Abstract
Gene copy number polymorphism was studied in a population of the arbuscular mycorrhizal fungus Glomus intraradices by using a quantitative PCR approach on four different genomic regions. Variation in gene copy number was found for a pseudogene and for three ribosomal genes, providing conclusive evidence for a widespread occurrence of macromutational events in the population.
For a long time, gene duplication events were considered to occur relatively rarely among closely related genomes and it was assumed that individuals of extant populations are unlikely to differ in gene copy number. This is surprising given that the presence of gene copy number and length polymorphism in ribosomal DNA (rDNA) genes and pathogen and herbivore resistance genes has long been recognized to occur both within and among populations of fruit flies (Drosophila melanogaster and Drosophila mercatorum) and wild barley (Hordeum vulgare) (2, 15, 16). Recently, large-scale among-individual copy number polymorphisms for coding genes have been discovered in humans by surveying completely sequenced genomes or by using molecular hybridization techniques (8, 13). In some cases, these genomic changes have been linked to the susceptibility of individuals to pathologies such as human immunodeficiency virus or glomerulonephritis (1, 5). In experimental populations of Candida albicans, copy number polymorphisms potentially evolved in as little as 330 generations (4).
Arbuscular mycorrhizal fungi (AMF) are important obligate symbionts of plants which improve plant nutrition and promote plant diversity in terrestrial ecosystems (14). Isolates of the AMF Glomus intraradices from one population were shown by fingerprinting to be genetically different (10), and the same isolates were also shown to differentially affect plant biomass (9). More recently, these G. intraradices isolates were found to harbor variable numbers of copies of genes that are potentially important in adaptation to environmental stress (P-type IID ATPases) (3). Considering that variation in gene copy number has been shown to be important for the adaptation to certain environmental or pathological conditions in other species, determining the presence and amount of such macromutational events in natural AMF populations is certainly warranted. From an evolutionary point of view it is also interesting to determine whether copy number polymorphisms are frequent enough within natural populations to be an important source of genetic variability. The issue which has been addressed here is whether variations in gene copy number are more widespread in a population of AMF or just restricted to the P-type IID ATPases. To address this question, we determined and compared the relative copy number of three rDNA genes (18S, 5.8S, and 25S) and a BiP gene (a chaperone gene) and pseudogenes among isolates of G. intraradices harvested from the same field.
Isolates of G. intraradices DAOM181602, A4, B3, C2, and C3 (the last four having been harvested from the same field in Switzerland and named according to the work of Koch et al. [9]) were grown with Ri T-DNA-transformed Daucus carrota roots. DNA was extracted from AMF using the DNeasy plant minikit (QIAGEN). The BiP gene cloning procedure was performed according to the work of Kuhn et al. (11). PCR amplification of specific variants of the BiP pseudogene presented in Fig. 1C was performed using the primer BiP2.F (5′-AAGACAAGCCACAAAAGATGCTGG-3′) in combination with BiPT1.R (5′-TGAATATCATTGGTATATCCGTATATCT-3′) or BiPT2.R (5′-TGAATGTCATTGGTATATCTCCGG-3′). Southern blot analyses were performed for the BiP genes of the isolate DAOM181602 using 3 μg of genomic DNA digested with each of the endonucleases EcoRV and XbaI. The hybridization was carried out using a digoxigenin-labeled probe obtained using the primers BiP2.F (5′-AAGACAAGCCACAAAAGATGCTGG-3′) and BiP2.R (5′-AGTAGGGATTACAGTGTTACGAGG-3′).
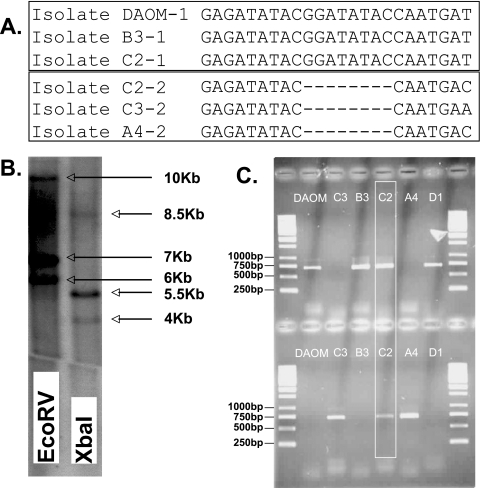
FIG. 1.
Evidence for the segregation of BiP pseudogene variants among isolates of G. intraradices. A. Partial nucleotide sequence alignments of two BiP pseudogene variants isolated from five isolates of G. intraradices (DAOM181602, A4, B3, C2, and C3). The two different sequence types are shown in separate boxes. The number following each isolate code represents the pseudogene sequence type. B. Southern blot hybridization on genomic DNA from the isolate DAOM181602 of a probe that hybridizes to all BiP gene and pseudogene variants. Results with genomic DNA digested with EcoRV and XbaI are shown. The sizes of the bands are shown on the right. C. PCR amplification of the two BiP pseudogene variants using specific primers with DNA from the different G. intraradices isolates. The analysis shows results from an additional isolate (D1) from the same population that was not included in the cloning and sequencing experiments. The upper gel shows amplification of the BiP pseudogene type 1, using the reverse primer BiPT1.R. The lower gel shows amplification of the BiP pseudogene type 2, using the reverse primer BiPT2.R. The isolate C2 is shown to harbor both types 1 and 2 of the BiP pseudogene. The size standard is in the far left and far right lanes of both gels.
Real-time quantitative PCR (qPCR) was performed on three G. intraradices isolates to compare relative copy numbers of the BiP, 18S, 5.8S, and 25S genes. For each gene we designed primers and probes that annealed to a region that was conserved among all known sequence variants and among the isolates that we studied (Table 1). The control assays for real-time qPCR were performed according to the work of Corradi and Sanders (3) and showed that cycle threshold (CT) values did not vary significantly between independent DNA extractions. The average difference in CT values among DNA extractions was <0.1 when qPCR was performed. In addition, control real-time qPCR was performed on the Rad15 and Rad32 genes. These two genes were chosen as controls as they showed no variation in sequence both among and within isolates and because Rad32 had already been shown to be present in a single copy in isolate DAOM181602 by a dot blot hybridization assay (6). Probes were labeled with 6-carboxyfluorescein at the 5′ end and 6-carboxytetramethylrhodamine at the 3′ end (except for Rad15 and Rad32, where Black Hole Quencher 1 was used at the 3′ end). The 6-carboxyfluorescein-real-time PCR amplification was performed and the relative copy number of the genes was calculated according to the work of Corradi and Sanders (3).
TABLE 1.
List of primers and probes used in the real-time quantitative PCR experiments for amplification of BiP genes and pseudogenes; 18S, 5.8S, and 25S ribosomal genes; and the Rad15 and Rad32 genes
| Primer name | Sense | 5′-3′ sequence |
|---|---|---|
| 5.8S.real.F | Forward | ACA ACG GAT CTC TTG GCT CT |
| 5.8S.probe | Probe | TGA AGA ACG TAG CGA AGT GCG |
| 5.8S.real.R | Reverse | AAT TTG CGT TCA AAG ATT CG |
| 18S.real.F | Forward | ATG CTA AAA CCT CCG ACT TC |
| 18S.probe | Probe | TGA TTC ATA ATA ACT TTT CGA ATC GTA TGA |
| 18S.real.R | Reverse | TAG GGC AGA AAT TTG AAT GA |
| 25S.real.F | Forward | GAG AGA CCG ATA GCG AAC AA |
| 25S.probe | Probe | CCG TGA GGG AAA GAT GAA AAG AA |
| 25S.real.R | Reverse | GAA GGT ACG ACT GGC TTC AA |
| BiP.real.F | Forward | TTG GTG GTT CCA CAC GTA TTC |
| BiP.probe | Probe | ATG GTG CCG CCA TAC AAG |
| BiP.real.R | Reverse | TGT CAT AAC GCC ACC AGT TGT |
| Rad15 F | Forward | AGA GAG AAT TAT CGA ATA CGA GAA AAT GAT |
| Rad15 probe | Probe | TTT TGA CTT TTG ATG CCA TGC GCC AT |
| Rad15 R | Reverse | AGC CCA TAA TCT GTT TTC CCT CTT |
| Rad32 F | Forward | TCA ATA ATG ATC GCG ACT GAT ATA CA |
| Rad32 probe | Probe | TAT CAG GAG CAA GAT CCA ACA CGC GG |
| Rad32 R | Reverse | TAT TTC TCT GAA AGT GTT GAA GCT ATC G |
Three different variants of the BiP gene were found in the isolates DAOM181602, A4, B3, and C3. One of these variants harbored a frameshift mutation and should, therefore, be considered a pseudogene, while the two other sequence variants carried putative functional genes. The pseudogene of isolates A4 and C3 was found to harbor an additional frameshift mutation that did not occur in the other isolates. Isolate C2 was found to harbor both of the pseudogenes that were found in the other isolates (Fig. 1A). Therefore, we isolated four different BiP gene and pseudogene sequence variants from isolate C2, instead of three as for the other isolates. Consistent with the number of sequence variants isolated, Southern blotting on genomic DNA from isolate DAOM181602 confirmed that these genes are present in three different regions of the genome (Fig. 1B) and most likely appeared by gene duplications. PCR cloning with specific primers confirmed that isolate C2 harbored two BiP pseudogene variants, instead of one (Fig. 1C).
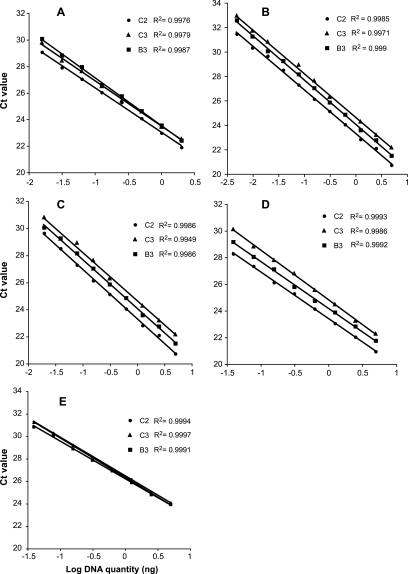
Relative quantification of BiP gene copy number was carried out on three G. intraradices isolates and showed that isolate C2 harbors, on average, 39% more copies of the BiP genes than do other isolates (Fig. 2A). This value is consistent with isolate C2 harboring four BiP genes instead of three. No significant variation in gene copy number was found between isolates B3 and C3. The search for copy number polymorphisms using qPCR in the G. intraradices population was extended to three rDNA genes (18S, 5.8S, and 25S). Estimates of the relative copy numbers of the three rDNA genes in isolates B3, C2, and C3 all showed the same pattern, with isolate C2 having the highest number of copies, followed by isolate B3, and with isolate C3 having the lowest number of copies. The differences in CT value showed that in the G. intraradices population, the number of rDNA genes can vary from two- to fourfold among isolates (Fig. 2B to D). If the relative copy numbers of the three rRNA genes among isolates had been different from one another, then this could have represented independent duplication events. However, because all the rDNA genes showed the same relative differences among isolates, it is likely that the whole rDNA tandem array (comprising the three rRNA genes) has been subject to deletion and duplication events in the genomes of the three isolates. Using the same replicate DNA extractions from isolates B3, C2, and C3, we found no significant variation in CT values (<0.1 CT) for the amplification of either of the two control genes Rad15 (Fig. 2E) and Rad32 (data not shown).
FIG. 2.
Results of real-time quantitative PCR showing linear regressions of the cycle threshold (CT values) and the log concentration of genomic DNA of G. intraradices isolates B3, C2, and C3. The analysis was performed with primers amplifying all previously found variants of the BiP gene and pseudogenes (A); ribosomal genes 18S (B), 5.8S (C), and 25S (D); and Rad15 (E). Real-time quantitative PCR was also performed on Rad32, but the data are not shown as they showed exactly the same pattern as that of Rad15. For all real-time quantitative PCR experiments, two replicate amplifications for each isolate were performed. Data points shown in the graphs represent the average CT value of the two replicates.
In the present study we provide additional evidence that gene copy number polymorphisms can occur within an AMF population and that these involve several different regions of the genome. Together with the study by Corradi and Sanders (3), this evidence shows that copy number polymorphism in this population occurs in rDNA, protein-encoding genes, and also pseudogenes. Obviously, copy number polymorphisms in protein-encoding genes could directly affect the phenotype. However, pseudogene expression is also known to regulate expression of homologous coding genes (7), and even changes in rDNA copy number can affect gene expression and epigenetic gene silencing (12). Therefore, none of these copy number polymorphisms in the AMF population should be assumed to be neutral a priori, and these macromutational events could have potential consequences for the ecology of these fungi (12).
Finally, the presence of copy number polymorphisms in AMF populations has direct implications for the study of AMF community structure. A main focus of many groups is currently to develop qPCR-based methods for assessing AMF community structure in plant roots, using the relative abundance of species-specific rDNA markers. The large within-population variation in rDNA gene copy number renders the results of such approaches largely uninterpretable.
Acknowledgments
This work was supported by a grant from the Swiss National Science Foundation (no. 3100A0-105790/1), and Gerrit Kuhn was supported by a grant from the Roche Foundation, from which support is gratefully acknowledged.
We thank all members of our research group for help in cultivation of G. intraradices isolates.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Aitman, T. J., R. Dong, T. J. Vyse, P. J. Norsworthy, M. D. Johnson, J. Smith, J. Mangion, C. Roberton-Lowe, A. J. Marshall, E. Petretto, M. D. Hodges, G. Bhangal, S. G. Patel, K. Sheehan-Rooney, M. Duda, P. R. Cook, D. J. Evans, J. Domin, J. Flint, J. J. Boyle, C. D. Pusey, and H. T. Cook. 2006. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439:851-855. [DOI] [PubMed] [Google Scholar]
- 2.Coen, E. S., J. M. Thoday, and G. Dover. 1982. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature 295:564-568. [DOI] [PubMed] [Google Scholar]
- 3.Corradi, N., and I. Sanders. 2006. Evolution of the P-type II ATPase gene family in the fungi and presence of structural genomic changes among isolates of Glomus intraradices. BMC Evol. Biol. 6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez, G. Catano, R. J. Nibbs, B. I. Freedman, M. P. Quinones, M. J. Bamshad, K. K. Murthy, B. H. Rovin, W. Bradley, R. A. Clark, S. A. Anderson, R. J. O'Connell, B. K. Agan, S. S. Ahuja, R. Bologna, L. Sen, M. J. Dolan, and S. K. Ahuja. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:1434-1440. [DOI] [PubMed] [Google Scholar]
- 6.Hijri, M., and I. R. Sanders. 2004. The arbuscular mycorrhizal fungus Glomus intraradices is haploid and has a small genome size in the lower limit of eukaryotes. Fungal Genet. Biol. 41:253-261. [DOI] [PubMed] [Google Scholar]
- 7.Hirotsune, S., N. Yoshida, A. Chen, L. Garrett, F. Sugiyama, S. Takahashi, K. Yagami, A. Wynshaw-Boris, and A. Yoshiki. 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423:91-96. [DOI] [PubMed] [Google Scholar]
- 8.Iafrate, A. J., L. Feuk, M. N. Rivera, M. L. Listewnik, P. K. Donahoe, Y. Qi, S. W. Scherer, and C. Lee. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36:949-951. [DOI] [PubMed] [Google Scholar]
- 9.Koch, A. M., D. Croll, and I. R. Sanders. 2006. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol. Lett. 9:103-110. [DOI] [PubMed] [Google Scholar]
- 10.Koch, A. M., G. Kuhn, P. Fontanillas, L. Fumagalli, J. Goudet, and I. R. Sanders. 2004. High genetic variability and low local diversity in a population of arbuscular mycorrhizal fungi. Proc. Natl. Acad. Sci. USA 101:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn, G., M. Hijri, and I. R. Sanders. 2001. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745-748. [DOI] [PubMed] [Google Scholar]
- 12.Michel, A. H., B. Kornmann, K. Dubrana, and D. Shore. 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19:1199-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebat, J., B. Lakshmi, J. Troge, J. Alexander, J. Young, P. Lundin, S. Maner, H. Massa, M. Walker, M. Chi, N. Navin, R. Lucito, J. Healy, J. Hicks, K. Ye, A. Reiner, T. C. Gilliam, B. Trask, N. Patterson, A. Zetterberg, and M. Wigler. 2004. Large-scale copy number polymorphism in the human genome. Science 305:525-528. [DOI] [PubMed] [Google Scholar]
- 14.Van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 15.Williams, S. M., R. DeSalle, and C. Strobeck. 1985. Homogenization of geographical variants at the nontranscribed spacer of rDNA in Drosophila mercatorum. Mol. Biol. Evol. 2:338-346. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Q. F., M. A. Saghai Maroof, and R. W. Allard. 1990. Effects on adaptedness of variations in ribosomal DNA copy number in populations of wild barley (Hordeum vulgare ssp. spontaneum). Proc. Natl. Acad. Sci. USA 87:8741-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]