Abstract
If the acquisition of virulence genes (VGs) for pathogenicity were not solely acquired through horizontal gene transfers of pathogenicity islands, transposons, and phages, then clonal clusters of enterotoxigenic Escherichia coli (ETEC) would contain few or even none of the VGs found in strains responsible for extraintestinal infections. To evaluate this possibility, 47 postweaning diarrhea (PWD) ETEC strains from different geographical origins and 158 commensal E. coli isolates from the gastrointestinal tracts of eight group-housed healthy pigs were screened for 36 extraintestinal and 18 enteric VGs using multiplex PCR assays. Of 36 extraintestinal VGs, only 8 were detected (fimH, traT, fyuA, hlyA, kpsMtII, k5, iha, and ompT) in the ETEC collection. Among these, hlyA (α-hemolysin) and iha (nonhemagglutinating adhesin) occurred significantly more frequently among the ETEC isolates than in the commensal isolates. Clustering analysis based on the VG profiles separated commensal and ETEC isolates and even differentiated serogroup O141 from O149. On the other hand, pulsed-field gel electrophoresis (PFGE) successfully clustered ETEC isolates according to both serotype and geographical origin. In contrast, the commensal isolates were heterogeneous with respect to both serotype and DNA fingerprint. This study has validated the use of VG profiling to examine pathogenic relationships between porcine ETEC isolates. The clonal relationships of these isolates can be further clarified by PFGE fingerprinting. The presence of extraintestinal VGs in porcine ETEC confirmed the hypothesis that individual virulence gene acquisitions can occur concurrently against a background of horizontal gene transfers of pathogenicity islands. Over time, this could enable specific clonotypes to respond to host selection pressure and to evolve into new strains with increased virulence.
Enterotoxigenic Escherichia coli (ETEC) is the major cause of postweaning diarrhea (PWD) in weaned pigs. ETEC strains colonize the small intestine with the aid of adhesion factors (e.g., F4 or F18 fimbriae). Once attached, the release of either heat-labile or heat-stable (STa or STb) enterotoxins induces diarrhea by affecting the electrolyte balance of the small intestine. The presence of virulence genes (VGs) encoding these determinants is a key requirement for pathogenicity in ETEC and is generally used to distinguish these pathogens from the nonpathogenic or commensal E. coli normally carried in the intestine (37).
Characterization of isolates from outbreaks of PWD has shown that ETEC strains lacking recognized fimbriae, such as F4 and F18, are becoming more common (19, 26, 44). This may be related to the widespread use of vaccines incorporating fimbriated strains, providing selection pressure for the acquisition and carriage of novel, unrecognized VGs. For example, Noamani et al. (37) found that most of the PWD isolates recovered in the period 1998 to 2001 in Ontario, Canada, were associated with a new type of O149 ETEC, which possessed VGs for STa and an enteroaggregative heat-stable enterotoxin (EAST1) not found previously in O149 isolates from the same region. Over time, porcine ETEC acquisition of new VGs associated with PWD may therefore change their VG profile and, potentially, their pathogenicity.
Numerous genes encoding virulence factors such as adhesins, host cell surface-modifying factors, invasins, toxins, and secretion systems are involved in mechanisms of pathogenicity in E. coli. Strains of the same pathotype and serotype normally carry a defined set of VGs that are critical for infection. For instance, enteropathogenic E. coli (EPEC) is associated with virulence factors encoded by the locus of enterocyte effacement, including tir, eaeA, and esp genes, while extraintestinal pathogenic E. coli (ExPEC) that causes urinary tract infections (UTI) or septicemia carries pap, afa/draBC, sfa/focDE, kpsMTI, and iutA genes (29).
However, using recent technological advances such as microarrays and multiplex PCR to explore the global virulence pattern of strains, combinations of VGs predictive of different pathotypes have been observed in unusual pathogenic and commensal E. coli strain backgrounds (2, 31). For example, Bekal et al. (2) found a human enterohemorrhagic (EHEC) strain harboring a number of VGs typically involved in extraintestinal infections. Additionally, a bovine ETEC strain was found to contain ExPEC-associated genes (traT, ompT, and fimH) and an EHEC-associated gene (etpD) as well as genes for F5 fimbriae and STa. A human ETEC strain (H10407) was found to contain two invasion genes (tia and tib) that direct the bacterium to invade human intestinal cell lines (17).
Population analysis of the autochthonous E. coli bacteria inhabiting the gastrointestinal tract (GIT) of pigs has shown them to be very diverse, with greater diversity being demonstrated between the different regions of the gut than between different animals (12). In terms of microbial fitness, a potential advantage that porcine ETECs have over commensal E. coli strains is greater amplification and dispersion in pigs with diarrhea compared to healthy pigs with normal feces and, thus, greater opportunity for acquisition of VGs from other strains (37). Characterization and comparison of VG profiles between ETEC and gut commensal E. coli isolates can provide important insights into the evolution and spread of VGs in potentially pathogenic lineages. The aims of this work were to determine whether PWD ETECs also carry VGs that are representative of other pathotypic E. coli strains and to assess whether combinations of VGs are associated with ETEC strains belonging to different serotypes. It was also our intention to examine if VG combinations could distinguish clinical isolates from different geographic origins and to explore the presence of VGs in commensal isolates. Additionally, molecular variation between the PWD ETEC and commensal isolates has been characterized by pulsed-field gel electrophoresis (PFGE) and genomic fingerprinting and compared with VG profiling as a predictor of genotype.
MATERIALS AND METHODS
E. coli strains used in this study.
A total of 205 porcine E. coli strains were analyzed in this study. The collection consisted of 47 isolates recovered from pigs with PWD and 158 E. coli isolates obtained from different parts of the GIT of healthy pigs. These isolates have been described in a previous publication (12) and are referred to as nonidentical commensal isolates or strains based on DNA fingerprinting. The PWD isolates were obtained from diagnostic submissions (fecal swabs or intestinal contents) from animals of different geographic origins. Diagnostic criteria for designation of an isolate as a PWD strain have been given previously (13). Seventeen isolates were from southern New South Wales (NSW), Australia, 23 from Queensland (Qld), Australia, and 7 from Vietnam. The isolates were represented by two major serogroups, O149 (n = 39) and O141 (n = 8). The 158 commensal E. coli strains were isolated from eight healthy 13-week-old male pigs (hybrids of Large White and Landrace), weighing 35 to 45 kg each, from eight different litters at Elizabeth Macarthur Agricultural Institute, Australia. On average, four to five isolates were taken from three different gut regions (duodenum, ileum, and colon) and fecal samples for each individual pig. The isolates were confirmed as E. coli by indole test (positive), minimal lactose agar growth (positive), and Simmons citrate agar growth (negative), as described by Dixit et al. (12). Strains used as reference sources of VGs for PCR analysis are listed in Table S1 in the supplemental material. All strains were grown on Luria-Bertani (LB) media and routinely stored at −80°C in 15% (vol/vol) glycerol. PCR template DNA was prepared by boiling 1 ml of overnight LB broth culture for 10 min, and 2-μl aliquots of the supernatant were subjected to PCR.
Serotyping porcine E. coli isolates.
E. coli isolates were serotyped by the Microbiological Diagnostic Unit, University of Melbourne, Australia, using slide agglutination tests as described previously (5, 6). Strains which failed to achieve motility on semisolid medium were considered nonmotile and designated H−.
Detection of virulence genes and rapid phylogenetic analysis by PCR.
ExPEC-associated virulence factors were detected by multiplex PCR testing of 36 different VGs associated with extraintestinal diseases. In addition to the 30 extraintestinal VGs that have been published already (28), 6 additional genes (univcnf, iha, ironec, ompT, iss, and ireA) were added as shown in Table 1 (J. R. Johnson, University of Minneapolis, personal communication). Primers were sorted into six pools based on primer compatibility and amplicon size. The multiplex PCR assay was modified from Johnson et al. (29). Briefly, PCR was performed with 0.2-ml PCR tubes (INTERPATH, Australia) on a PC-960 thermal cycler (CorbettResearch, Australia) with a reaction volume of 25 μl. The DNA template (2 μl) was added to a mixture containing 0.6 mM of each dATP, dGTP, dCTP, and dDTP (Astral Scientific, Australia), 1× buffer solution (QIAGEN, Australia), 4 mM MgCl2, 0.6 μM of each primer, and 1.5 U of HotStar Taq polymerase (QIAGEN). Each PCR program was preceded by a step of 15 min at 95°C to activate the HotStar Taq polymerase, followed by 25 cycles of 94°C for 30 s, 63°C for 30 s, and 68°C for 3 min, with an extension step at 72°C for 10 min. MilliQ H2O was included in each PCR run as a negative control. DNA from each strain was also subjected to PCR for 18 different VGs (Table 2), which represent various E. coli pathotypes causing enteric disease, including ETEC, EPEC, enteroaggregative E. coli (EaggEC), enteroinvasive E. coli (EIEC), and EHEC. The designations of all the primers in both Table 1 and 2 are described in Table S2 in the supplemental material.
TABLE 1.
Virulence genes and PCR sizes for 36 ExPEC genes
| Gene name | Description/function | Amplicon size (bp) |
|---|---|---|
| PAI | PAI marker malX from strain CFT073 | 930 |
| papAH | Major structural subunit of pilus associated with pyelonephritis (P fimbriae); defines F antigen | 720 |
| fimH | d-Mannose-specific adhesin, type 1 fimbriae | 508 |
| kpsMTIII | Group III capsular polysaccharide synthesis (e.g., K3, K10, and K54) | 392 |
| papEF | Minor tip pilins; connect PapG to shaft (PapA) | 336 |
| ibeA | Invasion of brain endothelium | 170 |
| fyuA | Yersinia siderophore receptor (ferric yersiniabactin uptake) | 880 |
| bmaE | M-agglutinin subunit | 507 |
| sfa/focDE | Central region of sfa (S fimbriae) and foc (F1C fimbriae) operons | 410 |
| iutA | Ferric aerobactin receptor (iron uptake; transport) | 300 |
| papG allele III | Cystitis-associated (prs or pap-2) papG variant | 258 |
| kpsMT K1 | Specific for K1 (group II) kpsMT | 153 |
| hlyA | α-Hemolysin | 1,177 |
| rfc | O4 lipopolysaccharide synthesis | 788 |
| nfaE | Nonfimbrial adhesin I assembly and transport | 559 |
| papG I (internal) | (Rare) J96-associated papG variant | 461 |
| kpsMTII | Group II capsular polysaccharide synthesis (e.g., K1, K5, and K12) | 272 |
| papC | Pilus assembly; central region of pap operon | 200 |
| gafD | N-Acetyl-D-glucosamine-specific (G) fimbriae adhesin | 952 |
| cvaC | Colicin V; conjugative plasmids | 680 |
| cdtB | Cytolethal distending toxin | 430 |
| focG | Pilus tip molecule, F1C fimbriae (sialic acid specific) | 360 |
| traT | Surface exclusion, serum survival | 290 |
| papG II | Pyelonephritis-associated papG variant | 190 |
| papG I | (Rare) J96-associated papG variant | 1,190 |
| papG II and III | J96-associated papG variant | 1,070 |
| afa/draBC | Central region of Dr antigen-specific fimbrial and afimbrial adhesin operons | 559 |
| cnf1 | Cytotoxic necrotizing factor 1 | 498 |
| sfaS | Pilus tip adhesin, S fimbriae (sialic acid specific) | 240 |
| kpsMT K5 | Specific for non-K1 and non-K2 group II kpsMT | 159 |
| univcnf | Universal primer for cytotoxic necrotizing factor 1 | 1,105 |
| iha | Novel non-hemagglutinin adhesin (from O157:H7 and CFT073) | 827 |
| iroNE.coli | Novel catecholate siderophore | 665 |
| ompT | Outer membrane protein T (protease) | 559 |
| iss | Serum survival gene | 323 |
| ireA | Iron-regulated element, a siderophore receptor | 254 |
TABLE 2.
Virulence genes and PCR sizes for 18 enteric pathogenic E. coli virulence genes and 3 Clermont PCR genes
| Gene name | Description | Amplicon size (bp) | Reference |
|---|---|---|---|
| Enterotoxigenic E. coli (ETEC) | |||
| faeG | F4 fimbrial adhesin (K88) | 86 | 22 |
| fedA | F18 fimbrial adhesin | 128 | 22 |
| fanC | F5 fimbrial adhesin (K91) | 450 | 13 |
| fasA | F6 fimbrial adhesin | 333 | 13 |
| F41 | Fimbrial adhesin | 431 | 13 |
| estI | Heat-stable enterotoxin (STa) | 166 | 13 |
| estII | Heat-stable enterotoxin (STb) | 172 | 13 |
| eltA | Heat-labile toxin | 696 | 13 |
| Enteroaggregative E. coli (EaggEC) | |||
| east-1 | EaggEC heat-stable enterotoxin (EAST1) | 111 | 52 |
| aggC | Fimbrial antigen-specific gene | 528 | 52 |
| Enteropathogenic E. coli (EPEC) | |||
| bfpA | Type IV bundle-forming pili | 326 | 23 |
| Enteroinvasive E. coli (EIEC) | |||
| ipaH | Invasion plasmid antigen | 600 | 41 |
| Shiga toxin E. coli (STEC) | |||
| stx1 | Shiga toxin I | 180 | 39 |
| stx2 | Shiga toxin II | 255 | 39 |
| stx2e | Shiga toxin 2e | 139 | 22 |
| eae | Intimin | 384 | 39 |
| ehxA | Enterohemolysin | 534 | 39 |
| Cytolethal distending toxin-producing E. coli | |||
| cdt | Cytolethal distending toxin | 108 | 11 |
| Clermont phylogeny PCR markers | |||
| chuA | A heme transport gene found in O157:H7 | 279 | 10 |
| yjaA | An unknown-function gene in K-12 | 211 | 10 |
| TspE4C2 | An anonymous DNA fragment associated with neonatal meningitis E. coli | 152 | 10 |
The phylogenetic grouping for each isolate was determined based on the PCR method described by Clermont et al. (10). A two-step triplex PCR was performed using three gene primers (chuA, yjaA, and DNA fragment TspE4C2) under the following conditions: denaturation for 4 min at 94°C, followed by 30 cycles of 5 s at 94°C and 10 s at 59°C, and a final extension step of 5 min at 72°C. The following criteria were used for assignment of strains to phylogenetic groups: group B2, chuA+, yjaA+; group D, chuA+, yjaA mutant; group B1, chuA mutant, TspE4C2+; and group A, chuA mutant, TspE4C2 mutant (15).
PCR products were separated by agarose gel electrophoresis. Five microliters of amplified product was mixed with 3 μl of loading dye containing 0.25% bromophenol blue in 15% Ficoll solution and loaded onto 2.5% agarose gels for electrophoresis (Ultrapure Agarose; Life Technology, Australia) with ethidium bromide (1 μg/ml), using 0.5× Tris-borate-EDTA (TBE) as running buffer. DNA in the gel was visualized by exposing the gel to UV light and was photographed with a digital capture system (Gel Doc; Bio-Rad). PCR product lengths were verified by comparison with a 100-bp DNA ladder (Promega, Australia).
Pulsed-field gel electrophoresis (PFGE).
Genomic DNA for PFGE was prepared using the technique of Barrett et al. (1), with modifications. In brief, bacterial colonies from overnight LB agar were embedded in 2% of low-melting-point agarose (Bio-Rad). Genomic DNA was extracted in situ by treatment with lysozyme (0.5 mg/ml; Sigma, Australia) and proteinase K (0.5 mg/ml; Sigma) lysis buffers. The agarose-packaged DNA was then digested with the endonuclease NotI or XbaI (New England Biolabs, Australia). The resulting fragments were separated using 1.2% PFGE-grade agarose gel (Bio-Rad) in a GeneNavigator system (Amersham Pharmacia Biotech), with a 5- to 35-s pulse time and 200 V for 25 h in 0.5× TBE buffer at 12°C. Gel images were saved in TIFF format and processed using GelCompar software (version 4.2; Applied Maths, Kortrijk, Belgium) for computer analysis. PFGE chromosomal fingerprints were compared by use of the criteria of Tenover et al. (46). Similarity between fingerprints was further determined on the basis of the Dice coefficient. A band position tolerance of 1.0% and a maximal optimization shift of 0.5% were set, and dendrograms were generated by the unweighted pair group method with arithmetic mean analysis.
DNA sequencing.
PCR fragments were partially sequenced for gene verification. The DNA band of the target PCR product was excised from the agarose gel and purified using a QIAGEN Gel Purification kit. A total of 70 ng of purified DNA was subjected to AmpliTaq cycle-sequencing reactions with the Prism ready reaction dye dideoxy terminator cycle-sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Electrophoresis of amplified products was performed on 7% polyacrylamide gels with an automated sequencer (model 377A; Applied Biosystems). The nucleotide sequences were analyzed with the ANALYSIS program (Applied Biosystems) and the ANGIS program (Australian National Genomic Information Service, University of Sydney).
Statistical analysis.
The chi-squared test of deviance was used to analyze the correlation between the presence of virulence genes and the distribution of virulence genes within different serogroups.
RESULTS
Prevalence of virulence genes in commensal and clinical porcine E. coli isolates.
Using multiplex PCR screening of 36 ExPEC-associated VGs, 8 genes were identified in the porcine isolates: fimH, fyuA, hlyA, kpsMtII, k5, iha, traT, and ompT. The prevalence of these VGs in PWD isolates and the commensal isolates is shown in Table 3. Two genes, hlyA and iha, occur significantly more frequently among the PWD isolates (P < 0.01). fimH and traT genes were present in most of the isolates, but fimH was not found in any PWD isolates belonging to serogroup O149. A number of commensal isolates possessed fyuA or ompT (23 and 13 out of 158 isolates, respectively), which was rarely found in PWD isolates.
TABLE 3.
Prevalence of VGs detected in porcine PWD and commensal isolates
| Gene | % Prevalence in serotype:
|
||||
|---|---|---|---|---|---|
| O149 (NSW; n = 9) | O149 (Qld; n = 23) | O149 (Vietnam; n = 7) | O141 (NSW; n = 8) | Commensals (n = 158) | |
| ExPEC virulence gene | |||||
| fimH | 0 | 0 | 0 | 100 | 84.8 (134/158) |
| hlyA | 100 | 74 (17/23) | 85.7 (6/7) | 88 (7/8) | 0.6 (1/158) |
| traT | 100 | 61 (14/23) | 100 | 12 (1/8) | 48.7 (77/158) |
| iha | 100 | 100 | 100 | 88 (7/8) | 1.2 (2/158) |
| fyuA | 0 | 0 | 0 | 12 (1/8) | 14.5 (23/158) |
| ompT | 0 | 0 | 0 | 12 (1/8) | 8.2 (13/158) |
| kpsMTII | 0 | 0 | 0 | 0 | 1.2 (2/158) |
| k5 | 0 | 0 | 0 | 0 | 1.2 (2/158) |
| Enteric pathogenic virulence gene | |||||
| eltA | 100 | 100 | 100 | 0 | 0 |
| estI | 100 | 83 (19/23) | 100 | 88 (7/8) | 5.7 (9/158) |
| estII | 100 | 100 | 100 | 88 (7/8) | 0 |
| stx2e | 0 | 0 | 0 | 88 (7/8) | 1.9 (3/158) |
| F4 | 100 | 100 | 100 | 0 | 0 |
| F18 | 0 | 0 | 0 | 88 (7/8) | 0 |
| east-1 | 100 | 100 | 100 | 0 | 5.1 (8/158) |
| stx2 | 0 | 0 | 0 | 88 (7/8) | 1.9 (3/158) |
| eae | 0 | 0 | 0 | 0 | 3.2 (5/158) |
| Clermont PCR gene | |||||
| chuA | 0 | 0 | 0 | 12 (1/8) | 0.6 (1/158) |
| yjaA | 0 | 0 | 0 | 88 (7/8) | 51.3 (81/158) |
| TspE4C2 | 0 | 0 | 0 | 0 | 31.6 (50/158) |
Nine of 18 enteric pathogenic VGs were also detected in the isolates (eltA, estI, estII, stx2e, faeG, fedA, east-1, eae, and stx2). Most PWD isolates contained a typical porcine ETEC fimbrial gene, either faeG or fedA, and at least one enterotoxin gene, such as eltA, estI, or estII, with the exception of one Qld serogroup O149 isolate, which was faeG+ and mutant for eltA, estI, and estII. The F4 fimbrial gene (faeG) was exclusively associated with serogroup O149 isolates, and all of the O141 isolates harbored the F18 fimbrial gene (fedA), except for one isolate in which only the type 1 fimbriae gene (fimH) was found. The east-1 gene, which encodes the heat-stable toxin EAST1 that is unrelated to STa and STb, was found to be particularly associated with isolates of serogroup O149 (Table 3). Notably, east-1 was only present in one O141 strain and 8 out of 158 commensal isolates. Moreover, none of the O149 isolates possessed any Shiga toxin E. coli (STEC) genes (stx1, stx2, eae, or exhA), whereas stx2 was of reasonably high prevalence in serotype O141. Comparably, the isolates designated commensals were found on the whole to lack enteric VGs. For example, faeG and fedA were not detected, only three strains contained stx2, and estI and east-1 were only detected in 9 and 8 out of 158 isolates, respectively, confirming that isolates from healthy pigs generally do not carry VG combinations to enable pathogenicity.
It is important to note that in the present study, different primer pairs were used for the detection of stx2 and stx2e allelic variations within the stx2 gene. The majority of strains which possessed stx2 were found to have the stx2e allele (six out of eight O141 and three out of three commensal isolates). This result corresponds to the finding by Fratamico et al. that confirmed that stx2e was the most frequent stx2 variant found in E. coli isolated from pig feces (20). In addition, three cdt gene primer pairs were used (a1/s1, a2/s2, and 3A/3B) for the PCR detection of three different cdt gene alleles: cdtIIB, cdtIB, and cdtIIIB, respectively. None of these three alleles were present in the ETEC or commensal isolates.
Phylogenetic group analysis of porcine PWD and commensal isolates.
Phylogenetic analysis using the triplex PCR technique developed by Clermont et al. (10) revealed that group A E. coli isolates were predominant in the collection of isolates, with a prevalence of 100% in serotype O149, 92.9% in O141, and 70.5% in commensal isolates. Group B1 was only found in commensals (29.5%). Only one O141 isolate was identified as group B2, and one commensal isolate was found in group D. Interestingly, none of the O149 isolates contained any of the three Clermont PCR gene markers, and most of the O141 and commensal isolates harbored yjaA or TspE4C2.
Clustering analysis of porcine E. coli based on virulence gene attributes.
Two major VG combinations were identified among the clinical isolates in this study: O149 (hlyA, iha, traT, east-1, estI, estII, eltA, and faeG) and O141 (fimH, hlyA, iha, stx2, estI, estII, fedA, and yjaA). Thirty VG combinations were present in the 158 commensal isolates, of which 13 were represented only by single isolates. The VG combinations of high prevalence were the following: 20/158 (12.7%; fimH and yjaA), 19/158 (12.0%; fimH, traT, and TspE4C2), 18/158 (11.4%; fimH, fyuA, and TspE4C2), 17/158 (10.8%; fimH, traT, and yjaA), 13/158 (8.2%; fimH), and 10/158 (6.3%; yjaA). The VG profiles identified in the ETEC isolates were significantly different from the VGs possessed by commensal isolates (P < 0.01). Clustering analysis of the isolates based on VG profiles differentiated the commensals from ETEC isolates by segregation into two differentiated clusters (cluster 1 for all commensal isolates and cluster 2 for all ETEC isolates). The ETEC isolates were further separated into two subclusters in accordance with serogroups (O141 and O149), but the ETEC isolates from different geographic origins could not be differentiated based on their VG profiles. As presented in Fig. 1, commensal isolates were grouped separately into cluster 1, whereas isolates in O141 and O149 were separated into clusters 2a and 2b, respectively. There was one exception: a serogroup O141 strain was found to be more closely related to the commensal E. coli group, because it was the only ETEC strain lacking hlyA, iha, faeG, and fedA genes, although it did possess fimH, fyuA, traT, east-1, and estII genes.
FIG. 1.
Clustering analysis of the genetic variation between PWD and commensal porcine E. coli isolates based on the virulence gene attributes. Short lines represent the presence of specific PCR amplicons. Thirty selected commensal isolates representing 30 different VG combinations were grouped in cluster 1; PWD isolates in serogroup O141 were grouped in cluster 2a, except one in cluster 1; 11 in serogroup O149 isolates from different geographic origin which represented seven VG combinations were in a separate cluster, 2b. Viet, Vietnam.
PFGE analysis of PWD isolates.
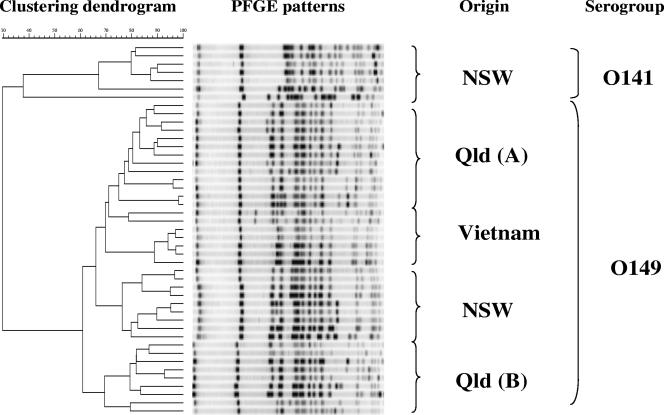
Forty-five PWD isolates were further analyzed by PFGE fingerprinting after restriction enzyme digestion with NotI and XbaI. These isolates included 7 serogroup O141 isolates (NSW), 9 serogroup O149 isolates (NSW), 7 serogroup O149 isolates (Vietnam), and 22 serogroup O149 isolates (Qld). Relatedness of each DNA fingerprint was clustered using the GelCompar software, as presented in Fig. 2. Based on the PFGE pattern, 45 isolates were separated into two main clusters (I and II) in accordance with serogroups (O141 and O149). There are three subclusters in serogroup O149, IIa, IIb, and IIc, primarily based on geographical origin. Queensland isolates fell into two distinct groups, with one group (Qld A in IIa) more closely related to Vietnamese isolates than the second Queensland group (Qld B in IIc). These groups were both distinct from the New South Wales isolates, which also fell into a single subcluster (IIb).
FIG. 2.
Clustering analysis of porcine PWD isolates from different geographic origins (NSW, Qld, and Vietnam) based on PFGE patterns obtained by digestion of bacterial genomic DNA with NotI. The dendrogram was generated using the Dice coefficient and unweighted-pair group method using average linkages.
Analysis of porcine commensal isolates by PFGE and serotyping.
PFGE profiles and serotype were obtained for 158 pig commensal isolates. A total of 45 different serotypes were identified, comprising 29 O groups and 19 H types, as well as 58 nonmotile (H−) strains. Seventeen serotypes were identified as nontypeable O antigens (Ont). The predominant commensal serotype was Ont 63 (39.9%), followed by this breakdown: O40, 19 (12.0%); O5, 9 (5.7%); O8, 5 (3.2%); O82, 5 (3.2%); O25, 4 (2.5%); O51, 4 (2.5%); O105, 3 (1.9%); O2, 3 (1.9%) and O71, 3 (1.9%). The typical PWD ETEC serogroups, such as O141, O149, and O139, were not identified in any of 158 commensal isolates. Serogroups O8 and O20, which are regarded as potentially associated with neonatal diarrhea in piglets, were detected with very low prevalence (5 and 2 out of 158 commensal isolates, respectively). Both of the O20 isolates possessed stx2 and east-1 genes, and one isolate of O8 harbored both hlyA and iha genes.
Based on the interpretive criteria for PFGE patterns (46), isolates differing by a single genetic event, reflected as a difference of one or two bands, are considered closely related and were therefore defined as a distinct PFGE pattern in this study. A total of 62 PFGE patterns were distinguished in the 158 commensal isolates, with 30 of the isolates showing a unique PFGE pattern. The predominant PFGE pattern (pattern 20#) was displayed by 16 isolates, followed by pattern 30#, which was found in 8 isolates. The typical PFGE patterns obtained for the PWD isolates, as shown in Fig. 2, were not identified in any of the commensals.
Isolates within the same serotype normally possessed identical PFGE patterns. For example, 16 out of 18 isolates within serotype O40:H25 displayed PFGE pattern 20# regardless of sampling animal or location. There were some exceptions: five isolates with serotype O114:H− had four distinct PFGE patterns, and 23 isolates belonging to Ont:H− had 15 PFGE patterns.
DISCUSSION
Porcine ETEC isolates associated with PWD belong to a limited number of serogroups, with O8, O138, O139, O141, O147, O149, and O157 being the most commonly reported worldwide (22, 36). Serotyping of porcine ETECs for epidemiological purposes utilizes surface-associated determinants of the bacterium, such as lipopolysaccharide, capsular antigens, and fimbrial antigens (3). Nonetheless, different serogroups still cause similar pathology and clinical disease and should therefore share many features that contribute to their virulence. An important and yet unclear issue addressed in this study is whether these clonal ETEC isolates have in common virulence genes typical of enteric E. coli or whether they also possess additional virulence genes or virulence gene combinations that can be found in other pathotypes, such as EHEC, EPEC, EIEC, EaggEC, and ExPEC.
It is likely that the acquisition of certain VGs by transferable genetic elements (plasmids and transposons) endows ETECs with the potential ability to broaden their host range or evolve into new serogroups over time (19, 33). The deployment of a multivirulence gene panel assembled from different E. coli pathotypes may provide some insight into the acquisition of VGs in different serogroups of porcine ETECs and whether such diversity may endow better survival attributes because of selection pressure. In this study, both PWD and nonidentical E. coli strains from healthy pigs (commensals) were screened by PCR for a panel of 54 VGs assembled from seven major human pathotypes (ExPEC, ETEC, EPEC, EHEC, STEC, EIEC, and EaggEC). The PWD strains were confirmed to be ETEC due to the possession of at least one of the typical porcine ETEC virulence genes encoding fimbriae and/or enterotoxins. VGs such as east-1 and stx2e were also identified in this PWD isolate collection, mirroring previous findings (9, 22, 38). stx2e is normally possessed by porcine STEC strains that cause edema disease, but some porcine E. coli strains were found to produce stx2e together with enterotoxins capable of causing diarrhea (42). The east-1 gene was initially found in human pathogenic EaggEC and encodes the heat-stable toxin termed EAST1, associated with persistent, watery diarrhea in young children (48, 52). Recent surveys have shown that east-1 is widely distributed among porcine diarrhea-related strains, although the role of EAST1 in diarrhea caused by porcine ETEC has still not been determined (37, 49). The results of this study showed a significantly higher incidence of east-1 in O149 ETEC isolates, while the majority of porcine commensal E. coli isolates are east-1 negative, providing incentive for further investigating the pathogenic significance of this gene in porcine PWD.
In 1985, Levine et al. suggested that factors in addition to enterotoxin production may be important in the pathogenesis of ETEC infection in animals (34). Later efforts have focused on searching for additional factors shown to be important for virulence in other intestinal pathotypes, but the extraintestinal virulence factors have been largely ignored. In the present study, eight ExPEC-associated VGs were found in the porcine E. coli isolate collection. Among these, fimH, which encodes type 1 fimbriae, was frequently detected in both PWD and commensal isolates, with the exception of serogroup O149 isolates. fimH was present in the majority of commensal porcine isolates, which may indicate a potential role for type 1 fimbriae in the adherence of resident E. coli to pig GIT via a mannose-resistant mechanism. Interestingly, contrary to the other PWD and neonatal diarrhea serogroups (data not shown) in which fimH prevalences were significantly high, none of the isolates in serogroup O149 contained fimH, which concurs with the results of a previous study (47).
In this investigation, O149 differed from O141, O8, and commensal strains because of the absence of fimH, fyuA, ompT, chuA, yjaA, and TspE4C2. However, all these serogroups did have in common faeG, hlyA, iha, and east-1 virulence genes. O149 is reported to be the most prevalent serogroup isolated from pigs with PWD in Europe and North America (22). The reason for this dominance is still unclear. According to Hampson (24), somatic O antigen or associated factors confers the ability to establish and proliferate in the pig intestine, thus, strains expressing O149 antigen possess certain advantages over other types. O149 dominance could also be related to virulence factor composition if these factors enhance competitiveness in the swine gastrointestinal tract. Larsen (32) has shown that the PWD O149 strains differed from other serotypes in carbohydrate fermentation, urease activity, and colicinogenicity tests. In the present study, PFGE analysis demonstrated that the DNA fingerprint of O149 isolates differed markedly from other ETEC and non-ETEC isolates, indicating that possession of the unique O149 VG profile is linked with genotype. Interestingly, the PFGE analysis showed that isolates from different geographical origins were highly clonal, and the O149 strains from Vietnam were more related to a group of Qld isolates than NSW isolates, confirming that there has been relatively little movement of E. coli populations in pigs between Qld and NSW (13). This also suggests that the Vietnamese strains probably originated from stock imported from Queensland, most probably during agricultural development programs sponsored by the Australian government (D. Trott, University of Queensland, Australia, personal communication). It is also in agreement with the previous report from Hampson et al. (25), which demonstrated that isolates of serogroup O149 from suckling piglets in Scandinavia, Australia, and Indonesia belonged to a single clonal grouping based on multilocus enzyme electrophoresis analysis and shared a close genetic relationship with O149 isolates associated with PWD. The clonal nature of O149 suggests that the deployment of commensal E. coli strains may provide an effective measure to control PWD via competitive exclusion mechanisms. However, the possibility that the data and conclusions may only represent a restricted geographical distribution of genotypes cannot be excluded. To support this hypothesis, a broad range of PWD E. coli strains from a wide variety of sources and serotypes will need to be analyzed.
Two virulence genes, hlyA and iha, were present in significantly higher proportions in PWD isolates than in the commensals. hlyA encodes α-hemolysin, which is a gram-negative bacterial membrane pore-forming cytolytic toxin of the RTX family with glycine-rich repeats (50). α-Hemolysin is predominantly detected in ExPEC strains. It is also a major marker of most porcine ETEC and STEC strains, causing PWD and edema disease in pigs. Porcine ETEC strains associated with PWD are almost universally hemolytic, and α-hemolysin in these isolates is considered to enhance virulence and colonization (43). The cytotoxic necrotizing factor (cnf-1) was reported to be strongly associated with α-hemolysin and virulence in both human ExPEC and pig diarrheal isolates. The cnf1 gene is closely linked with hlyA on a pathogenicity island (7, 16). This cooccurrence, however, was not shown in our porcine isolates. It has been demonstrated that porcine cnf-1-producing E. coli also expresses P, S, and F1C fimbriae in strains causing septicemia and/or diarrhea (14). Our results indicate that cnf-1-producing strains are more likely to be ExPEC strains which possess a repertoire of VGs that differ from the enteric PWD strains.
To our knowledge, this is the first report of the presence of the iha gene in porcine PWD isolates with significantly higher carriage rates than gut commensal isolates. iha encodes a novel nonhemagglutinating adhesin found in E. coli O157:H7, which facilitates the adherence of O157:H7 strains to epithelial cells in environment such as the GIT of animals (45). iha also facilitates adherence to HeLa cells when transformed into nonadherent E. coli K-12. Prior to this study, iha has only been found in STEC and ExPEC pathotypes. In addition to O157:H7, other STEC strains also contain iha, including strains of serotype O111:H− and O91:H− (21). Additionally, iha occurs significantly more frequently in UTI or bacteremia isolates than human commensal E. coli isolates (29). The role of iha in colonization of both humans and animals is still being elucidated. Porcine ETEC strains harboring additional fimbrial adhesins, such as iha and eae, could possibly exploit several alternative pathways for colonization of the host. The association of iha and hlyA with porcine PWD suggests that these VGs could be used as additional markers of pathogenicity in PWD isolates. More importantly, due to the fact that the use of vaccines against fimbriated strains may allow the emergence of other ETEC strains which lack recognized fimbriae such as F4 and F18, iha might be a potential target for anti-ETEC intervention. Future work will be focused on investigating the role of iha in pig gut colonization by ETEC strains, the mechanisms underlying iha expression, and the presence of this gene in other PWD serogroups.
It is widely believed that a positive correlation exists between environmental stability, community stability, and high microbial diversity (40). A highly diversified microflora may reflect a stable community with greater “colonization resistance” to enteric pathogens. In the suckling piglet, coliforms isolated from normal feces differ from diarrheal coliforms, because the latter is dominated by pathogenic clones with reduced diversity, and these express only limited combinations of virulence-associated factors (30). The present study investigated serotype, VG profile, and DNA fingerprint patterns for 158 E. coli isolates designated commensals on the basis of their isolation from healthy pigs. The study confirmed that none of the healthy pigs examined were colonized with typical ETEC strains. Very few isolates were of a serotype that is associated with porcine-pathogenic E. coli strains, and fewer still contained virulence genes such as estI, stx2, and east-1, which may be associated with enteric disease in swine. The majority of commensals only possessed “fitness or adaptive” genes such as fimH, traT, and fyuA, which are involved in colonization, serum resistance, and iron utilization, respectively. This result corresponds to a previous report by Hinton et al. (27), in which a complex E. coli microbiota was demonstrated in the GIT of healthy weaned pigs in the absence of ETEC serotypes such as O149. The present study also established a remarkable clonal diversity among the commensal isolates between pigs. This was an interesting finding, considering that pigs all shared the same environment and feed and water sources. A high level of diversity among commensal E. coli (12) has also been observed in cattle and human feces based on serotype identities (4, 6). By comparing E. coli biochemical fingerprints from five healthy piglets from 7 to 63 days of age, Melin et al. (35) suggested that a disturbed (i.e., decreased diversity) fecal coliform microbiota close to weaning may reflect a situation contributing to an increased susceptibility to various enteric diseases. This information, together with previous work on the diversity of E. coli isolates from different intestinal compartments of the gastrointestinal tract of pigs (12), supports the contention that a highly diverse spectrum of commensal E. coli strains that have colonized the gut epithelial surface can provide a competitive barrier against incoming pathogenic E. coli. The deployment of a large panel of virulence genes from a wide range of E. coli pathotypes has revealed that porcine ETECs have, in the course of their evolution, also acquired extraintestinal virulence genes. Consequently, virulence gene combinations selected from both the intestinal and extraintestinal pathotypic panel can be used to characterize particular ETEC serogroups. The absence of virulence gene combinations can also be used as an epidemiological tool to characterize clonal clusters of commensal E. coli. The advantage of such an approach over serotyping is its potential to evaluate the population structure of entire E. coli communities without the need for multiple analysis of many individual and nonrepresentative clones (8).
Supplementary Material
Acknowledgments
Xi-Yang Wu is a recipient of a CRC Beef Quality Postgraduate Research Scholarship, funded through International Animal Health, Pty. Ltd., Australia.
We thank Idris Barchia (special biometrician, EMAI, Australia) for support of the statistical analyses and Alexander Kuzevski for help with the serotyping.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Barrett, T., H. Lior, J. Green, R. Khakhria, J. Wells, B. Bell, K. Greene, J. Lewis, and P. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bekal, S., R. Brousseau, L. Masson, G. Prefontaine, J. Fairbrother, and J. Harel. 2003. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J. Clin. Microbiol. 41:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertschinger, H. 1968. On the technics of serotyping of E. coli cultures from swine with coli-enterotoxemia. Pathol. Microbiol. (Basel) 32:91-97. [PubMed] [Google Scholar]
- 4.Bettelheim, K., M. Faiers, and R. Shooter. 1972. Serotypes of Escherichia coli in normal stools. Lancet ii:1223-1224. [PubMed] [Google Scholar]
- 5.Bettelheim, K., and C. Thompson. 1987. New method of serotyping Escherichia coli: implementation and verification. J. Clin. Microbiol. 25:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettelheim, K. A., A. Kuzevski, R. A. Gilbert, D. O. Krause, and C. S. McSweeney. 2005. The diversity of Escherichia coli serotypes and biotypes in cattle faeces. J. Appl. Microbiol. 98:699-709. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, J., M. Blanco, M. P. Alonso, J. E. Blanco, E. A. Gonzalez, and J. I. Garabal. 1992. Characteristics of haemolytic Escherichia coli with particular reference to production of cytotoxic necrotizing factor type 1 (CNF1). Res. Microbiol. 143:869-878. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, T., X. Y. Wu, I. Barchia, K. Bettelheim, S. Driesen, D. Trott, M. Wilson, and J. Chin. 2006. A comparison of virulence gene profile between E. coli strains isolated from healthy and diarrheic swines. Appl. Environ. Microbiol. 72:4782-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, C., W. Cho, H. Chung, T. Jung, J. Kim, and C. Chae. 2001. Prevalence of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene in isolates in weaned pigs with diarrhea and/or edema disease. Vet. Microbiol. 81:65-71. [DOI] [PubMed] [Google Scholar]
- 10.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Da Silva, A. S., and D. Da Silva Leiteqq. 2002. Investigation of putative CDT gene in Escherichia coli isolates from pigs with diarrhea. Vet. Microbiol. 89:195-199. [DOI] [PubMed] [Google Scholar]
- 12.Dixit, S. M., D. M. Gordon, X. Y. Wu, T. Chapman, K. Kailasapathy, and J. J.-C. Chin. 2004. Diversity analysis of commensal porcine Escherichia coli - associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150:1735-1740. [DOI] [PubMed] [Google Scholar]
- 13.Do, T., C. Stephens, K. Townsend, X. Wu, T. Chapman, J. Chin, B. McCormick, M. Bara, and D. J. Trott. 2005. Rapid identification of virulence genes in enterotoxigenic Escherichia coli isolates associated with diarrhoea in Queensland piggeries. Aust. Vet. J. 83:293-299. [DOI] [PubMed] [Google Scholar]
- 14.Dozois, C. M., S. Clement, C. Desautels, E. Oswald, and J. M. Fairbrother. 1997. Expression of P, S, and F1C adhesins by cytotoxic necrotizing factor 1-producing Escherichia coli from septicemic and diarrheic pigs. FEMS Microbiol. Lett. 152:307-312. [DOI] [PubMed] [Google Scholar]
- 15.Duriez, P., O. Clermont, S. Bonacorsi, E. Bingen, A. Chaventre, J. Elion, B. Picard, and E. Denamur. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671-1676. [DOI] [PubMed] [Google Scholar]
- 16.Elliott, S. J., S. Srinivas, M. J. Albert, K. Alam, R. M. Robins-Browne, S. T. Gunzburg, B. J. Mee, and B. J. Chang. 1998. Characterization of the roles of hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect. Immun. 66:2040-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elsinghorst, E., and D. Kopecko. 1992. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 60:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Fairbrother, J., S. Lariviere, and W. Johnson. 1988. Prevalence of fimbrial antigens and enterotoxins in nonclassical serogroups of Escherichia coli isolated from newborn pigs with diarrhea. Am. J. Vet. Res. 49:1325-1328. [PubMed] [Google Scholar]
- 20.Fratamico, P. M., L. K. Bagi, E. J. Bush, and B. T. Solow. 2004. Prevalence and characterization of shiga toxin-producing Escherichia coli in swine feces recovered in the National Animal Health Monitoring System's Swine 2000 Study. Appl. Environ. Microbiol. 70:7173-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich, A. W., J. Borell, M. Bielaszewska, A. Fruth, H. Tschape, and H. Karch. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85:169-182. [DOI] [PubMed] [Google Scholar]
- 23.Gunzburg, S., N. Tornieporth, and L. Riley. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampson, D. 1994. Postweaning E. coli diarrhoea in pigs, p. 171-191. In C. Gyles (ed.), Escherichia coli in domestic animals and humans. CABI Int., Walingfors, United Kingdom.
- 25.Hampson, D., J. Woodward, and I. Connaughton. 1993. Genetic analysis of Escherichia coli from porcine postweaning diarrhoea. Epidemiol. Infect. 110:575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harel, J., H. Lapointe, A. Fallara, L. Lortie, M. Bigras-Poulin, S. Lariviere, and J. Fairbrother. 1991. Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J. Clin. Microbiol. 29:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinton, M., D. Hampson, E. Hampson, and A. Linton. 1985. A comparison of the ecology of E. coli in the intestine of healthy unweaned pigs and pigs after weaning. J. Appl. Bacteriol. 58:471-478. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J., and A. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2003. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn, I., M. Katouli, A. Lund, P. Wallgren, and R. Mollby. 1993. Phenotypic diversity and stability of the intestinal coliform flora in piglets during the first 3 months of age. Microb. Ecol. Health Dis. 6:101-107. [Google Scholar]
- 31.Kuhnert, P., P. Boerlin, and J. Frey. 2000. Target genes for virulence assessment of Escherichia coli isolates from water, food and the environment. FEMS Microbiol. Rev. 24:107-117. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, J. L. 1976. Differences between enteropathogenic Escherichia coli strains isolated from neonatal E. coli diarrhoea (N.C.D.) and post weaning diarrhoea (P.W.D.) in pigs. Nord. Vet. Med. 28:417-429. [PubMed] [Google Scholar]
- 33.Lee, C., S. Hu, P. Swiatek, S. Moseley, S. Allen, and M. So. 1985. Isolation of a novel transposon which carries the Escherichia coli enterotoxin STII gene. J. Bacteriol. 162:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 35.Melin, L., M. Jensen-Waern, A. Johannisson, M. Ederoth, M. Katouli, and P. Wallgren. 1997. Development of selected faecal microfloras and of phagocytic and killing capacity of neutrophils in young pigs. Vet. Microbiol. 54:287-300. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, B., and P. Z. Fekete. 1999. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30:259-284. [PubMed] [Google Scholar]
- 37.Noamani, B. N., J. M. Fairbrother, and C. L. Gyles. 2003. Virulence genes of O149 enterotoxigenic Escherichia coli from outbreaks of postweaning diarrhea in pigs. Vet. Microbiol. 97:87-101. [DOI] [PubMed] [Google Scholar]
- 38.Osek, J. 2002. Identification of Escherichia coli O157:H7-strains from pigs with postweaning diarrhoea and amplification of their virulence marker genes by PCR. Vet. Rec. 150:689-692. [DOI] [PubMed] [Google Scholar]
- 39.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pielou, E. 1975. Ecological diversity. Wiley Interscience, New York, N.Y.
- 41.Rich, C., A. Alfidja, J. Sirot, B. Joly, and C. Forestier. 2001. Identification of human enterovirulent Escherichia coli strains by multiplex PCR. J. Clin. Lab. Anal. 15:100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, H., and S. Halls. 1968. The production of oedema disease and diarrhoea in weaned pigs by the oral administration of Escherichia coli: factors that influence the course of the experimental disease. J. Med. Microbiol. 1:45-59. [DOI] [PubMed] [Google Scholar]
- 43.Smith, H., and M. Linggood. 1971. Observations on the pathogenic properties of the K88, Hly and Ent plasmids of Escherichia coli with particular reference to porcine diarrhoea. J. Med. Microbiol. 4:467-485. [DOI] [PubMed] [Google Scholar]
- 44.Soderlind, O., B. Thafvelin, and R. Mollby. 1988. Virulence factors in Escherichia coli strains isolated from Swedish piglets with diarrhea. J. Clin. Microbiol. 26:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandemaele, F. J., S. M. Hensen, and B. M. Goddeeris. 2004. Conservation of deduced amino acid sequence of FimH among Escherichia coli of bovine, porcine and avian disease origin. Vet. Microbiol. 101:147-152. [DOI] [PubMed] [Google Scholar]
- 48.Vila, J., M. Vargas, I. Henderson, J. Gascón, and J. Nataro. 2000. Enteroaggregative Escherichia coli virulence factors in traveler's diarrhea strains. J. Infect. Dis. 182:1780-1783. [DOI] [PubMed] [Google Scholar]
- 49.Vu-Khac, H., E. Holoda, and E. Pilipcinec. 2004. Distribution of virulence genes in Escherichia coli strains isolated from diarrhoeic piglets in the Slovak Republic. J. Vet. Med. Ser. B 51:343-347. [DOI] [PubMed] [Google Scholar]
- 50.Welch, R., C. Forestier, A. Lobo, S. Pellett, W. Thomas, and G. Rowe. 1992. The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5:29-36. [DOI] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.