Abstract
Listeria monocytogenes is a facultative intracellular pathogen responsible for food-borne disease with high mortality rates in humans and is the leading microbiological cause of food recalls. Lineage I isolates of L. monocytogenes are a particular public health concern because they are responsible for most sporadic cases of listeriosis and the vast majority of epidemic outbreaks. Rapid, reproducible, and sensitive methods for differentiating pathogens below the species level are required for effective pathogen control programs, and the CDC PulseNet Task Force has called for the development and validation of DNA sequence-based methods for subtyping food-borne pathogens. Therefore, we developed a multilocus genotyping (MLGT) assay for L. monocytogenes lineage I isolates based on nucleotide variation identified by sequencing 23,251 bp of DNA from 22 genes distributed across seven genomic regions in 65 L. monocytogenes isolates. This single-well assay of 60 allele-specific probes captured 100% of the haplotype information contained in approximately 1.5 Mb of comparative DNA sequence and was used to reproducibly type a total of 241 lineage I isolates. The MLGT assay provided high discriminatory power (Simpson's index value, 0.91), uniquely identified isolates from the eight listeriosis outbreaks examined, and differentiated serotypes 1/2b and 4b as well as epidemic clone I (ECI), ECIa, and ECII. In addition, the assay included probes for a previously characterized truncation mutation in inlA, providing for the identification of a specific virulence-attenuated subtype. These results demonstrate that MLGT represents a significant new tool for use in pathogen surveillance, outbreak detection, risk assessment, population analyses, and epidemiological investigations.
Listeria monocytogenes is the etiologic agent of listeriosis, an invasive food-borne disease that creates significant challenges for public health and the food industry. Clinical features of listeriosis include encephalitis, meningitis, septicemia, and abortion (11). Due to the severe clinical symptoms associated with listeriosis, L. monocytogenes has the second highest case mortality rate (20 to 30%) of any food-borne pathogen (30) and is responsible for over one-quarter of food-borne-disease-related deaths attributable to known pathogens (16). In addition, L. monocytogenes has been the leading cause of food recalls due to microbial adulteration (34, 50). L. monocytogenes is ubiquitously distributed in the environment, has the ability to associate with biofilms (54), demonstrates a high resistance to ionizing radiation (47), can tolerate high-salt and low-pH conditions, and is both microaerophilic and psychrotrophic (28). These traits allow L. monocytogenes to persist in food-processing environments and make L. monocytogenes a serious problem in ready-to-eat (RTE) meat products and cold-stored food that is eaten without significant heating (24).
DNA sequence and ribotype analyses have demonstrated that L. monocytogenes consists of at least three phylogenetically distinct lineages (41, 51, 53). Relative to their prevalence in animal listeriosis and food contamination isolates, lineage I isolates are overrepresented in human listeriosis cases (17, 22, 35). In addition, lineage I isolates are responsible for the vast majority of listeriosis outbreaks and 62.9% of human sporadic cases (22). Serotype 4b isolates from lineage I are of particular concern because they contribute significantly to sporadic listeriosis and include three previously defined epidemic clones responsible for multiple listeriosis outbreaks in Europe and North America (24).
Molecular subtyping methods are critical components of epidemiological investigation, outbreak detection, and source-tracking activities that are required for effective pathogen control programs. Due to the prevalence of lineage I isolates and, in particular, serotype 4b isolates among human listeriosis cases, significant attention has been devoted to differentiating these isolates below the lineage and serotype levels (4, 6, 9, 18). This is particularly problematic because L. monocytogenes lineage I appears to have experienced a population bottleneck that purged genetic variation, such that genetic distances between lineage I strains are significantly less than that for the other lineages of L. monocytogenes (51).
Pulsed-field gel electrophoresis (PFGE) is the current gold standard for subtyping most bacterial pathogens (13) and has been used for the molecular subtyping of L. monocytogenes as part of the PulseNet system since 1998 (48). However, PFGE patterns are complex and not always easy to interpret (13). In addition, PFGE is relatively labor-intensive and time-consuming, cannot be adapted to target specific polymorphisms of interest, and can be affected by relatively unstable genetic elements, such as plasmids and phages. As a result, PulseNet participants have expressed interest in the development and integration of new DNA sequence-based methods for subtyping food-borne pathogens (13, 48). Recently, a number of multilocus sequence typing (MLST) methods have been described for L. monocytogenes (42, 43, 58). However, MLST methods are expensive and time-consuming because they require numerous sequencing reactions per isolate and cannot be multiplexed. As the vast majority of sites sequenced for MLST are invariant, direct interrogation of single-nucleotide polymorphisms (SNPs) represents a more efficient alternative for DNA sequence-based subtyping. In this study, we describe the development and validation of the first single-well DNA sequence-based subtyping assay for L. monocytogenes lineage I isolates based on multilocus genotyping (MLGT) of SNP sites via flow cytometry.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 241 lineage I isolates of L. monocytogenes were used in this study. Lineage was determined by allele-specific oligonucleotide multiplex PCR as previously described (51). Serotypes were determined or confirmed by serotype-specific multiplex PCR assay as previously described (7). Multiplex PCR determined that serotypes correspond to one of the two major serotype complexes found in lineage I, serotype 1/2b complex (includes 3b isolates) and serotype 4b complex (includes 4d and 4e isolates) (7). Listeria isolates were maintained in the Agricultural Research Service Culture Collection (NCAUR, Peoria, IL) in liquid nitrogen vapor at −175°C. Isolates were cultured in brain heart infusion broth or tryptic soy agar supplemented with 0.6% (wt/vol) yeast extract (Difco) at 37°C.
Comparative DNA sequence analysis.
DNA isolation, PCR amplification, and DNA sequencing were performed as previously described (51). Reaction conditions are provided as Supplemental Material 1 (see the supplemental material) or were previously described (51). A panel of 65 lineage I isolates of L. monocytogenes, taken from clinical, veterinary, food, and environmental sources was selected to represent lineage I phylogenetic diversity identified in a previous evolutionary analysis of prfA virulence gene cluster sequences (51). Genetic polymorphisms were identified by obtaining 23,251 bp of DNA sequence from seven genomic regions encompassing 22 complete or partial genes (Table 1) and 15 intergenic regions. These regions were chosen to include genes responsible for virulence, stress response, and housekeeping functions. DNA sequences were edited and aligned with Sequencher (version 4.1.2; Gene Codes), variable sites were identified with MEGA (version 3.0 [25]), and unique multilocus haplotypes were identified with DAMBE (55).
TABLE 1.
Single nucleotide polymorphism and haplotype variation among 65 L. monocytogenes lineage 1 isolates
| Region | Positiona | Genesb | SNPs | Haplotypes |
|---|---|---|---|---|
| VGC | 209009-217550 | prfA, plcA, hly, mpl, actA, plcB | 102 | 21 |
| LMO | 341498-343450 | LMOf2365_0319-0321 | 107 | 23 |
| INL | 484557-488736 | inlA, inlB | 145 | 22 |
| SIG | 930033-931913 | rsbV, rsbW, sigB, rsbX | 11 | 10 |
| PDH | 1079888-1082533 | pdhA, pdhB, pdhC | 34 | 16 |
| AMI | 1533277-1535138 | ami, hisS | 6 | 7 |
| ACC | 1592519-1594571 | accD, dnaE | 8 | 10 |
Corresponding nucleotide positions in the genome sequence of L. monocytogenes strain 4b F2365 (NC002973.6).
Gene designations follow the annotation of the F2365 genome or homologous sequences from L. monocytogenes EGD-e (NC003210).
Phylogenetic reconstructions were performed under both distance and maximum parsimony frameworks. Prior to phylogenetic or haplotype analyses, ambiguously aligned characters were removed from the data set, and insertion or deletion (indel) polymorphisms that defined unique haplotypes were coded as single events. Distance analyses were performed using the neighbor-joining algorithm as implemented in MEGA. Maximum parsimony analyses were conducted using the tree bisection and reconnection method of branch swapping and the heuristic search algorithm of PAUP* (version 4.0B; Sinauer Associates). Relative support for individual nodes was assessed by bootstrap analysis with 1,000 replications (12, 39).
Multiplex amplification of template for MLGT assay.
Portions of the genomic regions listed in Table 1, totaling 18,832 aligned nucleotides, were coamplified in a single multiplex PCR using nine sets of amplification primers (Table 2), which enabled the simultaneous interrogation of polymorphisms identified in the comparative sequence analysis. Amplifications were performed in standard 50-μl reaction mixtures according to manufacturer specifications and included 2 mM MgSO4, 100 μM deoxynucleoside triphosphate, 300 nM primer, 1.5 U Platinum Taq DNA Polymerase High Fidelity (Invitrogen Life Technologies), and 100 ng of genomic DNA. PCR consisted of an initial denaturation of 90 s at 96°C, followed by 40 cycles of 30 s at 94°C, 45 s at 50°C, and 180 s at 68°C. Amplification products were purified using Montage PCR cleanup filter plates (Millipore) and stored at −20°C prior to use in extension reactions.
TABLE 2.
Primers used in multiplex amplification of MLGT assay
| Amplicon (size) | Primer | Region | Sequence (5′ to 3′)a |
|---|---|---|---|
| 1 (1,973 bp) | dnaE-F | ACC | GGATTTTCDCTTGGAGAAGCAG |
| dnaE-R | ACC | CRCCGACAACAGARCCCATAC | |
| 2 (1,782 bp) | hisS-F | AMI | CTTTCAGCRAGCGATTTATCAC |
| hisS-R | AMI | CGAACAACMGAAGCRGTCCC | |
| 3 (2,618 bp) | pdh-F | PDH | ACTTATCTGGCACGGACTTCC |
| pdh-R | PDH | TTTACGTACAGAAGGCATTGC | |
| 4 (2,479 bp) | hly-F2 | VGC | GAAGGAGAGTGAAACCCATG |
| mpl-R | VGC | CTCGCAACTTCYGGCTCAGC | |
| 5 (2,152 bp) | actA-F | VGC | CCAAYTGCATTACGATTAACC |
| plcB-R | VGC | CAAGCACATACCTAGAACCAC | |
| 6 (2,220 bp) | inlA-F2 | INL | ACGAAYGTAACAGACACGGTC |
| inlA-R2 | INL | CTACTTCTATTTACTAGCACG | |
| 7 (1,750 bp) | inlB-F2 | INL | AATCAAGGAGAGGATAGTGTG |
| inlB-R | INL | CTACCGGRACTTTATAGTAYG | |
| 8 (1,680 bp) | sigB-F | SIG | CATTACAACTTCCTGCCAAGC |
| sigB-R | SIG | GAACAAGTCCATCTGAATGCA | |
| 9 (2,052 bp) | lmo0300-F | LMO | GATTGGATTAAGTACGAGC |
| lmo0298-R | LMO | CTCTTATCAATGTGAAGGTGC |
See IUPAC codes for the definition of degenerate bases.
Probe design and extension reactions for the MLGT assay.
Combinations of polymorphic sites were selected for assay development in order to provide efficient discrimination of identified sequence types and to identify novel haplotypes from isolates that had not been sequenced. Oligonucleotide probes with 3′ nucleotides specific to individual SNP character states were designed from DNA sequence alignments. In addition, probes were designed for two indel polymorphisms as well as two SNP probes (1 truncation probe and 1 full-length probe) to examine a lineage I-specific nonsense mutation in inlA resulting in a virulence-attenuated phenotype. Each of the SNP, indel, or truncation probes was synthesized with a unique 24-bp sequence tag on the 5′ end that was specific to antitag sequences attached to the surface of individual sets of Luminex xMAP fluorescent polystyrene microspheres (Luminex Corporation). These probes served as allele-specific primers in multiplex extension reactions producing single-stranded DNA amplicons from multiplex-generated DNA that contained the probe sequence. Extension reactions were performed in standard 20-μl reaction mixtures according to manufacturer specifications and included 2 mM MgCl2, 5 μM biotin-dCTP, 5 μM dATP/dGTP/dTTP, 25 nM primer, 0.75 U Platinum GenoTYPE Tsp DNA polymerase (Invitrogen Life Technologies). Five microliters of purified multiplex PCR was added as the template for extension reactions, which were performed with an initial denaturation of 120 s at 96°C, followed by 40 cycles of 30 s at 94°C, 60 s at 52°C, and 150 s at 74°C.
Hybridization and detection for the MLGT assay.
Biotinylated extension products were hybridized with a mix of microsphere sets specific to each of the sequence tags appended to the 5′ end of the extension probes. Hybridization reactions were performed in 50-μl volumes with 1× TM buffer (0.2 M NaCl, 0.1 M Tris, 0.08% Triton X-100), pH 8.0, 10 μl of extension product, and 1,250 microspheres from each set. The samples were incubated for 90 s at 96°C, followed by 45 min at 37°C. Microspheres were twice pelleted by centrifugation (3 min at 3,700 × g) and resuspended in 70 μl 1× TM buffer. Following these washes, microspheres were pelleted and resuspended in 70 μl 1× TM buffer containing 2 μg/ml streptavidin R-phycoerythrin. Samples were incubated for 15 min at 37°C prior to detecting the microsphere complexes with a Luminex 100 flow cytometer (Luminex Corporation). Individual microsphere sets are labeled by the manufacturer with a specific mix of fluorescent dyes, creating a unique spectral address that enables extension products from different probes to be sorted and evaluated individually. The median fluorescence intensity (MFI) from biotinylated extension products attached to 100 microspheres was measured for each probe. The MFI of the average of three template-free control samples was also determined and subtracted from the raw MFI of each sample to account for background fluorescence.
Each probe was designed to match a specific SNP or indel character state, referred to as the target allele. Probe performance was assessed by comparing MFI values for isolates with target and nontarget alleles from the panel of 65 sequenced isolates. Each isolate was genotyped via two independent runs of the MLGT assay, and the results were combined to determine an index of discrimination (not related to the Simpson's index), defined as the ratio of the lowest target MFI to the highest nontarget MFI value for each probe. Probes with a ratio of less than 2.0 were redesigned. Minimum threshold values for discriminating between positive and negative genotypes for allele-specific probes were set at twice the value of the highest nontarget MFI that was observed in the two runs of the MLGT assay across the 65 sequenced isolates. In addition, positive control probes were designed to confirm the presence of each of the amplicons in the multiplex PCR. Minimum thresholds for positive control probes were set at 90% of the lowest MFI observed in the MLGT analysis of the 65 sequenced isolates. A step-by-step outline of the MLGT assay is provided in Table S1 in the supplemental material.
Comparative subtyping.
PFGE was performed following the PulseNet standardized protocol (15) implemented by the USDA Food Safety and Inspection Service (FSIS) using AscI. A Gel-Doc 2000 system (Bio-Rad Laboratories) was used for image acquisition. Image analysis was performed using BioNumerics software (Applied Maths) with manual inspection. Multilocus sequencing was carried out as previously described by Revazishvili et al. (42), and DNA sequences were edited and analyzed as described above. Simpson's discrimination index (SDI) values were calculated as previously described (20). SDI values are scored between 0 and 1. An SDI value approaching 1 indicates a greater discriminatory power of the method being analyzed.
Nucleotide sequence accession numbers.
DNA sequences were deposited in the GenBank database under accession numbers DQ812146 to DQ812517, DQ843664 to DQ844598, and AY512391 to AY512502.
RESULTS AND DISCUSSION
DNA sequence analysis.
A total of 413 SNPs, which defined 35 unique multilocus sequence types, were identified among the 23,251 bp of aligned DNA sequence from the 65 L. monocytogenes lineage I isolates listed in Table 3. Two additional sequence types were defined by indel variation within the LMO and SIG regions (Table 1). Individual analysis of the seven sequenced regions revealed substantial heterogeneity in SNP frequency and sequence type discrimination. The largest amount of variation was contained in the INL, LMO, and VGC regions, each of which contained more than 100 SNPs that defined at least 21 sequence types (Table 1). The remaining regions averaged 14.8 SNPs that defined an average of 10.8 sequence types. LMO was the most informative region sequenced and contained significantly greater numbers of SNPs (5.5; P < 0.001) and sequence types (1.2; P < 0.05) per 100 bp than did the other sequenced regions (average, 1.2 SNPs and 0.5 sequence types per 100 bp).
TABLE 3.
L. monocytogenes lineage 1 isolates analyzed by multilocus genotype analysis
| Isolatea | Equivalent | Origin | Outbreak | Serotypeb | PCR serotypec | STd | MLGT |
|---|---|---|---|---|---|---|---|
| 33010e | 4b complex | ST1-1a | Lm1.1 | ||||
| 33013e | ScottA | Human | MA 1983 | 4b | 4b complex | ST1-1a | Lm1.1 |
| 33049e | F4638 | Human | MA 1983 | 4b | 4b complex | ST1-1a | Lm1.1 |
| 33089 | F4637 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33093 | F4639 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33145e | F4645 | Human | MA 1983 | 4b | 4b complex | ST1-1a | Lm1.1 |
| 33146 | F4636 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33147 | F4644 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33149 | F4642 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33151 | F4641 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33153 | F4640 | Human | MA 1983 | 4b | 4b complex | Lm1.1 | |
| 33170 | 4b complex | Lm1.1 | |||||
| 33001e | RM2205 | Human | 4b | 4b complex | ST1-1b | Lm1.2 | |
| 33067 | 81-505 | Human | 4b | 4b complex | Lm1.2 | ||
| 33141e | 81-859 | Human | 4b | 4b complex | ST1-1b | Lm1.2 | |
| 33331 | Food | 4b complex | Lm1.2 | ||||
| 33334 | Food | 4b complex | Lm1.2 | ||||
| 33337 | Food | 4b complex | Lm1.2 | ||||
| 33355 | Food | 4b complex | Lm1.2 | ||||
| 33359 | Food | 4b complex | Lm1.2 | ||||
| 33413 | Ts45 | Food | UK 1988 | 4b | 4b complex | Lm1.2 | |
| 33414 | Ts38 | Human | UK 1988 | 4b | 4b complex | Lm1.2 | |
| 33224e | J0094 | 4d | 4b complex | ST1-1c | Lm1.3 | ||
| 33144e | 2112 | Food | 4b | 4b complex | ST1-1d | Lm1.4 | |
| 33007e | RM2218 | Food | 4b | 4b complex | ST1-2a | Lm1.5 | |
| 33217e | F113V | Animal | 4b | 4b complex | ST1-2a | Lm1.5 | |
| 33008e | RM2387 | Food | 4b | 4b complex | ST1-3a | Lm1.6 | |
| 33083e | F1109 | Food | 4b | 4b complex | ST1-4a | Lm1.7 | |
| 33098e | F5069 | Food | 4b | 4b complex | ST1-4a | Lm1.7 | |
| 33156 | V37CE | Food | 4b | 4b complex | Lm1.7 | ||
| 33158 | F1057 | Food | 4b | 4b complex | Lm1.7 | ||
| 33166e | 81-507 | Human | 4b | 4b complex | ST1-4a | Lm1.7 | |
| 33221e | LMB0347 | Human | 4b | 4b complex | ST1-4a | Lm1.7 | |
| 33252 | Food | 4b complex | Lm1.7 | ||||
| 33094e | 3889 | Animal | 4b | 4b complex | ST1-5a | Lm1.8 | |
| 33222e | F347S | Human | 4b | 4b complex | ST1-5a | Lm1.8 | |
| 33420 | F581E | Human | USA 1998 | 4b | 4b complex | Lm1.8 | |
| 33432 | Food | 4b complex | Lm1.8 | ||||
| 33453 | Food | 4b complex | Lm1.8 | ||||
| H7858e | Food | USA 1998 | 4b | 4b complex | ST1-5b | Lm1.9 | |
| 33386 | F470E | Human | USA 1998 | 4b | 4b complex | Lm1.9 | |
| 33179e | 25734-97 | Animal | 4b | 4b complex | ST1-5c | Lm1.10 | |
| F2365e | Food | CA 1985 | 4b | 4b complex | ST1-6a | Lm1.11 | |
| 33000 | F2379 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33004e | RM2215 | Food | CA 1985 | 4b | 4b complex | ST1-6a | Lm1.11 |
| 33050 | F7213 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33052 | F7243 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33055 | F7214 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33059 | F7231 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33060 | F7224 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33065 | F2385 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33066 | F7223 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33070 | F7225 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33071 | LALM-8 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33072 | F7150 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33084 | LALM-3 | Environment | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33086 | F7157 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33087 | F7206 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33091 | F2381 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33096 | LALM-5 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33097 | F2382 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33103 | F7215 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33104 | F2365 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33107 | LALM-7 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33108 | F2387 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33109 | LALM-1 | Environment | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33111 | F7149 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33112 | F2380 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33113 | LALM-4 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33117 | F7244 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33121 | F7207 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33122 | F7248 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33133 | LALM-6 | Environment | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33135 | F7226 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33137 | F7245 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33143e | DA-3 | Human | CA 1985 | 4b | 4b complex | ST1-6a | Lm1.11 |
| 33155 | LALM-10 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33157e | LALM-2 | Environment | CA 1985 | 4b | 4b complex | ST1-6a | Lm1.11 |
| 33159 | F2392 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33161 | F2386 | Food | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33410 | F4565 | Human | CA 1985 | 4b | 4b complex | Lm1.11 | |
| 33047e | 81-558 | Human | Halifax 1981 | 4b | 4b complex | ST1-6b | Lm1.12 |
| 33048 | 81-784 | Halifax 1981 | 4b | 4b complex | Lm1.12 | ||
| 33051 | 81-739 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33053 | 81-861 | Food | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33054 | 81-1087 | Food | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33056e | 81-884 | Human | Halifax 1981 | 4b | 4b complex | ST1-6b | Lm1.12 |
| 33057 | 81-509 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33058 | 81-619 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33061 | 81-678 | Halifax 1981 | 4b | 4b complex | Lm1.12 | ||
| 33062 | 81-501 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33063 | 81-511 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33075 | 81-498 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33079 | 81-515 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33081 | 81-711 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33082 | 81-618 | Halifax 1981 | 4b | 4b complex | Lm1.12 | ||
| 33099 | 81-590 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33101 | 81-923 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33102 | 81-694 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33110 | 81-679 | Halifax 1981 | 4b | 4b complex | Lm1.12 | ||
| 33118 | 81-591 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33119 | 81-516 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33129 | 81-512 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33131 | 81-637 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33134 | 81-682 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33136 | 81-592 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33138 | 81-502 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33139 | 81-886 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33142 | 81-680 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33150 | 81-499 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33163 | 81-712 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33411 | Ts50 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33412 | Ts27 | Human | Halifax 1981 | 4b | 4b complex | Lm1.12 | |
| 33011e | 4b complex | ST1-6c | Lm1.13 | ||||
| 33012e | 4b complex | ST1-6c | Lm1.13 | ||||
| 33078e | 7680 | Animal | 4b | 4b complex | ST1-6c | Lm1.13 | |
| 33085 | F7250 | Human | CA 1985 | 4b | 4b complex | Lm1.13 | |
| 33120e | ATCC 19118 | Animal | 4e | 4b complex | ST1-6c | Lm1.13 | |
| 33125e | 3869 | Animal | 4b | 4b complex | ST1-6c | Lm1.13 | |
| 33174 | 4b complex | Lm1.13 | |||||
| 33322 | Food | 4b complex | Lm1.13 | ||||
| 33323 | Food | 4b complex | Lm1.13 | ||||
| 33389 | FSL C1-122 | Human | 4b | 4b complex | Lm1.13 | ||
| 33415 | Ts21 | Food | Lausanne 1987 | 4b | 4b complex | Lm1.13 | |
| 33416 | Ts60 | Human | Lausanne 1987 | 4b | 4b complex | Lm1.13 | |
| 33140e | 2617 | Animal | 4b | 4b complex | ST1-6d | Lm1.14 | |
| 33095e | 7037 | Animal | 4b | 4b complex | ST1-6e | Lm1.15 | |
| 33116e | ATCC 19117 | Animal | 4d | 4b complex | ST1-7a | Lm1.16 | |
| 33015e | 12375 | 4b | 4b complex | ST1-8a | Lm1.17 | ||
| 33033e | OB001206 | Food | 1/2b | 1/2b complex | ST1-9a | Lm1.18 | |
| 33173 | 1/2b complex | Lm1.18 | |||||
| 33287 | Food | 1/2b complex | Lm1.18 | ||||
| 33291 | Food | 1/2b complex | Lm1.18 | ||||
| 33301 | Environment | 1/2b complex | Lm1.18 | ||||
| 33325 | Food | 1/2b complex | Lm1.18 | ||||
| 33394 | Human | 1/2b complex | Lm1.18 | ||||
| 33423 | G6003 | Food | IL 1994 | 1/2b | 1/2b complex | Lm1.18 | |
| 33424 | G6054 | Human | IL 1994 | 1/2b | 1/2b complex | Lm1.18 | |
| 33465 | Environment | 1/2b complex | Lm1.18 | ||||
| 33475 | Ts7 | Human | 3b | 1/2b complex | Lm1.18 | ||
| 33493 | Ts25 | Human | 3b | 1/2b complex | Lm1.18 | ||
| 33522 | Ts54 | Human | 3b | 1/2b complex | Lm1.18 | ||
| 33068e | 8058 | Animal | 1/2b | 1/2b complex | ST1-10a | Lm1.19 | |
| 33073e | 3883 | Animal | 1/2b | 1/2b complex | ST1-10a | Lm1.19 | |
| 33074e | 8054 | Animal | 1/2b | 1/2b complex | ST1-10a | Lm1.19 | |
| 33175 | 1/2b complex | Lm1.19 | |||||
| 33327 | Food | 1/2b complex | Lm1.19 | ||||
| 33038e | OB001385 | Food | 1/2b | 1/2b complex | ST1-11a | Lm1.20 | |
| 33315 | Food | 1/2b complex | Lm1.20 | ||||
| 33356 | Food | 1/2b complex | Lm1.20 | ||||
| 33037e | OB001350 | Food | 1/2b | 1/2b complex | ST1-12a | Lm1.21 | |
| 33251 | 1/2b complex | Lm1.21 | |||||
| 33358 | Food | 1/2b complex | Lm1.21 | ||||
| 33126e | 7034 | Animal | 1/2b | 1/2b complex | ST1-13a | Lm1.22 | |
| 33178e | 32736-96 | Animal | 1/2b | 1/2b complex | ST1-13a | Lm1.22 | |
| 33176e | 20240-954 | Animal | 1/2b | 1/2b complex | ST1-14a | Lm1.23 | |
| 33090e | 7675 | Animal | 1/2b | 1/2b complex | ST1-15a | Lm1.24 | |
| 33114e | 2613 | Animal | 1/2b | 1/2b complex | ST1-15a | Lm1.24 | |
| 33130e | 2071 | Food | 1/2b | 1/2b complex | ST1-15a | Lm1.24 | |
| 33162 | Animal | 1/2b complex | Lm1.24 | ||||
| 33164e | 5712 | Food | 1/2b | 1/2b complex | ST1-15a | Lm1.24 | |
| 33329 | Food | 1/2b complex | Lm1.24 | ||||
| 33392 | FSL J2-035 | Animal | 1/2b | 1/2b complex | Lm1.24 | ||
| 33028e | OB001102 | Food | 1/2b | 1/2b complex | ST1-16a | Lm1.25 | |
| 33308 | Food | 1/2b complex | Lm1.25 | ||||
| 33046e | OB000255J | Food | 1/2b | 1/2b complex | ST1-16b | Lm1.26 | |
| 33242 | Food | 1/2b complex | Lm1.26 | ||||
| 33309 | Food | 1/2b complex | Lm1.26 | ||||
| 33042e | OB000217B | Food | 1/2b | 1/2b complex | ST1-16c | Lm1.27 | |
| 33006 | Food | 1/2b complex | Lm1.28 | ||||
| 33030e | OB001171 | Food | 1/2b | 1/2b complex | ST1-16d | Lm1.28 | |
| 33239 | Food | 1/2b complex | Lm1.28 | ||||
| 33240 | Food | 1/2b complex | Lm1.28 | ||||
| 33245 | Environment | 1/2b complex | Lm1.28 | ||||
| 33250 | Food | 1/2b complex | Lm1.28 | ||||
| 33254 | Food | 1/2b complex | Lm1.28 | ||||
| 33262 | Food | 1/2b complex | Lm1.28 | ||||
| 33265 | Food | 1/2b complex | Lm1.28 | ||||
| 33269 | 1/2b complex | Lm1.28 | |||||
| 33272 | Food | 1/2b complex | Lm1.28 | ||||
| 33273 | Food | 1/2b complex | Lm1.28 | ||||
| 33284 | Food | 1/2b complex | Lm1.28 | ||||
| 33296 | Food | 1/2b complex | Lm1.28 | ||||
| 33303 | Environment | 1/2b complex | Lm1.28 | ||||
| 33304 | Food | 1/2b complex | Lm1.28 | ||||
| 33341 | Food | 1/2b complex | Lm1.28 | ||||
| 33347 | Food | 1/2b complex | Lm1.28 | ||||
| 33351 | Food | 1/2b complex | Lm1.28 | ||||
| 33434 | Environment | 1/2b complex | Lm1.28 | ||||
| 33445 | Food | 1/2b complex | Lm1.28 | ||||
| 33457 | Environment | 1/2b complex | Lm1.28 | ||||
| 33458 | Environment | 1/2b complex | Lm1.28 | ||||
| 33123e | 2110 | Environment | 1/2b | 1/2b complex | ST1-16e | Lm1.29 | |
| 33228e | ILSI09 | Human | 3b | 1/2b complex | ST1-16e | Lm1.29 | |
| 33248 | Food | 1/2b complex | Lm1.29 | ||||
| 33393 | FSL J1-169 | Human | 3b | 1/2b complex | Lm1.29 | ||
| 33459 | Environment | 1/2b complex | Lm1.29 | ||||
| 33461 | Environment | 1/2b complex | Lm1.29 | ||||
| 33463 | Environment | 1/2b complex | Lm1.29 | ||||
| 33466 | Environment | 1/2b complex | Lm1.29 | ||||
| 33581 | J1768 | Human | 3b | 1/2b complex | Lm1.29 | ||
| 33005e | RM2216 | Food | 1/2b | 1/2b complex | ST1-16f | Lm1.30 | |
| 33036e | Food | 1/2b complex | ST1-16f | Lm1.30 | |||
| 33080e | 7679 | Animal | 1/2b | 1/2b complex | ST1-16f | Lm1.30 | |
| 33154e | LALM-11 | Food | 1/2b | 1/2b complex | ST1-16f | Lm1.30 | |
| 33220e | B345S | Human | 1/2b | 1/2b complex | ST1-16f | Lm1.30 | |
| 33263 | Food | 1/2b complex | Lm1.30 | ||||
| 33293 | Food | 1/2b complex | Lm1.30 | ||||
| 33294 | Food | 1/2b complex | Lm1.30 | ||||
| 33300 | Environment | 1/2b complex | Lm1.30 | ||||
| 33302 | Environment | 1/2b complex | Lm1.30 | ||||
| 33305 | Food | 1/2b complex | Lm1.30 | ||||
| 33306 | Food | 1/2b complex | Lm1.30 | ||||
| 33312 | Food | 1/2b complex | Lm1.30 | ||||
| 33390 | FSL J2-064 | Animal | 1/2b | 1/2b complex | Lm1.30 | ||
| 33442 | Food | 1/2b complex | Lm1.30 | ||||
| 33444 | Food | 1/2b complex | Lm1.30 | ||||
| 33451 | Environment | 1/2b complex | Lm1.30 | ||||
| 33148e | 5713 | Environment | 1/2b | 1/2b complex | ST1-16g | Lm1.31 | |
| 33045e | Food | 1/2b complex | ST1-16h | Lm1.32 | |||
| 33032e | OB001186 | Food | 1/2b | 1/2b complex | ST1-17a | Lm1.33 | |
| 33124e | Food | 1/2b complex | ST1-17b | Lm1.34 | |||
| 33218e | LMB0338 | Human | 1/2b | 1/2b complex | ST1-17c | Lm1.35 | |
| 33186e | 20674-01 | Animal | 1/2b | 1/2b complex | ST1-18a | Lm1.36 | |
| 33369 | 24155-03 | Animal | 1/2b | 1/2b complex | Lm1.36 | ||
| 33160e | 3682 | Food | 1/2b | 1/2b complex | ST1-19a | Lm1.37 | |
| 33391 | FSL J1-177 | Human | 1/2b | 1/2b complex | Lm1.37 | ||
| 33462 | Environment | 1/2b complex | Lm1.37 | ||||
| 33464 | Environment | 1/2b complex | Lm1.37 | ||||
| 33313 | Food | 1/2b complex | Lm1.38 | ||||
| 33237 | Food | 1/2b complex | Lm1.39 | ||||
| 33258 | Food | 1/2b complex | Lm1.39 | ||||
| 33320 | Food | 1/2b complex | Lm1.39 | ||||
| 33343 | Food | 1/2b complex | Lm1.40 | ||||
| 33345 | Food | 1/2b complex | Lm1.41 | ||||
| 33346 | Food | 1/2b complex | Lm1.41 | ||||
| 33421 | J0144 | Food | NC 2000 | 4b | 1/2b complex | Lm1.42 | |
| 33422 | J0211 | Human | NC 2000 | 4b | 1/2b complex | Lm1.42 | |
| 33289 | Food | 1/2b complex | Lm1.43 | ||||
| 33429 | Food | 1/2b complex | Lm1.43 | ||||
| 33430 | Food | 1/2b complex | Lm1.43 |
With the exception of F2365 and H7858, isolates are identified with NRRL B numbers from the U.S. Department of Agriculture, Agricultural Research Service Culture Collection, Peoria, IL. F2365 and H7858 are equivalent to NRRL B-33232 and NRRL B-33233, respectively. However, sequence data for these isolates were obtained from GenBank accession numbers NC002973 and NZ_AADR00000000, respectively. Additional strain history information is available from the ARS Culture Collection website (http://nrrl.ncaur.usda.gov/cgi-bin/usda).
Serotypes were previously reported (by Schönberg et al. [45], Wesley and Ashton [52], Ward et al. [51], Borucki et al. [1, 2], and Zhang et al. [58]) or were previously determined by a reference laboratory.
Determined using the method of Doumith et al. (7).
ST, sequence type.
Isolates used in multilocus sequence analysis.
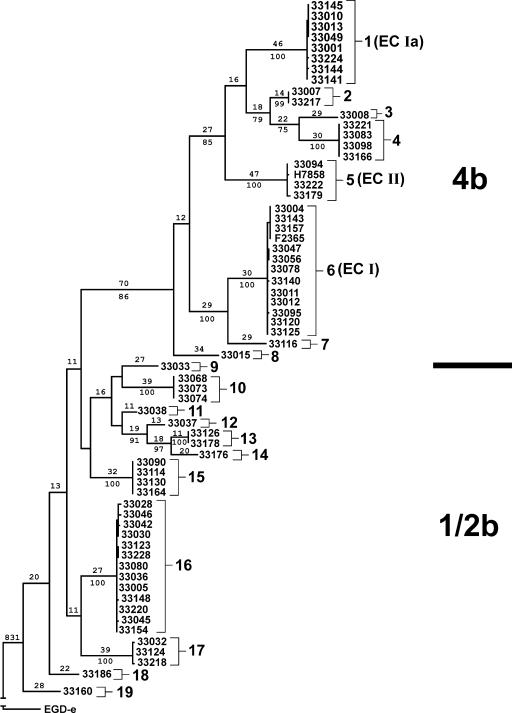
Phylogenetic analysis of the concatenated data from the seven sequenced regions indicated that the 37 observed sequence types were organized into 19 major sequence clusters that reflected serotype differences (Fig. 1). A monophyletic clade composed of isolates with serotype 4b, 4d, or 4e (serotype 4b complex) was strongly supported (86% bootstrap support) as distinct from sequence types composed of isolates with serotype 1/2b or 3b (serotype 1/2b complex), and there were no sequence types shared in common between 4b complex and 1/2b complex isolates (Table 3). Seventeen sequence types and eight major sequence clusters were observed within the monophyletic clade composed of 4b complex isolates (Fig. 1). Major sequence clusters 1, 5, and 6 corresponded to the previously reported epidemic clones ECIa, ECII, and ECI, respectively, which were responsible for numerous epidemic outbreaks of listeriosis in North America and Europe (10, 24, 40). We equate these major sequence clusters with epidemic clonal lineages on the basis that these clusters represent strongly supported monophyletic lineages that include isolates from one or more epidemic outbreaks and are readily distinguishable from other lineages identified in the phylogenetic analysis of the 22-gene DNA sequence data (Fig. 1). To confirm the utility of this phylogenetic approach, we determined that only the serotype 4b complex isolates within major sequence cluster 6 were resistant to Sau3AI digestion, which Zheng and Kathariou (59) described as a unique feature of ECI isolates (data not shown).
FIG. 1.
One of 72 most parsimonious trees inferred from analysis of the combined sequence data for 65 isolates. The tree was rooted with sequence data from the lineage II L. monocytogenes strain EGD-e (GenBank accession number NC003210). Branch lengths inferred by maximum parsimony are provided above individual branches. The frequency (%) with which a given branch was recovered in 1,000 bootstrap replications is shown below branches recovered in more than 70% of bootstrap replicates. Major sequence clusters are demarcated with numbered brackets. With the exception of F2365 and H7858, strains are identified by NRRL B numbers. Similar results were obtained with neighbor-joining analysis. The estimated length of the branch leading to EGD-e is not represented in order to permit visualization of the much smaller branches distinguishing lineage I isolates.
While there has been speculation that these epidemic clonal lineages may possess unique adaptations that explain their repeated involvement in listeriosis outbreaks, these three epidemic clones do not form a distinct monophyletic group within the 4b clade (Fig. 1). ECIa and ECII are more closely related to each other than to ECI (Fig. 1). However, all three groups represent distinct evolutionary lineages within the 4b clade. Therefore, any adaptations shared in common between the epidemic lineages were likely present in the ancestral 4b type and are unlikely to have resulted from a shared evolutionary history unique to these epidemic lineages. The evolutionary divergence between epidemic clones (Fig. 1) is consistent with previously published reports documenting a large number of ECI- and ECII-specific genes (10, 19, 32), raising the possibility that each of the epidemic lineages may possess features unique to that lineage, which account for their association with listeriosis outbreaks.
Isolates representing four listeriosis outbreaks associated with the three epidemic clonal lineages were included in the set of sequenced isolates (Table 3). ECI was represented by isolates from outbreaks in California (CA 1985) and Halifax (Halifax 1981), while ECIa was represented by isolates from an outbreak in Massachusetts (MA 1983). ECII was represented by the available genome sequence of strain H7858, which was associated with a multistate outbreak that occurred between 1998 and 1999 in the United States (USA 1998). Each of these common-source outbreaks, including the two representatives of ECI, were defined as unique sequence types with nucleotide data from the seven sequenced regions (Table 3). This result is consistent with previous subtyping studies demonstrating that individual common source outbreaks associated with ECI represent closely related, but genetically distinct, groups of isolates (52).
Design and validation of the MLGT assay.
Based on sequences from the 65 isolates in Fig. 1, 60 probes were designed for MLGT analysis of L. monocytogenes lineage I strains. This panel included 49 SNP probes, two probes designed to examine a pair of deletion mutations in the SIG (3-bp deletion in isolate 33218) and LMO (single base pair deletion in isolate 33140) regions, and nine positive control probes designed to confirm the presence of each of the amplicons in the multiplex PCR. Probe sequences and probe performance data are provided in Table 4. The index of discrimination values for the MLGT probes ranged from 2.4 to 28.2 with a mean of 9.3, which means that the MFI values for isolates with a negative genotype were always less than half of the MFI values for isolates with a positive genotype. In addition, the MFI values from each of the 60 probes were consistent with expectations based on sequence data. The single-well MLGT assay identified all 37 unique haplotypes among the 65 sequenced isolates, accurately reproducing 100% of the haplotype information contained in approximately 1.5 Mb of comparative DNA sequence (23,251 bp from each of the 65 isolates).
TABLE 4.
Probe performance among 65 sequenced isolatesa
| Probe | Probe sequence | Luminex microsphere | Target MFI range | Target sample sizeb | Nontarget MFI range | IDc |
|---|---|---|---|---|---|---|
| ACC1 | GTTTCAACCGAAGTCACGCT | 60 | 393-758 | 34 | 13-59 | 7.0 |
| ACC2 | CGTGTTATTCGTAAAGTGCT | 29 | 1,639-1,846 | 2 | 24-86 | 19.1 |
| ACC3 | CCGCGCCCATTTCGATTGAC | 30 | 2,104-3,374 | 11 | 97-298 | 7.1 |
| ACC4 | CAAATTAAGCCAGTTTTAACT | 70 | 2,588-3,170 | 10 | 45-161 | 16.1 |
| ACC5 | GTCTTCTTGTGGGAAAATGCGGTTGG | 82 | 545-757 | 3 | 33-137 | 4.0 |
| ACC6 | GTAATTATTGTGGTTTTCAT | 12 | 1,971-5,964 | 34 | 37-338 | 5.8 |
| ACCxd | GTTTCAGCGCATCCAGTATCG | 65 | 2,477-4,033 | 65 | N/A | N/A |
| AMI1 | GCCGTCAGAAGAAACGATAGCGACG | 77 | 463-882 | 17 | 15-87 | 5.3 |
| AMI2 | TTTGACGACTACGGAATTCT | 14 | 1,718-1,885 | 1 | 1-61 | 28.2 |
| AMIxd | AAGTTGTTTGCCCGCTTACC | 37 | 815-1,504 | 65 | N/A | N/A |
| INLa1 | CACGGTCTCGCAAACAGATC | 91 | 552-817 | 26 | 8-50 | 11.0 |
| INLa2 | ATATAGACCCGCTTAAAAAC | 4 | 345-562 | 12 | 20-69 | 5.0 |
| INLa3 | AAATTTAAATCGGCTAGAAT | 13 | 389-497 | 4 | 11-58 | 6.7 |
| INLa4 | GGTCTAACAAAACTAACTGG | 46 | 890-1,028 | 1 | 13-76 | 11.8 |
| INLa5 | CCCTAGCTGGTCTAACCGCC | 68 | 473-747 | 4 | 25-82 | 5.8 |
| INLa6 | AGGTAAGTGACGTAAGCTCA | 85 | 583-1,227 | 29 | 7-59 | 9.9 |
| INLa7 | GGCATAACCAAATTAGCGAT | 55 | 2,386-4,774 | 33 | 129-245 | 9.8 |
| INLa8 | TGATTGCACCAGCTACTATA | 26 | 870-1,139 | 5 | 22-93 | 9.4 |
| INLa9 | GGTTATACTTTYAAAGGCTGGTAT | 40 | 390-940 | 61 | 84-129 | 3.0 |
| INLa10 | ATACTTTCAAAGGCTGGTAA | 62 | 870-1,139 | 4 | 10-310 | 5.7 |
| INLa11 | GGTATGACGCAAAAACTGGC | 80 | 655-1,278 | 23 | 38-145 | 4.5 |
| INLa12 | ACAAGTGGGATTTTGCAACG | 78 | 474-849 | 20 | 32-112 | 4.2 |
| INLa13 | CGCTCAATTCACGAAAAATT | 10 | 697-1,042 | 4 | 22-64 | 10.9 |
| INLa14 | GACCCTTATAATTCAAAAGC | 87 | 2,556-3,891 | 20 | 26-101 | 25.3 |
| INLa15 | ATGACCCTTATAATTCAAAAGAAA | 9 | 1,956-2,801 | 1 | 23-167 | 11.7 |
| INLaxd | GGTGTCGGATATTAGTGTTCTGGC | 88 | 897-1,630 | 65 | N/A | N/A |
| INLb1 | AGCGGAGACTATCACCGTGT | 25 | 1,549-2,492 | 16 | 51-138 | 11.2 |
| INLb2 | ACGGATCTAGTGACACAAAA | 28 | 196-488 | 18 | 14-65 | 3.0 |
| INLb3 | ACCTAAGTTCGATCAAGGAC | 69 | 233-824 | 19 | -3-50 | 4.7 |
| INLb4 | GATTTCATGGGAGAGTAACG | 59 | 2,802-3,539 | 8 | 76-244 | 11.5 |
| INLb5 | AAATGGTGGACATGAGTGGG | 2 | 690-1,279 | 31 | 20-143 | 4.8 |
| INLb6 | CGACTGAAAAAGCGGTGAAC | 64 | 1,063-1,639 | 27 | 2-78 | 13.7 |
| INLb7 | TGCAAGAGTGAAAAATGCGT | 66 | 2,452-2,998 | 3 | 49-193 | 12.7 |
| INLb8 | CGAAACCATACAATACAGCT | 18 | 1,670-3,415 | 23 | 22-75 | 11.2 |
| INLbxd | ACTGCACCTAAACCTCCGAC | 90 | 1,670-3,415 | 65 | N/A | N/A |
| LMO1 | GGATGATGAAAGAAGGCGGA | 96 | 488-679 | 2 | 38-172 | 2.8 |
| LMO2 | GATATAAAATCGGCAACTACCCA | 16 | 501-917 | 26 | 9-101 | 5.0 |
| LMO3 | AATCGGCAACTACCCACCTA | 48 | 1,461-1,633 | 1 | 8-77 | 19.0 |
| LMO4 | GAAAACAGACGAACTACAAG | 98 | 742-845 | 1 | 18-154 | 4.8 |
| LMO5 | CTCGTTAAACCATACACTGC | 24 | 677-828 | 1 | 13-59 | 11.5 |
| LMO6 | ATAGCGAATCCGAGTATACT | 6 | 1,039-1,045 | 1 | 91-264 | 3.9 |
| LMO7 | TATGCCACAAATTAAACAGA | 89 | 2,614-2,655 | 1 | 21-207 | 12.6 |
| LMOxd | ATGTCTACAGGAATGCTTGCG | 99 | 378-1,822 | 65 | N/A | N/A |
| PDH1 | GTGAAGGCCCAACATTAATT | 3 | 1,379-1,834 | 1 | 35-122 | 11.3 |
| PDH2 | GGTGAAGGAATTCATGAAGGTA | 100 | 1,068-1,148 | 1 | 102-316 | 3.4 |
| PDHxd | GTGTTGCTGCTCCAGATAGCG | 31 | 1,329-2,703 | 65 | N/A | N/A |
| SIG1 | GTGATAAAACATGGAGTGTC | 76 | 651-950 | 13 | 15-76 | 8.6 |
| SIG2 | AGAAGAGCTGACGAGAGAAC | 50 | 2,144-3,289 | 20 | 75-175 | 12.3 |
| SIG3 | ATTTCATCGGTGTCACGGAG | 44 | 344-597 | 11 | 40-146 | 2.4 |
| SIG4 | GTGTCACGGAAGAAGTTT | 71 | 2,198-2,337 | 1 | 53-279 | 7.9 |
| SIG5 | CAACGTATGCTCTTAGAGAAG | 7 | 1,667-2,340 | 5 | 84-223 | 7.5 |
| SIG6 | TGCCATAAAAGAGGATATCT | 19 | 2,451-3,083 | 2 | 66-158 | 15.5 |
| SIGxd | GGCTCGAAGCTAATAGAGCT | 5 | 2,429-3,577 | 65 | N/A | N/A |
| VGCa1 | AAGAAAATTTAATTTCATCCATA | 42 | 334-562 | 4 | 9-51 | 6.6 |
| VGCa2 | CGCTCGCGCTAAGTTCTGAA | 72 | 1,089-1,538 | 14 | 119-391 | 2.8 |
| VGCaxd | CACTCTGGAGGATACGTTGCTC | 22 | 650-1,596 | 65 | N/A | N/A |
| VGCb1 | CGACATAATATTTGCAGCGG | 86 | 599-613 | 1 | 18-73 | 8.3 |
| VGCb2 | AGCGGGGATTTAGCTAGTTC | 35 | 1,165-1,804 | 4 | 13-55 | 21.4 |
| VGCb3 | TTAGTTCGCTGAATAGTGGC | 75 | 481-1,324 | 41 | 16-65 | 7.4 |
| VGCbxd | CAATTGATATGCCGAGCCTACC | 73 | 1,883-2,619 | 65 | N/A | N/A |
N/A, not applicable.
Out of 65 sequenced strains; the remainder comprises the nontarget sample size.
Index of discrimination (ID) value, determined as minimum target MFI/maximum nontarget MFI.
Positive control probe.
Replicate runs of the MLGT assay for the 65 sequenced isolates and 176 additional lineage I isolates were performed in order to assess the repeatability of the assay and to determine the ability of the assay to type a large panel of isolates. The 14,460 genotypes produced from application of the 60-probe assay to all 241 isolates were identical between replicate MLGT runs, demonstrating 100% reproducibility for the MLGT assay. In addition, all 241 isolates were successfully typed via the MLGT assay. Over 99.9% of the 2,169 amplicons targeted in each MLGT run were amplified successfully. Only the SIG region of isolate NRRL B-33138 (Halifax 1981 outbreak isolate) failed to amplify as indicated by a SIGx signal (MFI = 96) below the cutoff value for this positive control probe (MFI = 2,186). The absence of this amplicon was confirmed by visualization following agarose electrophoresis, demonstrating that the positive control probe performed as intended. Sequence determination confirmed that the SIG amplification primer sites were conserved in B-33138, and this region amplified normally outside of multiplex PCR. However, the consistent failure of the SIG region to amplify in multiplex PCR of B-33138 was confirmed by agarose electrophoresis and the MLGT assay. As a result of the amplicon failure, all SIG probes for NRRL B-33138 were scored as missing data. However, this did not effect the identification of this isolate as MLGT haplotype Lm1.12, a result consistent with the other Halifax 1981 outbreak isolates (Table 3).
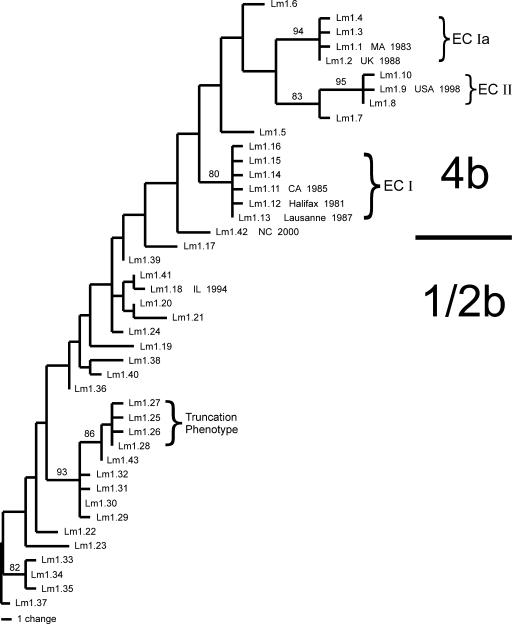
In total, 43 MLGT haplotypes were identified from the analysis of 241 lineage I isolates. Probe patterns for each haplotype are shown in Table S1 in the supplemental material. Thirty-seven of these haplotypes corresponded to the sequence types identified among the 65 sequenced isolates. In addition, six novel MLGT haplotypes were identified among the 176 isolates that had not been sequenced. The SNP and indel genotypes that defined the six novel haplotypes, labeled Lm1.38 through Lm1.43 (Table 3; see Table S1 in the supplemental material), were confirmed by sequence analysis. A comparison of MLGT results with strain histories, serotypes, and DNA sequence types (Table 3) indicated that the MLGT assay produced highly accurate subtype information. All 3,900 MLGT genotypes collected for the 65 sequenced isolates matched expectations based on DNA sequence data. There were no MLGT types shared in common between 4b complex and 1/2b complex isolates (Table 3), indicating that the major serotype complex could be predicted for unknown isolates based on MLGT type. For the six novel MLGT types that were not represented among the 65 sequenced isolates, the major serotype complex was accurately predicted based on their placement in the phylogenetic analysis of MLGT data (Fig. 2; Table 3).
FIG. 2.
One of six most parsimonious trees inferred from analysis of genotype data from the 43 unique haplotypes identified by application of the multilocus genotype assay to 241 lineage I L. monocytogenes isolates. Rooting was based on the results presented in Fig. 1. The frequency (%) with which a given branch was recovered in 1,000 bootstrap replications is shown above branches recovered in more than 70% of bootstrap replicates. Similar results were obtained with neighbor-joining analysis.
While MLGT types were unique to either the 4b or the 1/2b serotype complex, two MLGT types (Lm1.18 and Lm1.29) contained serotype 3b and 1/2b isolates, while MLGT type Lm1.13 contained serotype 4b and 4e isolates (Table 3). In addition, serotype 4d isolates were identified within two MLGT types that did not form a monophyletic group (94% bootstrap support) in the phylogenetic analysis of MLGT data (Fig. 2). These results are not surprising, as numerous molecular methods fail to discriminate these minor serotypes as independent groups, distinct from the more common 4b and 1/2b serotypes (1, 3, 7, 27, 31, 57, 58). In addition, our results suggest that serotypes 4d/4e and 3b arose from serotype 4b and 1/2b ancestors, respectively, while the polyphyletic distribution of 4d and 3b isolates indicate that these serotypes have multiple independent evolutionary origins and do not reflect distinct evolutionary groups (Fig. 2).
A total of 93 isolates from eight different outbreaks were included among the 241 lineage I isolates examined (Table 3). With two exceptions, all isolates from the same outbreak had identical MLGT haplotypes. NRRL B-33085, listed as a CA 1985 outbreak isolate, was classified as MLGT haplotype Lm1.13. However, the 39 other isolates examined from this outbreak were classified as MLGT haplotype Lm1.11, which is distinguished from Lm1.13 by a single-nucleotide difference in the hly gene from the VGC region. Similarly, an isolate (NRRL B-33420) from the USA 1998 outbreak was classified as MLGT haplotype Lm1.8. However, two additional isolates from that outbreak, including the genome sequence strain H7858, had MLGT haplotype Lm1.9, which is distinguished from Lm1.8 by a single-nucleotide difference in the LMO region. Sequence analysis of the regions containing these haplotype-defining SNPs confirmed that the MLGT genotypes were correct and that there were minor genetic differences between isolates within each of these outbreaks. This is not unexpected, as genetic or antigenic variation has been observed within multiple listeriosis outbreaks (reviewed by S. Kathariou in reference 24), including the CA 1985 and the USA 1998 outbreaks (5, 14, 57). Variation among isolates from a single outbreak has previously been interpreted as evidence that the outbreak could have resulted from a strain that was resident within the implicated food-processing facility long enough to produce minor variants (14). The fact that the intraoutbreak variants in our analyses differ by a mutation at a single SNP site is consistent with this hypothesis. However, we cannot discount the possibility that these isolates experienced mutations in culture following the outbreak investigations.
Excluding the two isolates that represented genetic variation within individual outbreaks, each of the eight examined outbreak strains had a different MLGT haplotype (Table 3). This is interesting because only four of these eight outbreaks were represented among the 65 sequenced isolates used for SNP discovery. Phylogenetic analysis of the MLGT haplotype data (Fig. 2) correctly identified the strains responsible for the CA 1985 (Lm1.11), Halifax 1981 (Lm1.12), and Lausanne 1987 (Lm1.13) outbreaks as members of a closely related lineage equivalent to the previously defined epidemic clone I (ECI) (10). The other lineage I epidemic clones (ECIa and ECII) were also resolved by phylogenetic analysis of the MLGT data, confirming the findings of De Cesare et al. (6), which stated that the strain responsible for an outbreak of listeriosis associated with contaminated pâté in the United Kingdom (UK 1988) (29) was a member of the ECIa lineage (Fig. 2). Phylogenetic analysis also indicated that the strain responsible for an outbreak associated with contaminated soft cheese in North Carolina (NC 2000) represented an evolutionary lineage distinct from previously described serotype 4b epidemic clonal lineages. Isolates from the NC 2000 outbreak had an MLGT haplotype (Lm1.42) that was not represented in the 65 sequenced isolates and was not associated with any other isolate examined in this study.
Overall, the phylogeny based on MLGT data (Fig. 2) is congruent with the phylogeny inferred from analysis of the 22-gene sequence data (Fig. 1). Both sets of data resolved 4b complex isolates as distinct from 1/2b complex isolates. In addition, both datasets resolved the individual epidemic clones within the serotype 4b clade as distinct monophyletic lineages and recovered the same evolutionary relationships between these epidemic clones. Additional probe development may be required to enhance bootstrap support for interior branches in the MLGT phylogeny. However, the fact that the MLGT data largely recapitulated the phylogenetic relationships inferred from analyses of more than 23 kb of DNA sequence confirmed the accuracy and epidemiological relevance of the subtype data produced by the MLGT assay.
Prevalence of epidemic clones in food.
Of the 241 isolates used in this study, 66 were part of an FSIS collection that consisted of isolates from RTE meat products and food-processing facilities (see Table S2 in the supplemental material). Of the 21 MLGT haplotypes identified among these isolates, only four MLGT types, representing nine isolates (13.6%), belonged to the serotype 4b complex. Combined with data from Ward et al. (51), indicating that lineage I accounted for approximately 47% of the L. monocytogenes isolates collected from RTE meat, this suggests that serotype 4b complex isolates are a rare (6.4%) contaminant of RTE meat products and food-processing facilities. This conclusion is consistent with the results of a recent study by Shen et al., which found that serotype 4b complex isolates accounted for approximately 7% of L. monocytogenes isolates collected from RTE foods in Florida (46).
While serotype 4b complex isolates may be rare in RTE foods, eight of the nine serotype 4b complex isolates from the FSIS panel had MLGT types specific to one of the three serotype 4b epidemic clones. Five of the nine serotype 4b complex isolates had an MLGT type associated with ECIa (Lm1.2; UK 1988), while ECI (Lm1.13; Lausanne 1987) and ECII (Lm1.8; USA 1998) were represented by one and two of the nine serotype 4b complex isolates, respectively. In a recent study of 34 serotype 4b isolates from RTE foods, 58.8% of isolates had ECI-specific genetic markers (56). These authors interpreted their findings as evidence that ECI strains may have a competitive edge over other 4b strains in food and food-processing environments, which may partially explain their repeated association with epidemic listeriosis in humans. While the small number of isolates included in our FSIS panel limits interpretation, our results suggest that this hypothesis could be extended to include ECIa and ECII strains. The ability to rapidly identify these epidemic subtypes using the MLGT assay will greatly facilitate additional surveys of RTE food products required to evaluate this hypothesis.
Examination of a virulence-attenuated subtype.
The ability to examine variation at individual nucleotide positions provides a mechanism for identifying genotypes that are directly responsible for specific phenotypes and is one of the key advantages of DNA sequence-based subtyping. The inlA gene encodes a membrane-anchored invasion protein that is critical for L. monocytogenes virulence (26). Analysis of the 65 sequenced isolates used to develop the MLGT assay revealed four serotype 1/2b isolates (NRRL B-33028, NRRL B-33030, NRRL B-33042, and NRRL B-33046) harboring a nonsense mutation in inlA equivalent to premature stop codon mutation type 1 (PMSC1) described by Nightingale et al. (33). This truncation occurs 5′ to the C-terminal LPXTG membrane-anchoring motif, which results in a secreted protein of 606 amino acids in length.
Previous studies have identified at least nine distinct mutations leading to InlA truncations occurring 5′ to the C-terminal LPXTG membrane-anchoring motif and documented that strains carrying these mutations display a virulence-attenuated phenotype in animal models and a significantly reduced ability to invade the Caco-2 human intestinal epithelial cell line (21, 23, 33, 37, 38). Only two of these nine truncation mutants have been confirmed among lineage I isolates: PMSC1 was previously reported to be the most frequent inlA truncation among lineage I isolates, and PMSC1 was the only inlA truncation observed among our panel of sequenced isolates. Phylogenetic analysis revealed that the four sequenced isolates we identified as harboring the PMSC1 mutation represent distinct but closely related sequence types within major sequence cluster 16 (Fig. 1). This cluster contained nine other sequenced isolates, comprising four additional sequence types, all of which had uninterrupted inlA open reading frames, suggesting that this InlA truncation had a very recent evolutionary origin.
In order to provide for the rapid identification of this specific set of virulence-attenuated subtypes, we developed an SNP probe (INLa10) specific to the nucleotide character state responsible for the truncated form of InlA. In addition, we developed a reciprocal probe (INLa9) specific to the alternate form of the inlA gene, which did not contain a stop codon at this location. MLGT analysis of all 241 lineage I isolates revealed 29 isolates that had an INLa9−/INLa10+ genotype, indicating an InlA truncation. These 29 isolates belonged to the serotype 1/2b complex and were characterized by MLGT haplotypes Lm1.25 (n = 2), Lm1.26 (n = 3), Lm1.27 (n = 1), and Lm1.28 (n = 23) (Table 3). These MLGT types were unique to isolates with the InlA truncation, and phylogenetic analysis of the MLGT data further indicated that this mutation had a recent evolutionary origin (Fig. 2).
Twenty-seven of the 29 isolates harboring the PMSC1 mutation were isolated from food or food-processing environments. The other two isolates were collected by FSIS, but the exact source of these isolates is unknown. In addition, we found isolates with this mutation at a 30.3% frequency among the panel of 66 isolates collected by FSIS from RTE meat and food-processing facilities (see Table S2 in the supplemental material). Given the frequency (47%) of lineage I isolates among RTE food products (51), these data suggest that approximately 14.2% of RTE meat isolates carry the InlA truncation identified by the INLa9−/INLa10+ genotype. Therefore, a substantial fraction of isolates from RTE meats may have reduced abilities to cause systemic listeriosis in humans. However, the extent to which the InlA truncations contribute to the attenuated virulence phenotypes of strains carrying these mutations needs to be more conclusively defined (33, 36). In addition, our estimate of the frequency of this particular InlA truncation is substantially higher than the frequency (5.3%) of ribotypes associated with PMSC1 described by Nightingale et al. (33). The Nightingale et al. (33) study included over 1,500 food isolates, but used ribotyping as an indirect assay for inlA truncations. Our results using the MLGT assay to directly type the PMSC1 inlA truncation suggest that this mutation may be particularly common in RTE meat products. However, additional studies that directly assay inlA truncation mutations among large numbers of L. monocytogenes isolates from different categories of food will be required to investigate this hypothesis.
Subtyping method comparisons.
The relative discriminatory power of the MLGT assay was assessed by comparison with that of PFGE and a recently published MLST assay that incorporated segments of four housekeeping genes and two virulence genes (42). In analyses performed using a panel of 62 isolates collected by FSIS, which were not part of the original set of 65 isolates used in SNP discovery and probe development, the MLGT assay identified 20 unique haplotypes, while MLST and PFGE identified 20 and 37 unique types, respectively (Table 5). Although the numbers of distinct haplotypes identified were comparable between the MLGT and MLST assays, an examination of SDI revealed that MLGT (SDI = 0.91) had discriminatory power approaching that of PFGE (SDI = 0.97) and above the level (SDI = 0.9) considered desirable for reliable subtyping (20). However, MLST provided substantially less discriminatory power (SDI = 0.80) than did MLGT or PFGE despite a previous report that this MLST approach provided greater discriminatory power than PFGE (122 sequence types versus 57 PFGE types) (42). The previous study was based on a survey of isolates from all three lineages, which would represent variation significantly greater than that available within lineage I alone. While it is possible that this MLST method has greater discriminatory power than PFGE among lineage II and lineage III isolates, the results of our analyses indicate that the MLST method of Revazishvili et al. (42) has less power to discriminate among the closely related strains within lineage I of L. monocytogenes than does PFGE or the MLGT assay described here.
TABLE 5.
Comparative subtyping analyses for 62 isolates from ready-to-eat food and food-processing facilities
| Isolatea | PCR serotypeb | Haplotype identified by:
|
||
|---|---|---|---|---|
| MLGT | MLSTc | PFGE | ||
| 33359 | 4b complex | Lm1.2 | 13 | 1 |
| 33337 | 4b complex | Lm1.2 | 8 | 1 |
| 33331 | 4b complex | Lm1.2 | 8 | 1 |
| 33355 | 4b complex | Lm1.2 | 13 | 2 |
| 33252 | 4b complex | Lm1.7 | 1 | 4 |
| 33432 | 4b complex | Lm1.8 | 14 | 3 |
| 33453 | 4b complex | Lm1.8 | 20 | 34 |
| 33323 | 4b complex | Lm1.13 | 5 | 29 |
| 33322 | 4b complex | Lm1.13 | 5 | 29 |
| 33301 | 1/2b complex | Lm1.18 | 17 | 17 |
| 33291 | 1/2b complex | Lm1.18 | 17 | 17 |
| 33287 | 1/2b complex | Lm1.18 | 17 | 17 |
| 33465 | 1/2b complex | Lm1.18 | 17 | 23 |
| 33325 | 1/2b complex | Lm1.18 | 17 | 32 |
| 33327 | 1/2b complex | Lm1.19 | 6 | 35 |
| 33356 | 1/2b complex | Lm1.20 | 11 | 30 |
| 33315 | 1/2b complex | Lm1.20 | 11 | 9 |
| 33251 | 1/2b complex | Lm1.21 | 12 | 6 |
| 33358 | 1/2b complex | Lm1.21 | 12 | 7 |
| 33329 | 1/2b complex | Lm1.24 | 7 | 26 |
| 33308 | 1/2b complex | Lm1.25 | 15 | 18 |
| 33242 | 1/2b complex | Lm1.26 | 15 | 10 |
| 33309 | 1/2b complex | Lm1.26 | 15 | 10 |
| 33254 | 1/2b complex | Lm1.28 | 15 | 11 |
| 33458 | 1/2b complex | Lm1.28 | 15 | 12 |
| 33240 | 1/2b complex | Lm1.28 | 15 | 15 |
| 33239 | 1/2b complex | Lm1.28 | 15 | 15 |
| 33304 | 1/2b complex | Lm1.28 | 15 | 16 |
| 33434 | 1/2b complex | Lm1.28 | 15 | 16 |
| 33303 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33296 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33284 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33262 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33250 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33265 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33347 | 1/2b complex | Lm1.28 | 15 | 18 |
| 33445 | 1/2b complex | Lm1.28 | 15 | 27 |
| 33341 | 1/2b complex | Lm1.28 | 15 | 33 |
| 33461 | 1/2b complex | Lm1.29 | 18 | 21 |
| 33248 | 1/2b complex | Lm1.29 | 18 | 28 |
| 33463 | 1/2b complex | Lm1.29 | 18 | 37 |
| 33466 | 1/2b complex | Lm1.29 | 18 | 37 |
| 33293 | 1/2b complex | Lm1.30 | 15 | 12 |
| 33300 | 1/2b complex | Lm1.30 | 15 | 12 |
| 33302 | 1/2b complex | Lm1.30 | 15 | 12 |
| 33294 | 1/2b complex | Lm1.30 | 15 | 12 |
| 33442 | 1/2b complex | Lm1.30 | 15 | 14 |
| 33306 | 1/2b complex | Lm1.30 | 2 | 16 |
| 33451 | 1/2b complex | Lm1.30 | 15 | 18 |
| 33312 | 1/2b complex | Lm1.30 | 19 | 19 |
| 33305 | 1/2b complex | Lm1.30 | 15 | 31 |
| 33462 | 1/2b complex | Lm1.37 | 16 | 24 |
| 33464 | 1/2b complex | Lm1.37 | 16 | 25 |
| 33313 | 1/2b complex | Lm1.38 | 3 | 22 |
| 33237 | 1/2b complex | Lm1.39 | 4 | 13 |
| 33258 | 1/2b complex | Lm1.39 | 4 | 13 |
| 33320 | 1/2b complex | Lm1.39 | 4 | 5 |
| 33343 | 1/2b complex | Lm1.40 | 9 | 20 |
| 33345 | 1/2b complex | Lm1.41 | 10 | 8 |
| 33346 | 1/2b complex | Lm1.41 | 10 | 8 |
| 33430 | 1/2b complex | Lm1.43 | 15 | 36 |
| 33429 | 1/2b complex | Lm1.43 | 15 | 36 |
PFGE continues to be the gold standard for L. monocytogenes subtyping, having higher discriminatory power than current DNA sequence-based methods. However, in addition to the technical challenges and limitations described above, PFGE does not always discriminate between related but distinct isolates (44, 49). For example, PFGE based on three enzymes failed to separate isolates of the Lausanne 1987 (MLGT haplotype, Lm1.13) and CA 1985 (MLGT haplotype, Lm1.11) ECI outbreaks (3). Conversely, the high discriminatory power of PFGE may be due, in part, to evolutionarily unstable genetic elements, such as plasmids and phages (8). This feature of PFGE may hamper long-term epidemiological tracking and can make it difficult to identify isolates associated with a common source outbreak. Analysis of more than 18,000 nucleotides of DNA sequence from the 62 isolates in the FSIS panel identified 29 unique haplotypes (data not shown). Compared with the 37 unique pulse types identified by PFGE (Table 5), our sequence data suggest that PFGE types can be unstable over very short evolutionary time scales.
In order to address the limitations of PFGE, the CDC PulseNet Task Force called for the development and validation of DNA sequence-based subtyping methods and indicated that SNP analyses could be readily incorporated into the PulseNet network for subtyping food-borne pathogens (13, 48). We have reported the design and validation of an SNP-based multilocus genotyping assay for rapid, accurate, and repeatable subtyping of lineage I L. monocytogenes. Strains within this lineage pose a unique threat to public health, and the relatively limited genetic diversity in this lineage poses unique challenges for molecular subtyping. However, the ability to perform multiplex interrogation of 60 probes in a single-well assay provided high discriminatory power (SDI = 0.91) and epidemiological relevance in differentiating serotype groups, epidemic clones, and all eight outbreaks examined. In addition, all 241 isolates examined were reproducibly (100%) typed with the MLGT assay, which recapitulated the phylogenetic relationships and 100% of the haplotype information identified in the analysis of over 23,000 nucleotides of DNA sequence and also provided for the identification of subtypes with a specific attenuated-virulence phenotype (33). The single-well MLGT assay also outperformed the MLST system developed by Revazishvili et al. (42), which required 12 sequencing reactions per isolate and the generation of 278,628 nucleotides of DNA sequence for the panel of 62 isolates examined. In addition, the cost of running the MLGT assay was approximately four times less than that of MLST per reaction (see Table S1 in the supplemental material).
Due to the flexibility of the microsphere-based SNP typing system, which currently permits multiplex analysis of up to 100 probes per well, additional SNP discovery could be used to increase the discriminatory power of the MLGT assay. For example, sequence analysis for the 62 isolates in the FSIS panel revealed nine novel haplotypes (data not shown) defined by single-nucleotide polymorphisms that could be incorporated into the current MLGT assay. The current probe set can also be modified to examine subsets of probes for targeted applications focused on discriminating between specific subtypes, such as the inlA truncation or epidemic clone types. In addition, the current probe set could be expanded to assay variation within lineages II and III by using a modular approach with individual MLGT assays developed to efficiently target SNP variation appropriate for differentiating subtypes within each of the major L. monocytogenes lineages. As such, MLGT represents a highly flexible DNA sequence-based tool for use in pathogen surveillance, outbreak detection, risk assessment, population analyses, and epidemiological investigations.
Supplementary Material
Acknowledgments
We thank Jody Robinson, Amy Morgan, and Jennifer Steele for excellent technical assistance. We are also indebted to Cletus Kurtzman for helpful discussions regarding SNP typing.
The mention of firm names or trade products does not imply that they are endorsed or recommended by the U.S. Department of Agriculture over other firms or similar products not mentioned.
Footnotes
Published ahead of print on 3 November 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Borucki, M. K., and D. R. Call. 2003. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borucki, M. K., S. H. Kim, D. R. Call, S. C. Smole, and F. Pagotto. 2004. Selective discrimination of Listeria monocytogenes epidemic strains by a mixed-genome DNA microarray compared to discrimination by pulsed-field gel electrophoresis, ribotyping, and multilocus sequence typing. J. Clin. Microbiol. 42:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosch, R., M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi, and J. Rocourt. 1996. Genomic fingerprinting of 80 strains from the WHO multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE). Int. J. Food Microbiol. 32:343-355. [DOI] [PubMed] [Google Scholar]
- 4.Brosch, R., C. Buchrieser, and J. Rocourt. 1991. Subtyping of Listeria monocytogenes serovar 4b by use of low-frequency-cleavage restriction endonucleases and pulsed-field gel electrophoresis. Res. Microbiol. 142:667-675. [DOI] [PubMed] [Google Scholar]
- 5.Clark, E. E., I. Wesley, F. Fiedler, N. Promadej, and S. Kathariou. 2000. Absence of serotype-specific surface antigen and altered teichoic acid glycosylation among epidemic-associated strains of Listeria monocytogenes. J. Clin. Microbiol. 38:3856-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Cesare, A., J. L. Bruce, T. R. Dambaugh, M. E. Guerzoni, and M. Wiedmann. 2001. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4b strains. J. Clin. Microbiol. 39:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doumith, M., C. Jacquet, V. Goulet, C. Oggioni, F. Van Loock, C. Buchrieser, and P. Martin. 2006. Use of DNA arrays for the analysis of outbreak-related strains of Listeria monocytogenes. Int. J. Med. Microbiol. 296:559-562. [DOI] [PubMed] [Google Scholar]
- 9.Eifert, J. D., P. A. Curtis, M. C. Bazaco, R. J. Meinersmann, M. E. Berrang, S. Kernodle, C. Stam, L. A. Jaykus, and S. Kathariou. 2005. Molecular characterization of Listeria monocytogenes of the serotype 4b complex (4b, 4d, 4e) from two turkey processing plants. Foodborne Pathog. Dis. 2:192-200. [DOI] [PubMed] [Google Scholar]
- 10.Evans, M. R., B. Swaminathan, L. M. Graves, E. Altermann, T. R. Klaenhammer, R. C. Fink, S. Kernodle, and S. Kathariou. 2004. Genetic markers unique to Listeria monocytogenes serotype 4b differentiate epidemic clone II (hot dog outbreak strains) from other lineages. Appl. Environ. Microbiol. 70:2383-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Gerner-Smidt, P., K. Hise, J. Kincaid, S. Hunter, S. Rolando, E. Hyytia-Trees, E. M. Ribot, and B. Swaminathan. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9-19. [DOI] [PubMed] [Google Scholar]
- 14.Graves, L. M., S. B. Hunter, A. R. Ong, D. Schoonmaker-Bopp, K. Hise, L. Kornstein, W. E. DeWitt, P. S. Hayes, E. Dunne, P. Mead, and B. Swaminathan. 2005. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 43:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 16.Gravesen, A., T. Jacobsen, P. L. Moller, F. Hansen, A. G. Larsen, and S. Knochel. 2000. Genotyping of Listeria monocytogenes: comparison of RAPD, ITS, and PFGE. Int. J. Food Microbiol. 57:43-51. [Google Scholar]
- 17.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grif, K., I. Heller, M. Wagner, M. Dierich, and R. Wurzner. 2006. A comparison of Listeria monocytogenes serovar 4b isolates of clinical and food origin in Austria by automated ribotyping and pulsed-field gel electrophoresis. Foodborne Pathog. Dis. 3:138-141. [DOI] [PubMed] [Google Scholar]
- 19.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacquet, C., M. Doumith, J. I. Gordon, P. M. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 22.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 23.Jonquières, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathariou, S. 2003. Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes, p. 243-256. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture: current topics. Iowa State University Press, Ames, IA.
- 25.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 26.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 27.Lei, X. H., N. Promadej, and S. Kathariou. 1997. DNA fragments from regions involved in surface antigen expression specifically identify Listeria monocytogenes serovar 4 and a subset thereof: cluster IIB (serotypes 4b, 4d, and 4e). Appl. Environ. Microbiol. 63:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClure, P. J., T. M. Kelly, and T. A. Roberts. 1991. The effects of temperature, pH, sodium chloride and sodium nitrite on the growth of Listeria monocytogenes. Int. J. Food Microbiol. 14:77-91. [DOI] [PubMed] [Google Scholar]
- 29.McLauchlin, J., S. M. Hall, S. K. Velani, and R. J. Gilbert. 1991. Human listeriosis and pate: a possible association. BMJ 303:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505-515. [PubMed] [Google Scholar]
- 35.Norton, D. M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olier, M., D. Garmyn, S. Rousseaux, J. P. Lemaitre, P. Piveteau, and J. Guzzo. 2005. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect. Immun. 73:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 38.Olier, M., F. Pierre, S. Rousseaux, J. P. Lemaitre, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penny, D., and M. D. Hendy. 1985. The use of tree comparison metrics. Syst. Zool. 34:75-82. [Google Scholar]
- 40.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 42.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauders, B. D., E. D. Fortes, D. L. Morse, N. Dumas, J. A. Kiehlbauch, Y. Schukken, J. R. Hibbs, and M. Wiedmann. 2003. Molecular subtyping to detect human listeriosis clusters. Emerg. Infect. Dis. 9:672-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schönberg, A., E. Bannerman, A. L. Courtieu, R. Kiss, J. McLauchlin, S. Shah, and D. Wilhelms. 1996. Serotyping of 80 strains from the WHO multicentre international typing study of Listeria monocytogenes. Int. J. Food Microbiol. 32:279-287. [DOI] [PubMed] [Google Scholar]
- 46.Shen, Y., Y. Liu, Y. Zhang, J. Cripe, W. Conway, J. Meng, G. Hall, and A. A. Bhagwat. 2006. Isolation and characterization of Listeria monocytogenes isolates from ready-to-eat foods in Florida. Appl. Environ. Microbiol. 72:5073-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommers, C. H., and G. Boyd. 2005. Elimination of Listeria monocytogenes from ready-to-eat turkey and cheese tortilla wraps using ionizing radiation. J. Food Prot. 68:164-167. [DOI] [PubMed] [Google Scholar]
- 48.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace, F. M., J. E. Call, A. C. Porto, G. J. Cocoma, and J. B. Luchansky. 2003. Recovery rate of Listeria monocytogenes from commercially prepared frankfurters during extended refrigerated storage. J. Food Prot. 66:584-591. [DOI] [PubMed] [Google Scholar]
- 51.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesley, I. V., and F. Ashton. 1991. Restriction enzyme analysis of Listeria monocytogenes strains associated with food-borne epidemics. Appl. Environ. Microbiol. 57:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong, A. C. 1998. Biofilms in food processing environments. J. Dairy Sci. 81:2765-2770. [DOI] [PubMed] [Google Scholar]
- 55.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 56.Yildirim, S., W. Lin, A. D. Hitchins, L. A. Jaykus, E. Altermann, T. R. Klaenhammer, and S. Kathariou. 2004. Epidemic clone I-specific genetic markers in strains of Listeria monocytogenes serotype 4b from foods. Appl. Environ. Microbiol. 70:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, W., B. M. Jayarao, and S. J. Knabel. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng, W., and S. Kathariou. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl. Environ. Microbiol. 63:3085-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.