Abstract
Bacterial inclusion bodies, while showing intriguing amyloid-like features, such as a β-sheet-based intermolecular organization, binding to amyloid-tropic dyes, and origin in a sequence-selective deposition process, hold an important amount of native-like secondary structure and significant amounts of functional polypeptides. The aggregation mechanics supporting the occurrence of both misfolded and properly folded protein is controversial. Single polypeptide chains might contain both misfolded stretches driving aggregation and properly folded protein domains that, if embracing the active site, would account for the biological activities displayed by inclusion bodies. Alternatively, soluble, functional polypeptides could be surface adsorbed by interactions weaker than those driving the formation of the intermolecular β-sheet architecture. To explore whether the fraction of properly folded active protein is a natural component or rather a mere contaminant of these aggregates, we have explored their localization by image analysis of inclusion bodies formed by green fluorescent protein. Since the fluorescence distribution is not homogeneous and the core of inclusion bodies is particularly rich in active protein forms, such protein species cannot be passively trapped components and their occurrence might be linked to the reconstruction dynamics steadily endured in vivo by such bacterial aggregates. Intriguingly, even functional protein species in inclusion bodies are not excluded from the interface with the solvent, probably because of the porous structure of these particular protein aggregates.
Procedures for in vitro protein refolding are under continuous development (22, 32, 35), since many proteins of industrial or pharmacological interest are produced in recombinant microorganisms, especially bacteria, as insoluble aggregates called inclusion bodies (IBs) (38). Recent insights into the structure and physiology of bacterial IBs have revealed that at least a significant fraction of the embedded protein occurs in a properly folded native-like form (36) and that for aggregates formed by enzymes, this fact is reflected by the occurrence of enzymatic activity associated with these particles (16, 34, 39). While for hormones or other drugs to be used in vivo, in vitro solubilization of IBs and refolding of IB proteins would still be required to allow their proper use (27), enzymes to be used in bioprocesses could be employed straight after production, skipping any refolding step. This is particularly appealing since the specific activity found in IB enzymes, although variable when comparing different protein species, is not dramatically different from that exhibited by the soluble counterparts (15, 16), and on the other hand, refolding procedures render yields of active protein that are usually far from 100%.
Apart from the obvious potential of enzyme IBs as catalyzers, the occurrence of properly folded, active enzymes poses intriguing structural questions. The conformational background sustaining the IB molecular structure lies on an extended, intermolecular β-sheet architecture (6) that coexists with various amounts of a population of native-like, correctly folded polypeptides (1-3, 26). Such a β-sheet pattern is progressively lost at the expense of native-like structure when the temperature at which IBs are formed decreases (19, 28), indicating the existence of several categories of protein aggregates in regard to their molecular organization and even global morphology (13). However, the process that selects in vivo the protein species to be deposited with regard to its conformational status, and especially the way in which both properly folded and β-sheet-rich species coexist, remains unexplored. It has been suggested that in single polypeptides, specific domains with a misfolded status could act as aggregating elements (and organize through intermolecular β-sheet interactions), while others might remain properly folded (and fully functional if containing the active site) (36). On the other hand, it is known that during IB isolation, contaminating cell proteins get attached to the IB surface (17), which is probably “sticky” from the exposure of hydrophobic patches (6). Therefore, active polypeptides also eventually could be surface trapped if they abound in the soluble cell fraction of the producing cell. In that case, the enzymatic activity displayed by IBs would lie on contaminant protein species rather than on structural elements. The experimental approaches to solving this issue are not obvious, since it is not possible to distinguish in situ conformational states within such nanoparticles.
To gain insights into the molecular organization of functional IBs, we have taken alternative approaches, namely, exploring the localization of the biological activity in actively catalyzing enzyme-based IBs and generating fluorescence emission maps of green fluorescent protein (GFP)-based IBs. Intriguingly, although an important part of the active protein species is easily released from catalyzing IBs, the core of such aggregates (but not the surface layers) is rich in functional protein. These results are discussed in the context of the porous structure of these protein aggregates, also considering the highly dynamic protein deposition and release processes that drive the in vivo building of bacterial IBs.
MATERIALS AND METHODS
Strain, plasmids, and culture conditions.
Escherichia coli MC4100 (29) was used for all the experiments. Plasmids pTVP1GFP and pTVP1LAC (16) encode engineered versions of GFP and β-galactosidase, respectively, both carrying the VP1 capsid protein of foot-and-mouth disease virus fused at the amino termini. This viral protein dramatically reduces the solubility of the whole fusions, resulting in aggregation of fusion proteins as IBs. All of the production processes were performed with shaker flask cultures growing at 37°C in LB rich medium (29) plus 100 μg/ml ampicillin for plasmid maintenance, and recombinant gene expression was induced when the optical density at 550 nm reached 0.4 by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cell samples were taken at 3 h after induction of gene expression. For the comparative analysis of IBs formed at different temperatures, samples were taken from IPTG-treated cultures at an optical density between 2.9 and 3.1, irrespective of the time taken for growth (3 h at 37°C but longer at lower temperatures).
IB-mediated catalysis conditions and determination of enzymatic activity.
IBs were purified by a detergent-washing protocol as described previously (8), resuspended in phosphate-buffered saline (PBS), and diluted either 5 or 50 times for VP1GFP or VP1LAC, respectively. For the analysis of VP1LAC, two aliquots of each sample were prepared and kept at 37°C in agitation. In one of them, 5 ml of 6 mM o-nitrophenyl-β-d-galactopyranoside (ONPG) (in PBS) was added, while the other was used as an internal control. To monitor the ONPG hydrolysis reaction, samples were taken every 5 min for 1 h and the enzymatic activity of VP1LAC was determined in 120-μl reaction mixtures in microplates, as described previously (14, 15). Also, to determine the localization of the enzymatic activity in such a reaction mixture, at different times of the catalysis process, 1-ml samples were taken at three different times (t0, just before the ONPG addition), t1, and t2 (2 min and 30 min after ONPG addition, respectively). These samples were centrifuged at a low speed (for 5 min at 15,000 × g), and the supernatant was used for the analysis of the soluble fraction of the enzymatic reaction (associated with protein released from IBs), while the resulting pellet, resuspended in 1 ml PBS, was used for the analysis of the protein still associated with IBs. VP1GFP IBs prepared as described were incubated at 37°C, and samples taken at different times were centrifuged at 15,000 × g for 5 min. Fluorescence of both soluble and insoluble fractions was determined in a Cary Eclipse fluorescence spectrophometer (Variant).
To analyze the enzymatic activity in these fractions and finally the specific activity, a second substrate, rendering red products upon hydrolysis by β-galactosidase (chlorophenol red β-d-galactopyranoside), was used to avoid the yellow background linked to the ONPG products already present in the samples. These assays were performed with 120-μl reaction mixtures in microplates with 6 mM chlorophenol red β-d-galactopyranoside, as described previously (14), for 16 h. The enzymatic activity was calculated by measuring the slope of the linear part of each graph plotted against the reaction time. All determinations were done with at least three independent experiments.
Quantitative protein analysis.
For protein quantification, supernatants and IB fractions were boiled for 15 or 25 min, respectively. Appropriate sample volumes were loaded onto denaturing gels for immunodetection. For Western blotting, polyclonal antibodies specific for β-galactosidase was used as previously described (12). Bands were quantified by means of the Quantity One software from Bio-Rad, using appropriate protein dilutions of known concentrations as controls. All of these analyses were done in at least three independent experiments. Protein amounts were finally employed to determine specific activities of the distinct samples.
Confocal microscopy analysis.
For image analysis, samples of VP1GFP-producing cells 3 h after IPTG addition were fixed with 0.1% formaldehyde and stored at 4°C until observed. Photographs were taken by using a Leica TCS SP2 AOBS confocal microscope (excitation wavelength at 488 nm and emission wavelength at 500 to 600 nm; optical lens magnification, 63×; 1,024 by 1,024 pixels; zooms between 4 and 8). For the analysis of the resulting images, we used the Adobe Photoshop software and two different lookup tables (tables of cross-references linking index numbers to output values) with coincident results. By this, we determined the colors and intensity values with which a particular image is to be displayed, producing color maps in which each pixel's value is treated as an index number instead of a definite color. The particular lookup table displayed in the Results section was “Metamorph.”
RESULTS
In vivo distribution of active IB polypeptides.
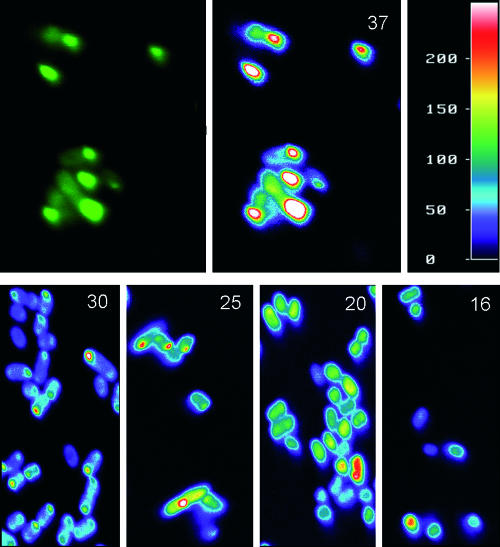
In a previous study, we analyzed in situ β-galactosidase protein material in sections of IB-bearing cells by immunodetection without noting any specific distribution of the enzyme in the aggregates (see Fig. 1 in reference 11). Since most of the well-formed, aged VP1LAC IBs are composed by VP1LAC (up to more than 90% of the IB protein material; see Fig. 4 in reference 7), this fact indicates that the density of IBs is rather homogeneous. Although specific IB protein density has not been directly investigated for other proteins and production conditions, the common aggregation mechanics (5), secondary structure pattern (2, 3, 6, 16, 19), and architectural data independently obtained from different IBs (5, 8) do not point out the homogenous distribution of IB polypeptides as being a particular, protein-restricted feature. Since it was not technically possible to map in situ the occurrence of active VP1LAC in VP1LAC IBs, we instead analyzed the fluorescence distribution in VP1GPF IBs to identify the localization of functional protein and any possible unbalanced distribution of fluorescent protein material. Like VP1LAC IBs, the aggregates formed by VP1GFP are highly active, and they are fluorescent (16). In this regard, confocal analysis of VP1GFP-producing cells through 0.04-μm virtual sections (note that IB diameter occurs between around 0.5 and 1 μm [7, 8, 11, 16]) rendered intriguing images in which there was a clear gradation in the emission intensity from low (external layer) to high (the IB core) (Fig. 1, top). The same pattern was consistently observed at suboptimal growth temperatures, namely, 30, 25, 20, and 16°C, known to favor both protein solubility (33, 37) and conformational quality of IB polypeptides (19, 37). Even at 16°C, when refractile IBs are hardly formed (37), the concentric fluorescence pattern was observed in some individual cells.
FIG. 1.
Top. Fluorescence microscopy (left) and Metamorph image analysis (right) of VP1GFP IBs formed at 37°C. The color scale is depicted at the right. Bottom. Metamorph image analysis of IBs formed at different growth temperatures (indicated by numbers at the upper right corners).
As indicated above, the obtained emission maps cannot be accounted for by any strong radial distribution of protein density. Therefore, functional, properly folded polypeptides are specifically found at the core of the aggregates, while their surface layer is poor in functional protein (note that the differences in the fluorescence emissions between such protein populations are at least twofold [Fig. 1]). This fact could be due to the dynamics of the in vivo IB building process that results from an unbalanced equilibrium between protein deposition and removal (9, 10). Disaggregating chaperones, namely DnaK, ClpB, and small heat shock proteins, act cooperatively on misfolded polypeptides at the aggregate interface (23, 25, 30, 31). Since, as derived from IB structural analysis (1, 2, 3, 26, 28), protein aggregation is not a highly selective process that involves functional polypeptides, a more selective removal of misfolded proteins at the IBs' surfaces (as suggested (30, 31) could enrich the nucleus with native-like, active species. This possibility is compatible with the gain of conformational homogeneity observed during the volumetric growth of IB (8), since the ratio between core and surface material increases with IB volume.
Distribution and release of active polypeptides in catalyzing IBs.
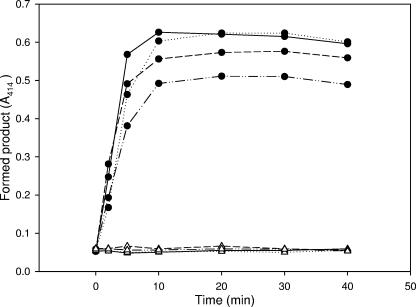
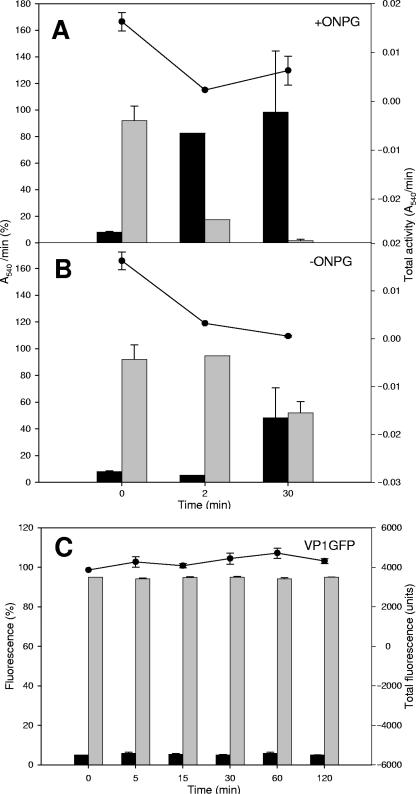
In a previous work (16), we suggested that enzyme-based IBs, since they contain functional proteins, could be useful catalyzers in enzymatic processes, and in fact, both β-galactosidase and human dihydrofolate reductase efficiently processed their respective substrates as embedded in IBs. To better understand how the reaction is performed by IBs in the context of the activity distribution seen in Fig. 1, we determined the occurrence of β-galactosidase enzymatic activity during substrate hydrolysis mediated by VP1LAC IBs. In the presence of an enzyme substrate (ONPG), resuspended VP1LAC IBs catalyzed the product formation kinetics with a very conventional profile (Fig. 2). Since IBs are highly porous and hydrated structures (5, 8), substrate diffusion to the core would not be unexpected. However, to explore to what extent such an enzymatic process was directed by enzyme molecules associated with or released from IBs, we determined the enzymatic activities in the insoluble and soluble fractions of the reaction mixture at different times of the process, as well as the enzyme present in each fraction. Intriguingly, a significant part of the enzymatic activity (between 7 and 8%) was found in the soluble fraction upon IB resuspension in the reaction buffer before substrate addition (Fig. 3A, time zero). Note that this occurred after IB isolation by a procedure that involves repeated detergent washing steps (8). Since the protein amount in the soluble fraction was very low (not shown; lower than 0.0002%), such a protein fraction must exhibit a specific activity higher than average for the aggregates and would not be linked to surface polypeptides. The immediate release of functional protein was also observed for VP1GFP IBs to an extent very similar to that for VP1LAC (5.0%, Fig. 3C). This fact suggested that fluorescent VP1GFP polypeptides, since they are not located at the IB surface layer, might be not completely excluded from the interface with the solvent because of the highly porous and hydrated IB architecture (5, 8). A similar situation could take place with VP1LAC IBs if their functional architecture is comparable to that of VP1GFP IBs. Interestingly, during substrate hydrolysis, an increasing fraction of the enzymatic activity is found not to be linked to IBs (Fig. 3A), and at 30 min, it essentially represents the total activity in the reaction mixture. In the absence of a substrate, VP1LAC IBs incubated under the same conditions also split the activity in soluble and insoluble fractions but to much lesser extent (up to around 50% after a 30-min incubation) (Fig. 3B). Despite the fact that this substrate-mediated modulation of the activity fractioning was clear in every individual experiment, we obtained only fairly significant differences (P = 0.057), probably because of the high variability found between experiments. It must be noted, however, that the total enzymatic activity decreased by more than sixfold in VP1LAC IBs when the substrate was absent (Fig. 3B) but only moderately in actively catalyzing reaction mixtures (with the substrate) (Fig. 3A). Also, 30 min after substrate addition, the specific activity of soluble VP1LAC was estimated to be 10-fold higher than the average for the remaining IB protein species (not shown). On the other hand, the fluorescence of VP1GFP remains associated to IBs for a long time (Fig. 3C), apart from the small fraction that remained constant after being immediately released.
FIG. 2.
Formation of the ONPG hydrolysis product as mediated by VP1LAC IBs (filled symbols) in four independent experiments. As a control, the absorbance of IB samples in the absence of ONPG is also shown (empty symbols).
FIG. 3.
Relative biological activity (measured by either absorbance or fluorescence; left scale) in suspended VP1LAC (A, B) or VP1GFP (C) IBs. Soluble (black bars) and insoluble (gray bars) fractions were analyzed. The total activity in absolute values is shown as dots (top, right scale).
DISCUSSION
Despite the structural similarities recently recognized between IBs and amyloids (6), bacterial aggregates are formed by an unbalanced, highly dynamic equilibrium between protein deposition and removal (10, 38), which implies a continuous reconstruction through exchange of polypeptides between the soluble and insoluble cell fractions (36). The occurrence of native-like structure in IB protein (1, 2, 3, 6, 26, 28) indicates that the aggregation process is not highly selective, involving polypeptides that at least to a significant extent are properly folded. As a consequence, instead of being inert structures, IBs, when formed by proteins with measurable biological activity, result in active nanoparticles with potential applications in catalytic bioprocesses (16, 34, 39). The extent of active (properly folded) protein in IBs is variable depending on the specific polypeptide (16), the environmental conditions under which IBs have been formed (such as temperature or the curve growth phase) (15, 19, 28), and the genetic background of the producer strain (15, 18). This conformational variability can be better understood in the context of a continuum of forms that aggregates formed in bacteria can adopt, including loose aggregates occurring in the soluble cell fraction, amyloid-like fibers, aggregates in the insoluble cell fraction, and conventional, refractile IBs (13). In true IBs, the coexistence of both active and inactive polypeptides has generated intriguing discussions about how such protein species could coexist and in particular if single polypeptides could exhibit both properly folded domains, accounting for the native-like structure observed in infrared spectroscopy analysis (1, 2, 3, 26) (and conferring biological activity if embracing the active site), and misfolded protein stretches, responsible for the intermolecular beta-sheet organization supporting the IB architecture (6, 28).
We have proved in this study that the localization of fluorescence emission in GFP-containing IBs is not homogeneous in all of the aggregate body but is concentrated in its core (Fig. 1). Although the approach used does not allow determination of the extent of misfolded portions of VP1GFP, which does not affect the fluorophore performance, this observation indicates that the active protein (with global proper folding) is not limited to the aggregate surface, which could have been eventually accounted for by in vivo sequence-specific association of soluble and functional polypeptides from the soluble cell fraction to the IB's surface. The fluorescence distribution pattern is consistent when observing IBs formed at different temperatures below 37°C, known to minimize aggregation (33) but enhance the conformational quality of the embedded protein (37). Therefore, the occurrence of functional protein is not an artifact from a weakly stringent purification process, but such active forms are a structural, natural component of IBs. Interestingly, a significant fraction of functional protein is immediately released to the solvent upon resuspension, indicating that despite their nuclear localization, active forms might be exposed to the IB-solvent interface. This can be accounted for by the highly porous architecture and hydrated nature of the IB (5, 8), which must be also supportive of substrate diffusion in IB-mediated catalysis (16) (Fig. 2). Obviously we cannot completely discard some an extent of spontaneous refolding of surface-attached inactive protein, but the progressive loss of activity in catalyzing VP1LAC IBs prompts us to favor the hypothesis of active protein release. In this context, the appearance of soluble functional protein in the reaction mixtures might be enhanced in catalyzing IBs, while it does not occur in VP1GFP IBs and occurs only moderately in VP1LAC IBs in the absence of the enzyme substrate (Fig. 3). Although the variability of the obtained data prevented robust significant support of this hypothesis, it is likely that the catalytic process itself would induce subtle conformational modifications in the active, aggregated polypeptides, promoting their release. Despite the fact that molecular chaperones are tightly associated to IBs (4, 11, 20, 21), more research is needed to know whether such protein release in vitro is modulated by such cell proteins or rather is mechanical process.
On the other hand, the core localization of functional protein could be due to different selectivities of aggregating and disaggregating polypeptides in vivo regarding the conformational status. If, as suggested, the chaperone (or protease)-mediated release from aggregates is surface restricted and specifically targeted to misfolded species (24, 30, 31) (while aggregation seem to be less selective regarding the folding status), the unbalanced equilibrium favoring the in vivo volumetric growth of IBs would progressively enrich the aggregates' core with properly folded polypeptides.
Acknowledgments
We are indebted to Mònica Roldan, from Servei de Microscopia (UAB), for helpful assistance with confocal microscopy.
This work was supported by grants BIO2004-0700 (MEC, Spain) and 2005SGR-00956 (AGAUR, Catalonia, Spain). E.G.-F. is a recipient of a doctoral fellowship from MEC, Spain.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Ami, D., L. Bonecchi, S. Cali, G. Orsini, G. Tonon, and S. M. Doglia. 2003. FT-IR study of heterologous protein expression in recombinant Escherichia coli strains. Biochim. Biophys. Acta 1624:6-10. [DOI] [PubMed] [Google Scholar]
- 2.Ami, D., A. Natalello, P. Gatti-Lafranconi, M. Lotti, and S. M. Doglia. 2005. Kinetics of inclusion body formation studied in intact cells by FT-IR spectroscopy. FEBS Lett. 579:3433-3436. [DOI] [PubMed] [Google Scholar]
- 3.Ami, D., A. Natalello, G. Taylor, G. Tonon, and D. S. Maria. 2006. Structural analysis of protein inclusion bodies by Fourier transform infrared microspectroscopy. Biochim. Biophys. Acta 1764:793-799. [DOI] [PubMed] [Google Scholar]
- 4.Boels, K., M. M. Carrió, A. Arís, J. L. Corchero, and A. Villaverde. 1999. Distinct chaperone affinity to folding variants of homologous recombinant proteins. Biotechnol. Lett. 21:531-536. [Google Scholar]
- 5.Bowden, G. A., A. M. Paredes, and G. Georgiou. 1991. Structure and morphology of protein inclusion bodies in Escherichia coli. Biotechnology (N. Y.) 9:725-730. [DOI] [PubMed] [Google Scholar]
- 6.Carrio, M., N. Gonzalez-Montalban, A. Vera, A. Villaverde, and S. Ventura. 2005. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 347:1025-1037. [DOI] [PubMed] [Google Scholar]
- 7.Carrio, M. M., J. L. Corchero, and A. Villaverde. 1998. Dynamics of in vivo protein aggregation: building inclusion bodies in recombinant bacteria. FEMS Microbiol. Lett. 169:9-15. [DOI] [PubMed] [Google Scholar]
- 8.Carrio, M. M., R. Cubarsi, and A. Villaverde. 2000. Fine architecture of bacterial inclusion bodies. FEBS Lett. 471:7-11. [DOI] [PubMed] [Google Scholar]
- 9.Carrio, M. M., and A. Villaverde. 2001. Protein aggregation as bacterial inclusion bodies is reversible. FEBS Lett. 489:29-33. [DOI] [PubMed] [Google Scholar]
- 10.Carrio, M. M., and A. Villaverde. 2002. Construction and deconstruction of bacterial inclusion bodies. J. Biotechnol. 96:3-12. [DOI] [PubMed] [Google Scholar]
- 11.Carrio, M. M., and A. Villaverde. 2005. Localization of chaperones DnaK and GroEL in bacterial inclusion bodies. J. Bacteriol. 187:3599-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazorla, D., J. X. Feliu, and A. Villaverde. 2001. Variable specific activity of Escherichia coli beta-galactosidase in bacterial cells. Biotechnol. Bioeng. 72:255-260. [PubMed] [Google Scholar]
- 13.de Marco, A., and A. Schroedel. 2005. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferraz, R. M., A. Aris, and A. Villaverde. 2004. Profiling the allosteric response of an engineered beta-galactosidase to its effector, anti-HIV antibody. Biochem. Biophys. Res. Commun. 314:854-860. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Fruitos, E., M. M. Carrio, A. Aris, and A. Villaverde. 2005. Folding of a misfolding-prone beta-galactosidase in absence of DnaK. Biotechnol. Bioeng. 90:869-875. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Fruitos, E., N. Gonzalez-Montalban, M. Morell, A. Vera, R. M. Ferraz, A. Aris, S. Ventura, and A. Villaverde. 2005. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Fact. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgiou, G., and P. Valax. 1999. Isolating inclusion bodies from bacteria. Methods Enzymol. 309:48-58. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Montalban, N., E. Garcia-Fruitos, S. Ventura, A. Aris, and A. Villaverde. 2006. The chaperone DnaK controls the fractioning of functional protein between soluble and insoluble cell fractions in inclusion body-forming cells. Microb. Cell Fact. 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jevsevar, S., V. Gaberc-Porekar, I. Fonda, B. Podobnik, J. Grdadolnik, and V. Menart. 2005. Production of nonclassical inclusion bodies from which correctly folded protein can be extracted. Biotechnol. Prog. 21:632-639. [DOI] [PubMed] [Google Scholar]
- 20.Jurgen, B., H. Y. Lin, S. Riemschneider, C. Scharf, P. Neubauer, R. Schmid, M. Hecker, and T. Schweder. 2000. Monitoring of genes that respond to overproduction of an insoluble recombinant protein in Escherichia coli glucose-limited fed-batch fermentations. Biotechnol. Bioeng. 70:217-224. [DOI] [PubMed] [Google Scholar]
- 21.Lethanh, H., P. Neubauer, and F. Hoffmann. 2005. The small heat-shock proteins IbpA and IbpB reduce the stress load of recombinant Escherichia coli and delay degradation of inclusion bodies. Microb. Cell Fact. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., Z. G. Su, and J. C. Janson. 2004. In vitro protein refolding by chromatographic procedures. Protein Expr. Purif. 33:1-10. [DOI] [PubMed] [Google Scholar]
- 23.Mogk, A., E. Deuerling, S. Vorderwulbecke, E. Vierling, and B. Bukau. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585-595. [DOI] [PubMed] [Google Scholar]
- 24.Mogk, A., D. Dougan, J. Weibezahn, C. Schlieker, K. Turgay, and B. Bukau. 2004. Broad yet high substrate specificity: the challenge of AAA+ proteins. J. Struct. Biol. 146:90-98. [DOI] [PubMed] [Google Scholar]
- 25.Mogk, A., C. Schlieker, K. L. Friedrich, H. J. Schonfeld, E. Vierling, and B. Bukau. 2003. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278:31033-31042. [DOI] [PubMed] [Google Scholar]
- 26.Oberg, K., B. A. Chrunyk, R. Wetzel, and A. L. Fink. 1994. Nativelike secondary structure in interleukin-1 beta inclusion bodies by attenuated total reflectance FTIR. Biochemistry 33:2628-2634. [DOI] [PubMed] [Google Scholar]
- 27.Panda, A. K. 2003. Bioprocessing of therapeutic proteins from the inclusion bodies of Escherichia coli. Adv. Biochem. Eng. Biotechnol. 85:43-93. [DOI] [PubMed] [Google Scholar]
- 28.Przybycien, T. M., J. P. Dunn, P. Valax, and G. Georgiou. 1994. Secondary structure characterization of beta-lactamase inclusion bodies. Protein Eng. 7:131-136. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Schlieker, C., I. Tews, B. Bukau, and A. Mogk. 2004. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 578:351-356. [DOI] [PubMed] [Google Scholar]
- 31.Schlieker, C., J. Weibezahn, H. Patzelt, P. Tessarz, C. Strub, K. Zeth, A. Erbse, J. Schneider-Mergener, J. W. Chin, P. G. Schultz, B. Bukau, and A. Mogk. 2004. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11:607-615. [DOI] [PubMed] [Google Scholar]
- 32.Singh, S. M., and A. K. Panda. 2005. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 99:303-310. [DOI] [PubMed] [Google Scholar]
- 33.Strandberg, L., and S. O. Enfors. 1991. Factors influencing inclusion body formation in the production of a fused protein in Escherichia coli. Appl. Environ. Microbiol. 57:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokatlidis, K., P. Dhurjati, J. Millet, P. Beguin, and J. P. Aubert. 1991. High activity of inclusion bodies formed in Escherichia coli overproducing Clostridium thermocellum endoglucanase D. FEBS Lett. 282:205-208. [DOI] [PubMed] [Google Scholar]
- 35.Vallejo, L. F., and U. Rinas. 2004. Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb. Cell Fact. 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ventura, S., and A. Villaverde. 2006. Protein quality in bacterial inclusion bodies. Trends Biotechnol. 24:179-185. [DOI] [PubMed] [Google Scholar]
- 37.Vera, A., N. Gonzalez-Montalban, A. Aris, and A. Villaverde. 29 September 2006, posting date. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol. Bioeng. doi: 10.1002/bit.21218. [DOI] [PubMed]
- 38.Villaverde, A., and M. M. Carrio. 2003. Protein aggregation in recombinant bacteria: biological role of inclusion bodies. Biotechnol. Lett. 25:1385-1395. [DOI] [PubMed] [Google Scholar]
- 39.Worrall, D. M., and N. H. Goss. 1989. The formation of biologically active beta-galactosidase inclusion bodies in Escherichia coli. Aust. J. Biotechnol. 3:28-32. [PubMed] [Google Scholar]