (100) | Sedimentibacter sp. strain B4 | 96-97 |
| AnS6 | 46 | 4.2 | Proteobacteria (100) | Pseudomonas sp. strain SKU | 99-100 |
| AnS7 | 46 | 4.1 | Bacteroidetes (100) | AY953168 | 97-98 |
| AnS8 | 33 | 3.0 | Bacteroidetes (100) | CR933150 | 98-99 |
| AnS9 | 30 | 2.7 | Firmicutes (100) | Eubacterium tenue | 99-100 |
| AnS10 | 28 | 2.6 | Actinobacteria (55) | AB092855 | 99-100 |
| US1 | 166 | 14.5 | Firmicutes (100) | Clostridium lituseburense | 98-100 |
| US2 | 48 | 4.2 | Firmicutes (100) | Turicibacter sanguinis | 99-100 |
| US3 | 36 | 3.2 | Firmicutes (100) | AF371787 | 99 |
| US4 | 35 | 3.1 | Firmicutes (100) | Eubacterium tenue | 99-100 |
| US5 | 30 | 2.6 | Spirochaetes (64) | AY228699 | 97-98 |
| US6 | 24 | 2.1 | Bacteroidetes (100) | AY438832 | 98-99 |
| US7 | 20 | 1.8 | Proteobacteria (100) | Pseudomonas sp. strain SKU | 99-100 |
| US8 | 19 | 1.7 | Firmicutes (100) | AY622268 | 94-95 |
| US9 | 19 | 1.7 | Firmicutes (100) | AY438851 | 98-99 |
| US10 | 17 | 1.5 | Bacteroidetes (100) | AY953229 | 96-98 |
Abbreviations are as follows: F, feed material; AR, aerobic reactor effluent; AnR, anaerobic reactor effluent; AS, aerobic reactor effluent storage tank; AnS, anaerobic reactor effluent storage tank; US, untreated storage tank.
The confidence threshold is an estimation of the classification reliability using bootstrapping.
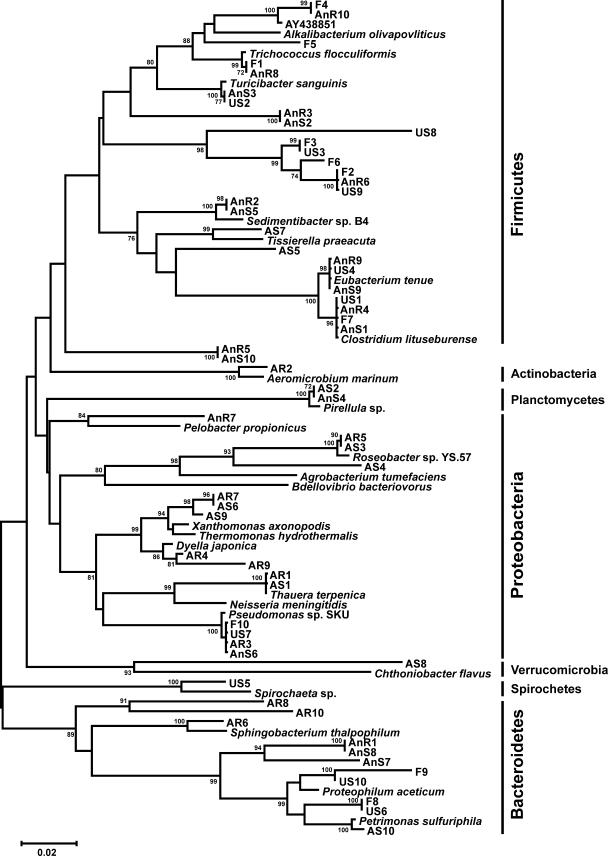
FIG. 2.
Phylogenetic relationship of the operational taxonomic units isolated from feed material (F), aerobic and anaerobic reactor effluent (AR and AnR, respectively), aerobic and anaerobic storage tanks (AS and AnS, respectively), and untreated storage (US). 16S rRNA gene sequences representing the 10 most prevalent OTUs from each environment and the most similar 16S rRNA gene sequences to each OTU from the NCBI nr database were aligned using CLUSTALX. The dendrogram was constructed using the neighbor-joining algorithm and the Kimura two-parameter distance estimation method. Bootstrap values of >70%, generated from 1,000 replicates, are shown at the nodes. The scale bar represents the number of substitutions per site. Phylum designations are indicated on the right.
Estimates of diversity, coverage, and rarefaction.
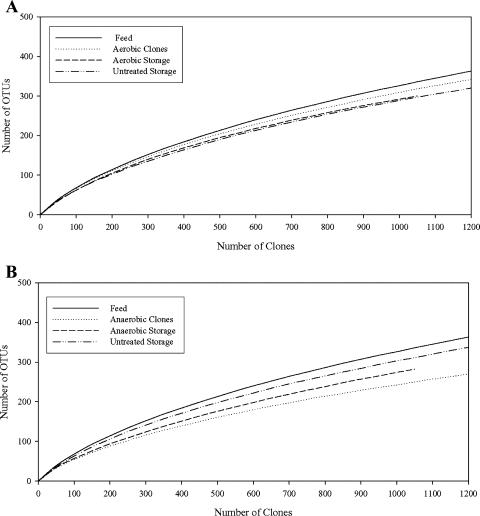
The diversity within the libraries, as measured by the Shannon-Wiener diversity index (H), is presented in Table 4. H was greatest for the feed material-derived library (5.28), indicating that this library contained the greatest diversity. H decreased in the aerobic reactor effluent-derived library (5.04) and decreased further in the aerobic effluent storage tank-derived library (4.69), indicating that both aerobic treatment and subsequent storage have negative effects on diversity. H also decreased in the anaerobic reactor-derived library (4.46), but increased slightly in the anaerobic effluent storage tank-derived library (4.60). The index of evenness (E), which is proportional to the number of individuals that belong to each OTU, was 0.82 for the feed material-derived library and declined in the anaerobic reactor effluent-derived library (0.77) but increased in the aerobic reactor-derived library (0.84). Reductions in evenness indices were observed in the aerobic reactor effluent storage material (0.82) but increased in the anaerobic storage tank-derived library (0.81). The libraries derived from untreated waste held in storage tanks had very similar H values (4.79 and 4.81) and the same E values (0.83). Analysis of the libraries revealed that the coverage within the feed material-derived library was the highest (82.0%), followed by those in the anaerobic reactor-derived library (80.8%), the aerobic reactor-derived library (74.8%), the anaerobic effluent storage tank-derived library (72.4%), and the aerobic effluent storage tank-derived library (69.7%). Both of the untreated control material-derived libraries had similar coverage levels (72.2 and 73.6%). Rarefaction analysis of the 16S rRNA libraries indicates that our sampling was not exhaustive, but that most predominant OTUs were likely identified as the slopes of all of the curves decrease greatly towards the end points (Fig. 3). These graphs are in agreement with the Shannon-Wiener index data, indicating diversity is lost after aerobic or anaerobic treatment and continued to decline during the storage of the aerobic reactor effluent while storage of the anaerobic reactor effluent resulted in increased diversity.
TABLE 4.
Estimation of diversity within rRNA gene libraries
| Library | No. of clones | Richness (no. of OTUs) | % Coverage | Eveness index (E) | Shannon index (H) |
|---|---|---|---|---|---|
| Feed material | 3,434 | 618 | 82.0 | 0.82 | 5.28 |
| Reactor | |||||
| Aerobic | 1,593 | 401 | 74.8 | 0.84 | 5.04 |
| Anaerobic | 1,693 | 325 | 80.8 | 0.77 | 4.46 |
| Storage | |||||
| Aerobic | 1,017 | 308 | 69.7 | 0.82 | 4.69 |
| Anaerobic | 1,047 | 289 | 72.4 | 0.81 | 4.60 |
| Untreated | |||||
| Storage 1 | 1,177 | 327 | 72.2 | 0.83 | 4.79 |
| Storage 2 | 1,221 | 323 | 73.6 | 0.83 | 4.81 |
FIG. 3.
Rarefaction curves for the (A) aerobic reactor and (B) the anaerobic reactor. Rarefaction analysis was performed using the approximation algorithm of Hurlbert (18) with 95% confidence intervals estimated as described by Heck et al. (16) using the freeware program aRarefactWin by Holland (http://www.uga.edu/strata/software/Software.html).
DISCUSSION
Modern high-intensity dairies generate copious amounts of waste that is usually stored in holding lagoons until it is applied to agricultural land as a fertilizer. This practice is becoming more problematic due to changes in agricultural demographics that concentrate large confined animal feeding operations in geographically limited regions like the San Joaquin Valley of California. These changes result in greater amounts of waste being deposited on crop fields with the potential to contribute to food-borne illness (4), surface and groundwater contamination (15), and poor air quality (17). A possible solution to these challenges is the treatment of waste before storage and subsequent land application. The objective of the present study was to model a typical dairy waste stream, monitor the chemical and bacterial population dynamics that occur during aerobic or anaerobic treatment and subsequent storage, and compare them to those of waste held without treatment in a simulated storage lagoon.
Our results indicate that both aerobic and anaerobic treatments followed by storage were superior to storage alone for the reduction of total solids, BOD5, and coliform bacteria. In addition to these reductions, each system had unique remediation properties. For example, aerobic treatment significantly reduced both total nitrogen and ammonia levels. These reductions are likely the result of the deamination of proteins and peptides and the hydrolysis of urea to ammonia by ruminant bacteria (12, 28). In the oxygen-rich environment of the aerobic reactor, ammonia likely became nitrified by ammonia-oxidizing bacteria of the genus Nitrosomonas, whose 16S rRNA gene sequences were only observed in the aerobic treatment system (data not shown). When the oxidized nitrogen species entered the anoxic conditions of the storage tank, they were denitrified to volatile nitrogen-containing gases that escaped into the atmosphere. In addition, some ammonia was likely volatized and assimilated by the bacteria. In the anaerobic system, significant reductions in sulfate and total sulfur were observed. This loss is likely explained by dissimilatory sulfate reduction to form hydrogen sulfide and other volatile sulfur-containing compounds (3, 5, 6) and, to a lesser extent, by assimilation.
At the phylum level, the feed material-derived 16S rRNA gene library was very similar to a library constructed from dairy waste reported previously (26). In both of these libraries, the greatest percentages of sequences were from members of the phylum Firmicutes (74% in this study versus 77% in the previous one), followed by the phyla Bacteroidetes (16% versus 7%), Actinobacteria (11% versus 9%), and Proteobacteria (3% versus 5%). The feed material library also possessed similarities to libraries derived from human feces (10), the gastrointestinal tracts of pigs (22), and, to a lesser extent, broiler chicken litter (23). The aerobic reactor effluent library had similarities to a library derived from a circulated dairy waste lagoon. In these libraries, the phylum Proteobacteria was most prominent followed by the Firmicutes, Bacteroidetes, and Actinobacteria (25). However, these libraries differed in the abundance of the phylum Firmicutes, which represented 26.8% of the circulated waste lagoon-derived library, as compared to only 9.5% in the aerobic reactor effluent-derived library. This difference may be explained by the growth inhibition of many of the obligate anaerobic members of the Firmicutes in the aerobic reactor which maintained an oxygen concentration of 2 mg liter−1 compared to the circulated waste lagoon, which was essentially anoxic. The predominance of Firmicutes 16S rRNA sequences increased to 21.7% after storage in a simulated waste lagoon, making it more closely resemble the library derived from the circulated dairy waste lagoon reported previously (25). The increased number of Firmicutes-like sequences may be explained by the anoxic conditions encountered in the simulated waste lagoon that support the growth of the obligate anaerobic species within this phylum. The library generated from the anaerobic reactor was similar to a library derived from a stagnant dairy waste lagoon (25); however, the relative levels of the Proteobacteria and Bacteroidetes were inverted. Subsequent storage of the anaerobic reactor effluent did little to change the bacterial community structure at the phylum level, with only a slight increase in the phylum Deinococcus-Thermus observed.
Of the 10 most prevalent OTUs in the waste-derived library, most have been recovered previously in dairy waste (F7) (26), wastewater lagoons (F2, -5, -6, and -8) (25), or the gastrointestinal tracts of swine (F3) (22). Storage without treatment does little to change the predominance of these OTUs, with the vast majority resembling those isolated previously in dairy waste (US1) (26), dairy wastewater (US2, -4, -6, -7, and -9) (25), or swine waste (US3 and -10) (22). Aerobic treatment and subsequent effluent storage resulted in the greatest changes in the most commonly observed OTUs, with only 3 of 20 OTUs previously associated with waste (AR3 and -6 and AS7), and the rest were similar to environmental isolates. Anaerobic treatment and subsequent storage resulted in fewer changes in the OTUs identified. Many of the 10 most prevalent OTUs have been recovered previously in manure or stagnant dairy waste lagoons (AnR4, -6, -8, -9, and -10 and AnS1, -3, -6, -8, and -9) (25, 26).
The results presented here were obtained from bench-scale (3 to 5 liters) reactors and thus may not exactly replicate the much larger systems needed for a 1,000-cow dairy farm. For example, the hydraulic retention times, reactor temperatures, and mixing methods will likely require modifications during scale up. However, the results obtained using this model provide insights as to how the full-scale reactors will perform. Ultimately, the type of waste treatment utilized on dairies and other confined animal feeding operations will depend on multiple factors, including cost, type and amount of nutrient reduction desired, and government-imposed emission regulations. Because nitrogen is usually the limiting nutrient in animal waste-based fertilizers, anaerobic digestion, which tends to conserve nitrogen and is also the least expensive method to employ, will likely be popular. In addition, methane can be collected from anaerobic reactors and used as a fuel to generate heat and electricity or to run farm equipment. Another key factor is volatile chemical emissions, which are becoming a major problem in agricultural regions such as the San Joaquin Valley of California, where dairy farming is intensive. We are currently examining the emissions of various gases, including volatile organic compounds, from these processes.
Acknowledgments
This research was supported in part by the U.S. Department of Agriculture, Agricultural Research Service, National Program 108, and grant no. 008826 from the California Environmental Protection Agency, State Water Resources Control Board, and the Merced County Department of Environmental Health, awarded to F.M.
We acknowledge the technical assistance of Jeremy Lathrop and Anna Korn and thank Jenn Brofft and Jenni Boonjakjuakul for critical reading of the manuscript.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, DC.
- 3.Arogo, J. Z., R. H. Zhang, and D. L. Day. 2000. Hydrogen sulfide production from stored liquid swine manure: a laboratory study. Trans. ASAE 43:1241-1245. [Google Scholar]
- 4.Beuchat, L. R., and J. H. Ryu. 1997. Produce handling and processing practices. Emerg. Infect. Dis. 3:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., M. J. Higgins, N. A. Maas, S. N. Murthy, W. E. Toffey, and D. J. Foster. 2005. Roles of methanogens on volatile organic sulfur compound production in anaerobically digested wastewater biosolids. Water Sci. Technol. 52:67-72. [PubMed] [Google Scholar]
- 6.Clanton, C. J., and D. R. Schmidt. 2000. Sulfur compounds in gases emitted from stored manure. Trans. ASAE 43:1229-1239. [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, M. T. 1997. Mycobacterium paratuberculosis: a potential food-borne pathogen? J. Dairy Sci. 80:3445-3448. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricke, K., H. Santen, and R. Wallmann. 2005. Comparison of selected aerobic and anaerobic procedures for MSW treatment. Waste Manag. 25:799-810. [DOI] [PubMed] [Google Scholar]
- 12.Fulghum, R. S., and W. E. C. Moore. 1963. Isolation, enumeration, and characteristics of proteolytic ruminal bacteria. J. Bacteriol. 85:808-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerba, C. P., and J. E. Smith, Jr. 2005. Sources of pathogenic microorganisms and their fate during land application of wastes. J. Environ. Qual. 34:42-48. [PubMed] [Google Scholar]
- 14.Hancock, D. D., T. E. Besser, and D. H. Rice. 1998. Ecology of E. coli O157:H7 in cattle and impact of management practices. ASM Press, Washington, DC.
- 15.Harter, T., H. Davis, M. C. Mathews, and R. D. Meyer. 2002. Shallow groundwater quality on dairy farms with irrigated forage crops. J. Contam. Hydrol. 55:287-315. [DOI] [PubMed] [Google Scholar]
- 16.Heck, K. L., Jr., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 17.Hughes, L. S., J. O. Allen, L. G. Salmon, P. R. Mayo, R. J. Johnson, and G. R. Cass. 2002. Evolution of nitrogen species air pollutants along trajectories crossing the Los Angeles area. Environ. Sci. Technol. 36:3928-3935. [DOI] [PubMed] [Google Scholar]
- 18.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 19.Juteau, P., D. Tremblay, C. B. Ould-Moulaye, J. G. Bisaillon, and R. Beaudet. 2004. Swine waste treatment by self-heating aerobic thermophilic bioreactors. Water Res. 38:539-546. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid sequencing techniques in bacterial systematics. Wiley and Sons, New York, NY.
- 22.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 25.McGarvey, J. A., W. G. Miller, S. Sanchez, C. J. Silva, and L. C. Whitehand. 2005. Comparison of bacterial populations and chemical composition of dairy wastewater held in circulated and stagnant lagoons. J. Appl. Microbiol. 99:867-877. [DOI] [PubMed] [Google Scholar]
- 26.McGarvey, J. A., W. G. Miller, S. Sanchez, and L. Stanker. 2004. Identification of bacterial populations in dairy wastewaters by use of 16S rRNA gene sequences and other genetic markers. Appl. Environ. Microbiol. 70:4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miner, R. J., F. J. Humenik, and M. R. Overcash. 2000. Managing livestock wastes to preserve environmental quality. Iowa State University Press, Ames.
- 28.Mobley, H. L. T., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohaibes, M., and H. Heinonen-Tanski. 2004. Aerobic thermophilic treatment of farm slurry and food wastes. Bioresour. Technol. 95:245-254. [DOI] [PubMed] [Google Scholar]
- 30.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 31.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie, N. J., M. E. Schutter, R. P. Dick, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarzenbeck, N., J. M. Borges, and P. A. Wilderer. 2005. Treatment of dairy effluents in an aerobic granular sludge sequencing batch reactor. Appl. Microbiol. Biotechnol. 66:711-718. [DOI] [PubMed] [Google Scholar]
- 34.Shima, E., I. F. Svoboda, S. Tsutsumi, and H. Ohkubo. 2002. Waste management systems of dairy cattle farms in Japan. Water Sci. Technol. 45:63-69. [PubMed] [Google Scholar]
- 35.U.S. Department of Agriculture, National Agricultural Statistics Service. 2005. 2005 agricultural statistics. U.S. Department of Agriculture, Washington, DC. http://www.usda.gov/nass/pubs/agstats.htm.
- 36.Van Horn, H. H., A. C. Wilkie, W. J. Powers, and R. A. Nordstedt. 1994. Components of dairy manure management systems. J. Dairy Sci. 77:2008-2030. [DOI] [PubMed] [Google Scholar]
- 37.Warnick, L. D., L. M. Crofton, K. D. Pelzer, and M. J. Hawkins. 2001. Risk factors for clinical salmonellosis in Virginia, USA cattle herds. Prev. Vet. Med. 49:259-275. [DOI] [PubMed] [Google Scholar]
- 38.Wesley, I. V., S. J. Wells, K. M. Harmon, A. Green, L. Schroeder-Tucker, M. Glover, and I. Siddique. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]