Abstract
Quantitative determination of IncP-1 plasmid loss from Escherichia coli cells colonizing the gastrointestinal tracts of germfree rats was achieved by flow cytometry. Results show that the plasmid's ability to conjugate counteracts plasmid loss and is thus an important mechanism for the stable maintenance of IncP-1 plasmids within the gastrointestinal environment.
Conjugative plasmids of the incompatibility group IncP-1 play an important role in bacterial adaptation in diverse environmental settings and have been studied intensively for the last few decades (1). Much information regarding their accessory elements (1), conjugational kinetics (4), regulatory networks (13), and evolution (5) has been reported, and additionally, several plasmids belonging to various subgroups have been fully sequenced (9, 11, 12). The intrinsic stability and natural existence conditions of these extrachromosomal elements, however, have attracted much less attention, and questions still remain unanswered as to whether IncP-1 plasmids may persist in natural bacterial populations or communities as genetic parasites in the absence of selective pressure (3). In the present study, we investigate the stability of the tetracycline-resistant IncP-1 plasmid pKJK5 (10) harbored by Escherichia coli cells colonizing the gastrointestinal (GI) environment of germfree rats and ask whether it may be maintained in this environment in the absence of evident plasmid-selective conditions. Furthermore, the importance of conjugative transfer as a plasmid-stabilizing mechanism will be evaluated.
Escherichia coli strain MC4100::gfp, which expresses a flow cytometry-optimized version of the green fluorescence protein gene (gfp) constitutively from the strong PA1-04/03 lac promoter, was used as a plasmid host strain. Plasmids pMIB4 and pMIB8 were constructed by insertion of the Entranceposon (Kmr lacIq1), encoding a Lac repressor protein and a kanamycin resistance determinant, into plasmid pKJK5 as previously described (2) and represent a conjugation-proficient (insertion not in an open reading frame) and a conjugative-deficient (insertion in traF) derivative of the plasmid, respectively (M. I. Bahl, L. H. Hansen, and S. J. Sørensen, submitted for publication). These plasmid derivatives apparently differ only in their ability to conjugate, and both repress the expression of gfp in the host cell, which is, however, derepressed following plasmid loss. Twelve female germfree Sprague-Dawley rats, 5 to 6 weeks old and bred at the Danish Institute for Food and Veterinary Research, were originally obtained from IFFA Credo, France. Living conditions were as previously described (6). The animals were caged individually and placed in two groups with six in each group. On day 0, all rats were inoculated by oral gavage with approximately 1.2 × 109 CFU of either E. coli MC4100::gfp/pMIB4 (group A) or E. coli MC4100::gfp/pMIB8 (group B). Fecal samples (0.5 to 1 g) excreted during the preceding 24 h were collected from the individual animal cages every 2 or 3 days. Samples were diluted in phosphate-buffered saline and plated on LB agar plates supplemented with kanamycin (50 μg/ml) or both kanamycin and tetracycline (20 μg/ml) for CFU counting or partially sedimented by centrifugation at 200 × g for 2 min to remove fecal debris. Subsequently 50 μl of the bacterial cell-containing supernatant was inoculated into 5 ml prewarmed LB broth and incubated on a rotary shaker (300 rpm) at 37°C for 2 h to allow sufficient enrichment of the bacterial populations for flow cytometry analysis. Parallel enrichments were also performed in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside, which induces the lac operon, in order to evaluate the flow cytometry detection system. A FACScalibur flow cytometer (Becton Dickenson), equipped with an argon ion laser (488-nm) capable of green fluorescent protein (GFP) excitation, was used for analysis. The voltages were set at 366 V for side scatter (SSC) and 730 V for fluorescence detectors FL1 and FL2, and the E01 setting (amplification, ×10) was used for forward scatter. A threshold of 200 was set on the SSC detector. The total population of exponentially growing plasmid sensor cells was well defined and easily distinguishable from background noise in a bivariate SSC/forward-scatter dot plot. A sort gate was established around this population, and only events within this were used for further analysis. A total of 100,000 events were acquired at every sampling. Green-fluorescing cells, i.e., plasmid-free cells, constituted a well-defined subpopulation of the plasmid sensor cells when plotted in a bivariate FL1/FL2 dot plot, and a second sort gate was established around this population for enumeration. No background events were observed within this gate during analysis of cell-free fecal material. In all cases, above 95% of the total populations achieved a GFP-positive phenotype in control samples induced with isopropyl-β-d-thiogalactopyranoside, and a correction for this small discrepancy was made in the calculations. Further information on the flow cytometry approach has been published elsewhere (2). Data analysis was performed with Cellquest software (BD Biosciences).
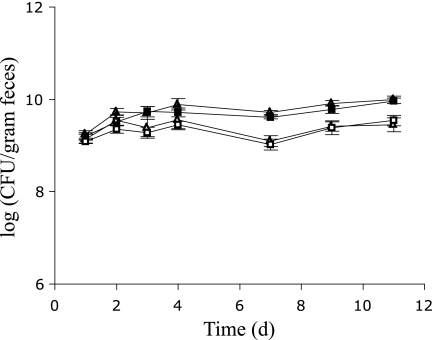
Both plasmid sensor strains colonized the GI tracts of the germfree rats equally well and reached constant CFU counts between 109 and 1010 CFU/g (wet weight) of feces on day 3 (Fig. 1). The ratio of CFU counts on plasmid-selective LB agar plates to counts on plates without tetracycline decreased during the study period. On day 9, the fractions of tetracycline-resistant CFU were between 30% and 50% in both animal groups (Fig. 1), which could give the impression of significant plasmid segregation. Replica plating of 200 colonies from non-plasmid-selective LB plates on day 9 from one animal in each group, however, revealed that all tested clones displayed a tetracycline-resistant phenotype, which is consistent with harboring the plasmid. This indicates that the large differences observed in CFU counts on plasmid-selective and nonselective plates were not caused by plasmid loss but rather were attributable to a phenotypic lag in expression of the plasmid-associated energy-dependent tetracycline efflux pump.
FIG. 1.
CFU counts determined on plasmid-selective LB agar plates containing tetracycline (open symbols) or on non-plasmid-selective LB agar plates (closed symbols) in fecal samples from rats colonized by E. coli MC4100::gfp cells harboring either the conjugation-proficient plasmid pMIB4 (triangles) or the conjugation-deficient plasmid pMIB8 (squares). Error bars indicate standard errors of the means. d, days.
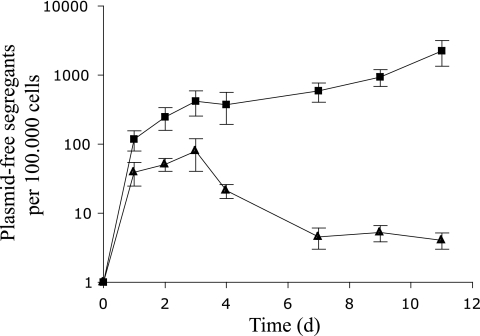
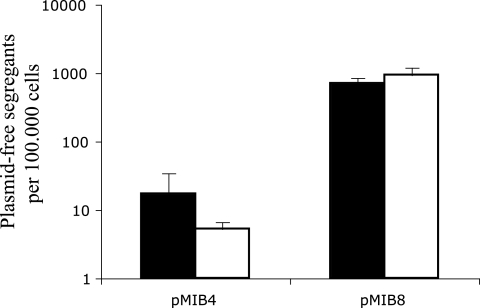
The stability of plasmids pMIB4 and pMIB8 harbored by E. coli MC4100::gfp cells in the GI tract was further assessed by a flow cytometry-based approach (Fig. 2). Prior to inoculation on day 0, both cell suspensions contained ≤0.001% GFP-positive (i.e., plasmid-free) cells. In group B (nonconjugative plasmid), the fraction of plasmid-free cells increased progressively throughout the entire period and reached 2.3% on day 11. This is consistent with low-level vegetative plasmid segregation during growth without an opportunity for reinfection by conjugation. A specific growth advantage of segregant cells could further accelerate the loss of plasmids from these populations. In contrast to this, the average fraction of plasmid-free segregant cells in group A animals (conjugative plasmid) reached a maximum value of 0.08% on day 3 and then decreased again to approximately 0.005%, which was maintained from day 7 until the end of the study period. Thus, during the colonization process, the plasmids' ability to conjugate was not sufficient to compensate for plasmid loss by vegetative segregation. This could be due to the high initial growth rate in the nutrient-rich gut environment of the germfree animals, resulting in a high rate of plasmid loss. Additionally, the density of bacterial cells within the gut environment reached a maximum level around day 3 (Fig. 1), thus providing the optimal conditions for plasmid transfer from this point in time (7). Phenotypic examination of approximately 10,000 single colonies derived from nonselective LB plates from samples retrieved on day 9 from both groups of animals was conducted under blue light (100-W mercury pressure short-arc lamp with a 450- to 490-nm filter). Thirty entirely GFP-positive colonies were all tetracycline sensitive, and the ratios of GFP-positive colonies were concordant with those obtained by flow cytometry analysis (Fig. 3). The apparent ability of the conjugative plasmid derivative to reinfect plasmid-free segregant cells in the GI tracts of germfree rats is consistent with the plasmid behaving as a genetic parasite in this environment. Our results highlight the importance of investigating the existence conditions of plasmids in natural environmental habitats and further demonstrate that flow cytometry is a powerful and extremely sensitive technique for such studies. It remains to be investigated whether IncP-1 plasmids could also be stably maintained in the intestinal tracts of conventional animals in the absence of selective pressure. The typically very broad host range of this plasmid group (8) could provide the necessary fluidity to allow them to occupy many different bacterial taxa, which in turn may be sufficient for a parasitic lifestyle.
FIG. 2.
The ratios of GFP-positive, i.e., plasmid-free cells to the total number of cells in fecal samples from rats colonized by E. coli MC4100::gfp cells harboring either the conjugation-proficient plasmid pMIB4 (triangles) or the conjugation-deficient plasmid pMIB8 (squares). A total of 100,000 cells were analyzed by flow cytometry for each sample. Error bars indicate standard errors of the means.
FIG. 3.
Comparison between ratios of GFP-positive, i.e., plasmid-free cells in fecal samples on day 9 as determined by phenotypic discrimination of single colonies (black bars) or flow cytometry analysis (white bars) in populations of E. coli MC4100::gfp cells harboring either the conjugation-proficient plasmid pMIB4 or the conjugation-deficient plasmid pMIB8. Error bars indicate standard errors of the means.
Acknowledgments
This work was partly funded by a grant from the Danish Natural Science Research Council, ref. 272-05-0325.
We thank Anne Ørngreen and her department for handling of the animals and Karin Vestberg for technical assistance.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Adamczyk, M., and G. Jagura-Burdzy. 2003. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50:425-453. [PubMed] [Google Scholar]
- 2.Bahl, M. I., S. J. Sørensen, and L. H. Hansen. 2004. Quantification of plasmid loss in Escherichia coli cells by use of flow cytometry. FEMS Microbiol. Lett. 232:45-49. [DOI] [PubMed] [Google Scholar]
- 3.Bergstrom, C. T., M. Lipsitch, and B. R. Levin. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155:1505-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Gelder, L., F. P. Vandecasteele, C. J. Brown, L. J. Forney, and E. M. Top. 2005. Plasmid donor affects host range of promiscuous IncP-1beta plasmid pB10 in an activated-sludge microbial community. Appl. Environ. Microbiol. 71:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis, J. J. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16:291-298. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsen, B. L., M. Skou, A. M. Hammerum, and L. B. Jensen. 1999. Horizontal transfer of the satA gene encoding streptogramin A resistance between isogenic Enterococcus faecium strains in the gastrointestinal tract of gnotobiotic rats. Microb. Ecol. Health Dis. 11:241-247. [Google Scholar]
- 7.Levin, B. R., F. M. Stewart, and V. A. Rice. 1979. The kinetics of conjugative plasmid transmission: fit of a simple mass action model. Plasmid 2:247-260. [DOI] [PubMed] [Google Scholar]
- 8.Musovic, S., G. Oregaard, N. Kroer, and S. J. Sørensen. 2006. Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among gram-positive and gram-negative bacteria indigenous to the barley rhizosphere. Appl. Environ. Microbiol. 72:6687-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 10.Sengeløv, G., K. J. Kristensen, A. H. Sørensen, N. Kroer, and S. J. Sørensen. 2001. Effect of genomic location on horizontal transfer of a recombinant gene cassette between Pseudomonas strains in the rhizosphere and spermosphere of barley seedlings. Curr. Microbiol. 42:160-167. [DOI] [PubMed] [Google Scholar]
- 11.Thorsted, P. B., D. P. Macartney, P. Akhtar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPbeta plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 12.Vedler, E., M. Vahter, and A. Heinaru. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zatyka, M., G. Jagura-Burdzy, and C. M. Thomas. 1997. Transcriptional and translational control of the genes for the mating pair formation apparatus of promiscuous IncP plasmids. J. Bacteriol. 179:7201-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]