Abstract
A study was conducted to understand the descriptive and molecular epidemiology of antimicrobial-resistant gram-negative enteric bacteria in the feces of healthy lactating dairy cattle. Gram-negative enteric bacteria resistant to ampicillin, florfenicol, spectinomycin, and tetracycline were isolated from the feces of 35, 8, 5, and 42% of 213 lactating cattle on 74, 39, 9, 26, and 82% of 23 farms surveyed, respectively. Antimicrobial-resistant gram-negative bacteria accounted for 5 (florfenicol) to 14% (tetracycline) of total gram-negative enteric microflora. Nine bacterial species were isolated, of which Escherichia coli (87%) was the most predominant species. MICs showing reduced susceptibility to ampicillin, ceftiofur, chloramphenicol, florfenicol, spectinomycin, streptomycin, and tetracycline were observed in E. coli isolates. Isolates exhibited resistance to ampicillin (48%), ceftiofur (11%), chloramphenicol (20%), florfenicol (78%), spectinomycin (18%), and tetracycline (93%). Multidrug resistance (≥3 to 6 antimicrobials) was seen in 40% of E. coli isolates from healthy lactating cattle. Of 113 tetracycline-resistant E. coli isolates, tet(B) was the predominant resistance determinant and was detected in 93% of isolates, while the remaining 7% isolates carried the tet(A) determinant. DNA-DNA hybridization assays revealed that tet determinants were located on the chromosome. Pulsed-field gel electrophoresis revealed that tetracycline-resistant E. coli isolates (n = 99 isolates) belonged to 60 subtypes, which is suggestive of a highly diverse population of tetracycline-resistant organisms. On most occasions, E. coli subtypes, although shared between cows within the herd, were confined mostly to a dairy herd. The findings of this study suggest that commensal enteric E. coli from healthy lactating cattle can be an important reservoir for tetracycline and perhaps other antimicrobial resistance determinants.
Widespread reliance on antimicrobials in food animal production has resulted in a considerable rise of antimicrobial-resistant strains of bacteria, complicating the treatment of infectious diseases in livestock, companion animals, and humans. This has led to important changes in the perceptions and priorities of federal agencies with regard to antimicrobial usage, in particular the use of antimicrobials as growth promoters and prophylactic agents (2, 37, 45).
Sawant et al. (47) conducted a study on antimicrobial usage in Pennsylvania dairy herds. They observed that antimicrobials were used frequently for treating enteritis (36%) and pneumonia (25%) in calves and foot infections in adult cattle (16%). Twenty-four antimicrobials, including beta-lactams, spectinomycin, florfenicol, and tetracyclines, were used on these farms. Beta-lactam antimicrobials were used mostly for dry cow therapy, for clinical mastitis, and, on some farms, for pneumonia and metritis. On 18% of the dairy herds surveyed, ceftiofur was used in an extra-label manner to treat mastitis in lactating cattle. On 70% of farms, calves were fed medicated milk replacers containing oxytetracycline and neomycin. The results of the study by Sawant et al. suggested that antimicrobials were used extensively on dairy herds for both therapeutic and prophylactic purposes. Beta-lactams and oxytetracyclines were the most widely used antimicrobials.
The selective pressure from the use of antimicrobial agents at subtherapeutic levels in dairy cattle could result in the selection of those strains that contain genes for antimicrobial resistance (37). Most of the antimicrobial susceptibility studies reported in the literature have examined pathogenic bacteria, such as Salmonella and enterotoxigenic Escherichia coli, or bacteria isolated from clinical cases (42, 54). Diagnostic laboratories focus primarily on the detection and characterization of specific pathogenic bacteria. The use of data compiled from routine diagnostic workups may prove to be insufficient to measure the relevance of antimicrobial-resistant normal gut flora of animals or humans (37). Recent studies have shown that commensal bacteria of humans and animals could serve as good indicators of antimicrobial selective pressure and reveal the potential for antimicrobial resistance emerging in enteric pathogens (29). The French Institute for Public Health Surveillance has recommended that studying antimicrobial resistance in commensal bacteria from healthy animals would be extremely valuable, as these organisms could serve as a reservoir for genes that encode antimicrobial resistance and, given the right conditions, could transfer these resistance genes to pathogenic bacteria (15).
The tetracyclines continue to be one of the most widely used antibiotics in human medicine and animal agriculture, as they are relatively inexpensive, can be administered orally, and have relatively few side effects (10). The growth-promoting properties of oxytetracyclines were first reported by Stockstad et al. (53), when young chicks fed with chlortetracycline showed improvement in growth rate. Following this report, chlortetracycline and oxytetracycline were extensively used as animal growth promoters in swine and cattle (16).
Resistance to tetracyclines was first detected in the 1950s and became more apparent by the 1970s when it was widely reported among Enterobacteriaceae, staphylococci, streptococci, and Bacteroides spp. (30). Tetracycline resistance determinants are now widely spread among bacterial species and have been identified in as many as 39 gram-negative and 22 gram-positive bacteria (10). In most gram-negative species, tetracycline resistance is due to the acquisition of an operon which consists of an efflux gene tet(A) and a repressor gene tet(R) that are divergently transcribed from overlapping operator regions. Nine types of efflux genes consisting of the above operon have been described so far for gram-negative bacteria [tet(A) to tet(E) and tet(G) to tet(J)] (48).
This study was conducted to determine (i) the prevalence and distribution of antimicrobial-resistant gram-negative enteric bacteria (GN-EB) from healthy lactating cattle, (ii) the MICs of commonly used antimicrobials against E. coli isolated from the feces of lactating dairy cattle, and (iii) genotypic characteristics of tetracycline-resistant E. coli. It is anticipated that the findings of this study will provide a comprehensive understanding of antimicrobial resistance in commensal bacteria isolated from the feces of healthy lactating cattle.
MATERIALS AND METHODS
Dairy cattle.
A total of 23 herds from central and south-central Pennsylvania (four counties) were surveyed for antimicrobial-resistant GN-EB in the feces of healthy lactating cattle. These herds had participated earlier in a study on food-borne pathogens in bulk tank milk (20) and antimicrobial usage (47). Dairy producers kept records on the type of antimicrobials that were used and the purpose for which they were administered. However, information on the dose, frequency of use, animals that received antimicrobials, and whether antimicrobial treatment was completed was not available for all of the farms. Therefore, no attempt was made to correlate the use of an antimicrobial with the occurrence of antimicrobial-resistant bacteria. All dairy herds used ampicillin, florfenicol, spectinomycin, and tetracycline. Fecal samples from 213 lactating cattle (n = 23 herds) were screened for antimicrobial resistance. From each herd, the number of lactating cattle that was sampled ranged from 4 to 12 animals (approximately 10% of the lactating cattle present in the milking herd). Fecal samples were randomly collected from lactating cattle that exited the milking parlor or those restrained in a tie stall for bacteriological analysis. Feces were collected from cows per rectal using a sterile disposable rectal sleeve. About 10 g of feces was transferred to a sterile 50-ml screw-cap centrifuge tube. The tube was transported to the laboratory on ice and processed the same day.
Isolation and enumeration of GN-EB and antimicrobial-resistant GN-EB.
The fecal samples were thoroughly mixed using a sterile spatula, and 1 g of feces was transferred to a 15-ml centrifuge tube containing 9 ml of sterile normal saline solution. The contents were mixed thoroughly and serially diluted 10-fold. MacConkey agar (MAC) was used for selective growth of GN-EB. From the 10−3 and 10−4 dilutions, 0.1 ml was plated on a MAC control plate without antimicrobials, while from the 10−1 and 10−2 dilutions, 0.1 ml of the sample was plated on a MAC plate containing one of the following antimicrobials: ampicillin (64 μg/ml), tetracycline (32 μg/ml), spectinomycin (256 μg/ml) (ICN Biomedicals, Aurora, OH), enrofloxacin (8 μg/ml) (Sigma Aldrich, St. Louis, MO), or florfenicol (16 μg/ml) (Sigma Aldrich, St. Louis, MO). The concentration of antimicrobials used in MAC media was one “twofold-higher concentration” than the MIC breakpoint values suggested by the CLSI (formerly NCCLS) (39) for bacteria isolated from animals. The inoculated plates were incubated at 37°C for 24 h. The numbers of colonies on MAC with antimicrobials and without antimicrobials were counted and expressed as antimicrobial-resistant GN-EB CFU/g of feces and total GN-EB CFU/g of feces, respectively.
Species identification of antimicrobial-resistant GN-EB.
A total of 258 isolates (n = 23 farms; 213 lactating cattle) from MAC with antimicrobials were identified to the species level. Based on colony morphology and lactose fermentation, two to three colonies were selected from each plate for species identification. Colonies were tested by Gram reaction, oxidase testing, and indole-methyl red-Voges-Proskauer-citrate battery testing as described by Harley and Prescott (18). The isolates were then identified to the species level using an API 20E/NE identification kit (bioMérieux, Hazelwood, MO) as described by the manufacturer.
Antimicrobial susceptibility assays.
All E. coli isolates (n = 223) out of 258 GN-EB were examined for susceptibility to antimicrobials by broth microdilutions as described by the CLSI (39). The isolates were screened, and the results were interpreted according to criteria established by the CLSI (39) for bacteria isolated from animals. MICs of E. coli for ampicillin, chloramphenicol, spectinomycin, streptomycin, tetracycline (MP Biomedicals, Aurora, OH), ceftiofur, and florfenicol (Sigma Aldrich, St. Louis, MO) were determined as described by the CLSI (39). The MIC for each isolate was read as the lowest dilution demonstrating no visible growth. Based on CLSI recommendations (39), Staphylococcus aureus ATCC 29213, E. coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Enterococcus faecalis ATCC 29212 were used as quality control strains for MIC assays. The MIC50 and MIC90 values were calculated and expressed as the percentage of isolates at each of the respective MICs.
Tetracycline resistance determinants.
Tetracycline-resistant enteric E. coli isolates (n = 113) from lactating dairy cattle were screened for tetracycline resistance genes tet(A), tet(B), tet(C), tet(D), tet(E), and tet(G) as described by Ng et al. (40). The PCR assays were optimized (template and primer concentration) for PuReTaq Ready-To-Go PCR beads (GE Healthcare, Piscataway, NJ).
Pulsed-field gel electrophoresis.
Tetracycline-resistant E. coli isolates (n = 113 isolates from 18 herds) were subtyped using the pulsed-field gel electrophoresis (PFGE) protocol described by Hegde et al. (19). The PFGE subtypes for 14 isolates could not be determined due to lack of clear discernible DNA banding patterns. The 14 isolates were all subjected to PFGE analysis on three separate occasions; all of the analyses failed to result in clear DNA banding patterns. The PFGE patterns were first compared visually and analyzed using Gel Doc 2000 Molecular Analyst Fingerprinting Plus, version 6.1, software (Bio-Rad, Hercules, CA). Salmonella enterica serovar Newport was used as a reference strain for PFGE analysis.
Southern blot hybridization assays.
Small- and large-sized plasmids were obtained using an alkaline lysis protocol with sodium dodecyl sulfate as described by Sambrook and Russell (46). Small-sized plasmids (≤12 kb) were electrophoresed on a 0.8% agarose gel, while plasmids of large sizes that may be difficult to obtain on regular gel electrophoresis were resolved by PFGE using a 1% agarose gel. Genomic DNA-PFGE was performed using the 1-day PFGE protocol developed by Hegde et al. (19). The bacterial plugs were digested with XbaI and run on the CHEF Mapper system (Bio-Rad). The running conditions for large plasmids and genomic DNA were as follows: initial switch time, 2.16 s; final switch time, 35.07 s; run time, 14 h; angle, 120°, gradient, 6.0 V/cm; temperature, 14°C (ramping factor was kept linear). Gels were blotted with a vacuum blotter according to the manufacturer's instructions (Bio-Rad Laboratories, CA). A positively charged Hybond N+ nylon membrane (GE Healthcare) was used for blotting. Small plasmids were blotted for 90 min, while large plasmids and genomic DNA were first depurinated (0.25 N HCl for 15 min) and then blotted for 180 min.
PCR-amplified products tet(A) and tet(B) were purified and used as probes for the Southern blotting assay. A digoxigenin (DIG) DNA-labeling and detection kit (Roche Molecular Biochemicals, Mannheim, Germany) was used to randomly label DNA probes. Briefly, tet(A) and tet(B) amplified products were denatured by heating in a boiling water bath for 10 min and immediately chilled on ice. Random hexanucleotides (10×, 2 μl) were added to hybridize to the denatured template. Klenow enzyme (1 μl) was added to create a complementary strand to the template, using the hybridized hexonucleotides as primers and a mix of deoxynucleoside triphosphates (2 μl) containing DIG-labeled dUTP for elongation. The reaction mixture was incubated overnight at 37°C and kept at −20°C until further use. The DIG-labeled probes were used to detect blotted nucleic acids by standard Southern blot methods described by the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). The hybridized probes were visually detected with anti-digoxigenin-alkaline phosphatase Fab fragments after the addition of the colorimetric substrate Nitro Blue Tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate).
RESULTS
Prevalence of antimicrobial-resistant GN-EB.
Ampicillin-resistant GN-EB were isolated from 72 of 211 (34%) of cows on 17 (74%) dairy herds. Tetracycline-resistant GN-EB were isolated from 89 of 212 (42%) cows on 19 (82%) dairy herds. Florfenicol- and spectinomycin-resistant GN-EB were also detected in the feces of healthy cattle; however, their prevalence was lower than that observed for tetracycline and ampicillin. Enrofloxacin-resistant GN-EB were not detected (Table 1). Due to laboratory errors related to the enumeration of bacterial count or the contamination of selective media, ampicillin and tetracycline counts could not be determined for two and one fecal sample, respectively. The number of total GN-EB (on MacConkey agar without antimicrobial agent) ranged from 1.2 × 106 to 6.9 × 106 CFU/g of feces. The mean CFU/g of ampicillin-resistant GN-EB (3.1 × 105 CFU/g) and tetracycline-resistant GN-EB (3.9 × 105 CFU/g) were 1 log lower that of the total GN-EB. On average, the ampicillin- and tetracycline-resistant bacteria accounted for 9 and 14% of total GN-EB, respectively. Florfenicol- and spectinomycin-resistant GN-EB were 3 and 2 log lower than total GN-EB, respectively (Table 1).
TABLE 1.
Prevalence and distribution of antimicrobial-resistant and total GN-EB in feces of healthy lactating dairy cattle from 23 farms
| Antimicrobial | Total no. of cows screened | Distribution of antimicrobial-resistant GN-EB
|
Prevalence of antimicrobial-resistant GN-EB
|
Prevalence of total GN-EB
|
% of resistant bacteria (% of total GN-EB)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| No. (%) of farms | No. (%) of cattle | Range (CFU/g of feces) | Mean (CFU/g of feces) | Range (CFU/g of feces) | Mean (CFU/g of feces) | Range | Mean | ||
| Ampicillin | 211 | 17 (74) | 72 (34) | 4 × 101-1.9 × 107 | 3.1 × 105 | 4.9 × 103-5.5 × 107 | 3.8 × 106 | 0.005-96 | 9 |
| Enrofloxacin | 213 | 0 (0.0) | 0 (0.0) | 40-8.5 × 107 | 3.3 × 106 | ||||
| Florfenicol | 213 | 9 (39) | 18 (8) | 1 × 102-1.2 × 104 | 2.9 × 102 | 1 × 104-1.1 × 107 | 1.2 × 106 | 0.004-63 | 5 |
| Spectinomycin | 213 | 6 (26) | 10 (5) | 1 × 102-3.2 × 105 | 6.2 × 104 | 3 × 104-1.1 × 107 | 1.9 × 106 | 0.004-89 | 10 |
| Tetracycline | 212 | 19 (82) | 89 (42) | 4 × 101-6.7 × 106 | 3.9 × 105 | 6 ×102-8.5 × 107 | 6.9 × 106 | 0.01-100 | 14 |
Antimicrobial-resistant GN-EB species.
A total of 258 antimicrobial-resistant GN-EB isolates from 213 cows on 23 dairy herds were identified to species (Table 2). The antimicrobial-resistant GN-EB (n = 258) belonged to 13 gram-negative bacterial species, including Citrobacter koseri, Enterobacter aerogenes, Escherichia coli, Morganella morganii, Klebsiella oxytoca, Klebsiella pneumoniae, Kluyvera spp., Providencia alcaligenes, Providencia stuartii, Pasteurella spp., and Pseudomonas spp. Escherichia coli (n = 223) was isolated from four MAC plates, each plate supplemented with ampicillin, tetracycline, florfenicol, and spectinomycin. A total of eight and five bacterial species were isolated from MAC supplemented with ampicillin and spectinomycin, respectively (Table 2). All of the isolates from MAC supplemented with tetracycline (32 μg/ml) were identified as E. coli. Two isolates from MAC supplemented with florfenicol (16 μg/ml) were identified as Pasteurella spp.
TABLE 2.
Antimicrobial-resistant GN-EB species isolated from lactating cattle
| Species | No. of bacteria isolated on MacConkey agar supplemented with:
|
|||
|---|---|---|---|---|
| Ampicillin (64 μg/ml) | Florfenicol (16 μg/ml) | Spectinomycin (256 μg/ml) | Tetracycline (32 μg/ml) | |
| Citrobacter koseri | 14 | |||
| Enterobacter aerogenes | 4 | |||
| Escherichia coli | 75 | 22 | 13 | 113 |
| Klebsiella oxytoca | 3 | |||
| Klebsiella pneumoniae | 1 | |||
| Kluyvera spp. | 1 | |||
| Morganella morganii | 1 | |||
| Pasteurella spp. | 2 | |||
| Providencia alcaligenes | 1 | |||
| Providencia stuartii | 1 | |||
| Pseudomonas aeruginosa | 1 | |||
| Pseudomonas fluorescens | 5 | |||
| Pseudomonas spp. | 1 | |||
| All isolates | 104 | 24 | 17 | 113 |
MICs for E. coli isolates.
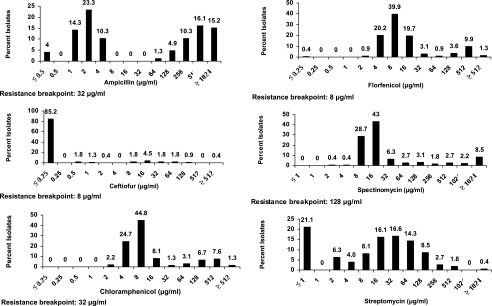
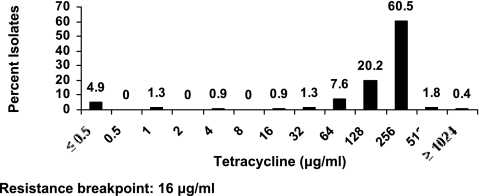
Interpretation of the MICs based on CLSI criteria (39) for E. coli isolates would result in 48, 11, 78, 20, 18, and 93% resistance to ampicillin, ceftiofur, florfenicol, chloramphenicol, spectinomycin, and tetracycline, respectively (Fig. 1 and Table 3). The MICs for E. coli showed a distinct bimodal distribution for ampicillin (MIC50, 4 μg/ml; MIC90, 1,024 μg/ml). Nearly 85% isolates showed an MIC of ≤0.25 μg/ml for ceftiofur, while 11% of the isolates exhibited MICs above the CLSI's (39) recommended resistance breakpoint for ceftiofur (Fig. 1 and Table 3). The MIC50 for both florfenicol and chloramphenicol was 8 μg/ml. The MIC90 values for florfenicol and chloramphenicol were 256 and 128 μg/ml, respectively (Table 3). Based on the CLSI's (39) recommended breakpoint values, 78 and 20% of the isolates were found to be resistant to florfenicol and chloramphenicol, respectively. The most frequently observed MICs for spectinomycin were 8 μg/ml (28.7%) and 16 μg/ml (43%). No clear trend was observed for susceptibility towards streptomycin. Ninety-three percent of isolates exhibited elevated MICs (≥16 μg/ml) for tetracycline. Both the MIC50 and MIC90 values for tetracycline were 256 μg/ml as 90% of the isolates were inhibited (Table 3).
FIG. 1.
MICs of 223 E. coli isolates for seven antimicrobials. The resistance breakpoints are based on CLSI interpretative criteria (39).
TABLE 3.
Summary of MICs for 223 E. coli isolates to antimicrobials
| Antimicrobial | MIC (μg/ml)
|
% Resistanta | |||
|---|---|---|---|---|---|
| Minimum | 50% | 90% | Maximum | ||
| Ampicillin | ≤0.5 | 4 | ≥1,024 | ≥1,024 | 48 |
| Ceftiofur | ≤0.25 | ≤0.25 | 8 | ≥512 | 11 |
| Chloramphenicol | 2 | 8 | 128 | ≥512 | 20 |
| Florfenicol | ≤0.25 | 8 | 256 | ≥512 | 78 |
| Spectinomycin | 2 | 16 | 1,024 | ≥2,048 | 18 |
| Streptomycin | ≤1 | 16 | 128 | ≥2,048 | |
| Tetracycline | ≤0.5 | 256 | 256 | ≥1,024 | 93 |
Based on CLSI interpretive criteria established for bacteria isolated from animals (39).
Resistance patterns of E. coli isolates.
The 223 E. coli isolates belonged to 21 antimicrobial resistance profiles (Table 4). Resistance to only tetracycline was observed in 8.97% of isolates, whereas only four isolates were ampicillin resistant. Florfenicol and tetracycline (35.87%) resistance was the most common pattern of resistance. Multidrug resistance (≥3 to 6 antimicrobials) was observed in 90 (40.4%) of E. coli isolates. The major multidrug resistance profile was ampicillin-florfenicol-tetracycline, which was observed in 13.9% of isolates. A significant number of isolates (8.07%) exhibited resistance to all of the six antimicrobials used for screening (Table 4).
TABLE 4.
Resistance patterns of E. coli isolates (n = 223) on the basis of MIC breakpointsa
| Resistance profile | No. of isolates with profile | % of isolates with profile |
|---|---|---|
| TET | 20 | 8.97 |
| AMP | 4 | 1.79 |
| FLO-TET | 80 | 35.87 |
| CHL-FLO | 3 | 1.35 |
| AMP-TET | 17 | 7.62 |
| AMP-SPT | 1 | 0.45 |
| AMP-FLO | 8 | 3.59 |
| AMP-FLO-TET | 31 | 13.90 |
| FLO-SPT-TET | 1 | 0.45 |
| CHL-FLO-TET | 9 | 4.04 |
| AMP-SPT-TET | 4 | 1.79 |
| AMP-CHL-TET | 2 | 0.90 |
| CHL-FLO-SPT-TET | 3 | 1.35 |
| AMP-SPT-TET-XNL | 1 | 0.45 |
| AMP-FLO-TET-XNL | 1 | 0.45 |
| AMP-FLO-SPT-TET | 9 | 4.04 |
| AMP-CHL-FLO-TET | 3 | 1.35 |
| AMP-CHL-FLO-TET-XNL | 4 | 1.79 |
| AMP-CHL-FLO-SPT-TET | 3 | 1.35 |
| AMP-FLO-SPT-TET-XNL | 1 | 0.45 |
| AMP-CHL-FLO-SPT-TET-XNL | 18 | 8.07 |
AMP, ampicillin; CHL, chloramphenicol; FLO, florfenicol; SPT, spectinomycin; TET, tetracycline; XNL, ceftiofur.
Tetracycline-resistant E. coli PFGE subtypes.
Of the 113 tetracycline-resistant E. coli isolates, the PFGE patterns of 99 isolates are presented in these data. Repeated attempts failed to obtain discrete PFGE patterns from 14 isolates. The tetracycline-resistant E. coli isolates (n = 99) belonged to 60 PFGE subtypes. The E. coli subtypes were frequently detected among cows within a farm but were rarely observed on more than one farm. The number of tetracycline-resistant E. coli PFGE subtypes within a dairy herd ranged from one to eight subtypes. Four subtypes were observed on at least one farm.
Characterization of tetracycline resistance determinants.
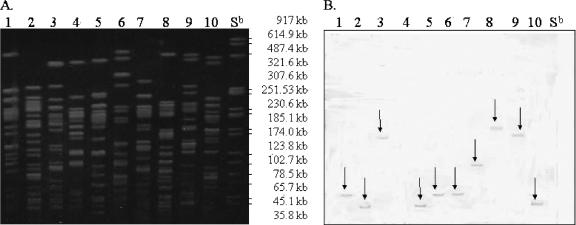
A total of 113 tetracycline-resistant E. coli isolates were analyzed for tet genes by PCR analysis. It was observed that 105 (93%) and 8 (7%) isolates encoded tet(B) and tet(A), respectively. Southern blot hybridizations of small- and large-sized plasmids and genomic DNA from 30 randomly selected isolates with tet(B) and all 8 isolates with tet(A) were probed with their respective gene probes. No hybridization was seen with either plasmid size. Hybridization of tet probes with genomic DNA was detected (Fig. 2). Only one fragment per genomic DNA PFGE lane exhibited a color reaction, indicating that a single copy of tet(B) was present in the genome (Fig. 2). The same was observed with tet(A) probe (data not shown). The result showed that the efflux genes were located on the genomic DNA.
FIG. 2.
Southern hybridization of genomic DNA with tet(B) probe. (A) Lanes 1 to 10 represent PFGE genomic DNA fragments. (B) Blot of PFGE genomic DNA probed with tet(B) probe. The Salmonella serovar Newport (Sb) PFGE reference strain (XbaI-digested genomic DNA fragments) is tetracycline sensitive and served as a negative control.
DISCUSSION
The study of the prevalence of antimicrobial resistance in commensal microflora can be very useful in monitoring and understanding the process of antimicrobial-mediated selection in individual hosts as well as in the general population (29). MacConkey media supplemented with antimicrobial agents can be effectively used for the isolation of antibiotic-resistant coliforms (25). The isolation of the flo gene in gram-negative bacteria with an MIC of ≥16 μg/ml for florfenicol has been more successful than in bacteria with an MIC of 8 μg/ml (breakpoint value recommended by the CLSI [39] for florfenicol) (54). We believed that by using concentrations of antimicrobials higher than those recommended by the CLSI, antibiotic-resistant enteric bacteria could be effectively isolated with fewer false-positive results.
The high prevalence of tetracycline-resistant (82% of farms, 42% of cows, and 14% of total GN-EB) and ampicillin-resistant (74% of farms, 35% of cows, and 9% of total GN-EB) GN-EB observed in our study suggests that lactating cattle can be a significant reservoir of tetracycline- and ampicillin-resistant GN-EB. Although the prevalences of florfenicol-resistant (39% of farms, 8% of cows, and 5% of total GN-EB) and spectinomycin-resistant (26% of farms, 5% of cows, and 10% of total GN-EB) GN-EB were lower than those of ampicillin- and tetracycline-resistant GN-EB, their role as reservoirs of antimicrobial resistance determinants should not be overlooked.
In an earlier study, Sawant et al. (47) reported that beta-lactams and tetracycline were the most widely used antimicrobials on these dairy herds. They concluded that the absence of antimicrobial treatment records, the lack of written plans for treating sick animals, the failure to consult a veterinarian for treating sick animals, and the failure to complete an antimicrobial treatment course are factors that could lead to the inappropriate use of antimicrobials and the emergence of antimicrobial-resistant bacteria. The variation in the number of farms and cows that shed antimicrobial-resistant GN-EB could be influenced by conditions such as (i) transition of animals from one environment to another, (ii) change in nutrition, (iii) severe weather conditions, and (iv) interaction with other animals in the herd (11, 23, 33, 38).
Lau and Ingham (26) showed that enteric E. coli in soil mixed with bovine manure could survive for at least 19 weeks at 9 to 21°C. Mobile genetic elements, such as plasmids, transposons, and integrons, have the ability to maintain and spread antimicrobial-resistant determinants (35). Aminov et al. (1) reported that antimicrobial-resistant GN-EB could serve as an antimicrobial resistance gene pool and facilitate the exchange of antimicrobial genetic determinants with other species in the environment.
Escherichia coli is the predominant bacterial species in the feces of dairy cattle (41). In our study, E. coli (86.4%) was the predominant species. Antimicrobial-resistant E. coli, Citrobacter, Enterobacter, Klebsiella, and Pseudomonas spp. are considered opportunistic pathogens, which can cause mastitis, enteritis, pneumonia, and urinary tract infections in dairy cattle (3, 12, 56). Recent studies have shown that Salmonella and E. coli isolates from humans and animals had the same antimicrobial resistance determinants (13, 31).
Interpretive criteria for E. coli isolated from cattle have not been established. In addition, human breakpoints have not been validated for the interpretation of antimicrobial resistance of veterinary isolates (44). The CLSI (39) and Schroeder et al. (49) advise caution in extrapolating the predicted response of a specific animal species to a particular antimicrobial drug, as it may lead to incorrect prediction of the clinical outcome. The caution for extension of interpretation of our data is less likely to be an issue, as the focus of this study was on commensal bacteria from healthy lactating cattle. Nearly 50% of E. coli isolates exhibited elevated MICs of ampicillin above the CLSI's (39) recommended resistant breakpoint. The MIC90 for ampicillin was as high as ≥1,024 μg/ml. Orden et al. (42) reported a similar observation with E. coli isolates from calves with diarrhea. Escherichia coli isolates with elevated MICs of ampicillin have been isolated from the feces of healthy lactating cattle and other food-producing animals (22, 44).
The MICs observed for florfenicol- and chloramphenicol-resistant E. coli in our study followed a bimodal distribution. Singer et al. (51) reported similar MIC patterns for both antimicrobials in fecal E. coli isolates from dairy cattle. They also observed that E. coli isolates with an MIC of ≥32 μg/ml harbored the flo gene responsible for florfenicol resistance and that isolates without the flo gene had an MIC of ≤8 μg/ml. The findings of this study also concur with the observations made by White et al. (54). They reported that the appearance of E. coli isolates with MICs of ≥128 μg/ml was a recent phenomenon. They also observed that E. coli isolates with elevated MICs for florfenicol (≥16 μg/ml) also exhibited high MICs for chloramphenicol (54).
Of the 43 isolates that showed an MIC of ≥32 μg/ml for florfenicol, an overwhelming majority (n = 37; 86%) exhibited MICs at or beyond the resistance breakpoint (≥32 μg/ml) defined by the CLSI (39) for chloramphenicol. Though chloramphenicol has been banned from use in food animals in the United States since the 1980s (14), florfenicol, a structural analog of chloramphenicol, approved by the Food and Drug Administration in 1996, is used for treating bovine respiratory pathogens. Resistance to florfenicol in E. coli attributed to the presence of flo is also capable of conferring cross-resistance to chloramphenicol (54).
In this study, E. coli isolates exhibited very high MICs (MIC90, 1,024 μg/ml) for spectinomycin. Orden et al. (42) also observed that E. coli isolates from dairy calves with diarrhea had very high MICs for spectinomycin. Kijima-Tanaka et al. (24) screened E. coli isolates from Japanese cattle for resistance to various antimicrobials, including streptomycin.
Researchers categorized 20.8% (>50 μg/ml) of isolates as resistant. At this breakpoint, as well as the resistance breakpoint for streptomycin (64 μg/ml) recommended by the U.S. Food and Drug Administration, the U.S. Department of Agriculture, and the Centers for Disease Control and Prevention standard, 27.7% of E. coli isolates from our study were classified as resistant to streptomycin. Bywater et al. (8) observed a similar MIC90 value (128 μg/ml) for E. coli isolated from cattle in Denmark and Italy. Escherichia coli isolates collected by this group from France, Germany, and United Kingdom exhibited a much lower MIC90 value (8 μg/ml) for the same antimicrobials (8).
Blake et al. (5) observed that E. coli isolates that carried the tet genes exhibited high MICs. They observed that E. coli isolates from animal feces devoid of tetracycline resistance genes exhibited an average MIC of 1.25 μg/ml, while E. coli isolates with tet genes had MICs of ≥132 μg/ml. Bryan et al. (7) observed an MIC of ≥93 μg/ml in E. coli isolates from humans and animals associated with one or more tetracycline resistance genes.
The majority of the isolates (85.3%) we screened were highly susceptible to ceftiofur. Choi et al. (9) and Hariharan et al. (17) made similar observations for pathogenic E. coli isolates from swine and calves. Escherichia coli isolates that grew at ≥8 μg/ml have been isolated from retail meat by Zhao et al. (57) and Schroeder et al. (49). Three E. coli isolates had MICs of ≥128 μg/ml for ceftiofur in our study. Similar MICs have been reported for E. coli isolates from calf scours, feces of cattle, and carcass swabs (6, 50). Resistance to ceftiofur, ampicillin, and the beta-lactamase inhibitor clavulanic acid in E. coli is associated primarily with the AmpC class of cephamycinases (blaCMY) (57). The multidrug resistance of ceftiofur-resistant E. coli isolates to other unrelated antimicrobials such as tetracycline and chloramphenicol observed in our study has previously been documented in Salmonella and E. coli isolated from animals (55, 54).
Blake et al. (5) observed that porcine E. coli isolates that are resistant to tetracycline also exhibited resistance to ampicillin and streptomycin, often in combination with chloramphenicol. Multidrug resistance patterns in our study included resistance to the above-mentioned antimicrobials. White et al. (54) reported similar multidrug resistance in E. coli isolates from bovine diarrhea that were selected for chloramphenicol and florfenicol resistance.
We used PFGE to study tetracycline-resistant E. coli isolates (n = 99) and found 60 subtypes. Other studies have reported similar levels of high diversity. Subtypes of E. coli isolated from sewage, gulls, and dairy cattle exhibited extreme diversity (4, 34). Even with the amount of diversity observed within a relatively small sample size (average, one to two isolates per cow), we were able to isolate the same clonal type from different cows repeatedly. Interestingly, each farm had a reservoir of E. coli clonal types that circulated within the animals of the farm but were distinct from isolates from other farms. Further studies are needed to understand the scope of diversity in E. coli to clarify whether each farm harbors a unique set of genotypes or whether our results are confounded due to small sampling size.
To understand the wide spread of tetracycline resistance among subtypes of E. coli, isolates were screened for tet determinants. The predominance of the tet(B) efflux gene observed in our study has previously been documented in coliforms (73%) of human and animal origins by Marshall et al. (32). Schnabel and Jones (48) showed that tet efflux determinants were carried predominantly by plasmids and that tet(A), tet(B), and tet(C) efflux determinants were associated with transposons. Lee et al. (28) observed that in E. coli isolates from domestic pigs, tet(B) and tet(A) were carried on plasmids. Others have shown that tet efflux genes are part of plasmid-borne transposons in gram-negative bacteria (21, 36). The wide distribution of tet(B) across gram-negative genera, including Escherichia, Enterobacter, Proteus, Salmonella, Actinobacillus, Haemophilus, Moraxella, and Treponema spp., indicates that it is highly likely that horizontal transfer of tetracycline resistance occurs (10, 43, 52). Conjugative plasmids have undoubtedly contributed to the spread of efflux gene classes A to E within gram-negative bacteria and classes K and L within gram-positive bacteria (27). The majority of gram-negative efflux genes are normally associated with large plasmids, most of which are conjugative and belong to different incompatibility groups (21, 36). The presence of tet(A) and tet(B) on the chromosome, rather than on a plasmid, as frequently reported in the literature, was an intriguing observation in our study.
In summary, we found tetracycline- and ampicillin-resistant GN-EB in the feces of healthy lactating dairy cattle. Nine bacterial species were identified from MAC plates supplemented with four commonly used antimicrobial agents, of which E. coli was the most frequently isolated organism. The MICs for E. coli were comparable to those for E. coli isolated from clinical and food samples reported previously. The majority of the tetracycline-resistant E. coli isolates encoded the tet(B) determinant, which was determined to be located on the chromosome and not on plasmids, as reported by other researchers. PFGE analysis revealed that tetracycline-resistant E. coli isolates were highly diverse and, on most occasions, were localized to a given dairy herd. Routine screening of commensal GN-EB, in particular E. coli, could facilitate the study of current and prospective concerns related to the emergence of antimicrobial-resistant GN-EB in dairy cattle.
Acknowledgments
This study has been supported in part by a grant from the USDA NRI (2002-02589; Oxytetracycline resistance of gram-negative bacteria in dairy cattle: risk factors and implications on food safety).
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo, F. J., V. N. Nargund, and T. C. Chiller. 2004. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B 51:374-379. [DOI] [PubMed] [Google Scholar]
- 3.Aslan, V., M. Maden, O. Erganis, F. M. Birdane, and M. Corlu. 2002. Clinical efficacy of florfenciol in the treatment of calf respiratory tract infections. Vet. Q. 24:35-39. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, M., E. J. Threlfall, and E. Liebana. 2005. Cephalosporin resistance among animal-associated Enterobacteria: a current perspective. Expert Rev. Anti-Infect. Ther. 3:403-417. [DOI] [PubMed] [Google Scholar]
- 5.Blake, D. P., R. W. Humphry, K. P. Scott, K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Influence of tetracycline exposure on tetracycline-resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J. Appl. Microbiol. 94:1087-1097. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, P. A., P. J. Petersen, I. M. Fingerman, and D. G. White. 1999. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J. Antimicrob. Chemother. 44:607-610. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, A., N. Shapir, and M. J. Sadowsky. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70:2503-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bywater, R., H. Deluyker, E. Deroover, A. de Jong, H. Marion, M. McConville, T. Rowan, T. Shryock, D. Shuster, V. Thomas, M. Valle, and J. A. Walters. 2004. European survey of antimicrobial susceptibility among zoonotic and commensal bacteria isolated from food-producing animals. J. Antimicrob. Chemother. 54:744-754. [DOI] [PubMed] [Google Scholar]
- 9.Choi, C., H. J. Ham, D. Kwon, J. Kim, D. S. Cheon, K. Min, W. S. Cho, H. K. Chung, T. Jung, K. Jung, and C. Chae. 2002. Antimicrobial susceptibility of pathogenic Escherichia coli isolated from pigs in Korea. J. Vet. Med. Sci. 64:71-73. [DOI] [PubMed] [Google Scholar]
- 10.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edrington, T. S., C. L. Schultz, K. J. Genovese, T. R. Callaway, M. L. Looper, K. M. Bischoff, J. L. McReynolds, R. C. Anderson, and D. J. Nisbet. 2004. Examination of heat stress and stage of lactation (early versus late) on fecal shedding of E. coli O157:H7 and Salmonella in dairy cattle. Foodborne Pathog. Dis. 1:114-119. [DOI] [PubMed] [Google Scholar]
- 12.Erskine, R. J., R. D. Walker, C. A. Bolin, P. C. Bartlett, and D. G. White. 2002. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J. Dairy Sci. 85:1111-1118. [DOI] [PubMed] [Google Scholar]
- 13.Fey, P. D., T. J. Safranek, M. E. Rupp, E. F. Dunne, E. Ribot, P. C. Iwen, P. A. Bradford, F. J. Angulo, and S. H. Hinrichs. 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242-1249. [DOI] [PubMed] [Google Scholar]
- 14.Gilmore, A. 1996. Chloramphenicol and the politics of health. Can. Med. Assoc. J. 134:423-435. [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemot, D., P. Courvalin, and the French Working Party to Promote Research to Control Bacterial Resistance. 2001. Better control of antibiotic resistance. Clin. Infect. Dis. 33:542-547. [DOI] [PubMed] [Google Scholar]
- 16.Gustafson, R. H., and J. S. Kiser. 1985. Nonmedical uses of the tetracyclines, p. 405-446. In J. J. Hlavka and J. H. Boothe (ed.), Handbook of experimental pharmacology, vol. 78. Springer-Verlag KG, Berlin, Germany. [Google Scholar]
- 17.Hariharan, H., M. Coles, D. Poole, and R. Page. 2004. Antibiotic resistance among enterotoxigenic Escherichia coli from piglets and calves with diarrhea. Can. Vet. J. 45:605-606. [PMC free article] [PubMed] [Google Scholar]
- 18.Harley, J. P., and L. M. Prescott (ed.). 1993. Laboratory exercises in microbiology, 2nd ed. Wm. C. Brown Publishers, Dubuque, IA.
- 19.Hegde, N. V., M. L. Cook, D. R. Wolfgang, B. C. Love, C. C. Maddox, and B. M. Jayarao. 2005. Dissemination of Salmonella enterica subsp. enterica serovar Typhimurium var. Copenhagen clonal types through a contract heifer-raising operation. J. Clin. Microbiol. 43:4208-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayarao, B. M., S. C. Donaldson, B. A. Straley, A. A. Sawant, N. V. Hegde, and J. L. Brown. 2006. A survey of foodborne pathogens in bulk tank milk and raw milk consumption among farm families in Pennsylvania. J. Dairy Sci. 89:2451-2458. [DOI] [PubMed] [Google Scholar]
- 21.Jones, C. S., D. J. Osborne, and J. Stanley. 1992. Enterobacterial tetracycline-resistance in relation to plasmid incompatibility. Mol. Cell. Probes 6:313-317. [DOI] [PubMed] [Google Scholar]
- 22.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2006. Antimicrobial drug resistance genes do not convey a secondary fitness advantage to calf-adapted Escherichia coli. Appl. Environ. Microbiol. 72:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kijima-Tanaka, M., K. Ishihara, A. Morioka, A. Kojima, T. Ohzono, K. Ogikubo, T. Takahashi, and Y. Tamura. 2003. A national surveillance of antimicrobial resistance in Escherichia coli isolated from food-producing animals in Japan. J. Antimicrob. Chemother. 51:447-451. [DOI] [PubMed] [Google Scholar]
- 25.Langlois, B. E., K. A. Dawson, T. S. Stahly, and G. L. Cromwell. 1984. Antibiotic resistance of fecal coliforms from swine fed subtherapeutic and therapeutic levels of chlortetracycline. J. Anim. Sci. 58:666-674. [DOI] [PubMed] [Google Scholar]
- 26.Lau, M. M., and S. C. Ingham. 2001. Survival of faecal indicator bacteria in bovine manure incorporated into soil. Lett. Appl. Microbiol. 33:131-136. [DOI] [PubMed] [Google Scholar]
- 27.LeBouguenec, C., G. de Cespedes, and T. Horaud. 1990. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J. Bacteriol. 172:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, C., B. E. Langlois, and K. A. Dawson. 1993. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl. Environ. Microbiol. 59:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin, B. R., M. Lipsitch, V. Perrot, S. Schrag, R. Antia, L. Simonsen, N. M. Walker, and F. M. Stewart. 1997. The population genetics of antibiotic resistance. Clin. Infect. Dis. 24:S9-S16. [DOI] [PubMed] [Google Scholar]
- 30.Levy, S. B. 1984. Resistance to the tetracyclines, p. 191-240. In L. E. Bryan (ed.), Antimicrobial drug resistance. Academic Press, Orlando, FL.
- 31.Maidhof, H., B. Guerra, S. Abbas, H. M. Elsheikha, T. S. Whittam, and L. Beutin. 2002. A multiresistant clone of Shiga-toxin-producing Escherichia coli O118:[H16] is spread in cattle and humans over different European countries. Appl. Environ. Microbiol. 68:5834-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, B., C. Tachibana, and S. B. Levy. 1983. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob. Agents Chemother. 24:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew, A. G., D. B. Arnett, P. Cullen, and P. D. Ebner. 2003. Characterization of resistance patterns and detection of apramycin resistance in Escherichia coli isolated from swine exposed to various environmental conditions. Int. J. Food Microbiol. 89:11-20. [DOI] [PubMed] [Google Scholar]
- 34.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations from host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medeiros, A. A. 1997. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin. Infect. Dis. 24:S19-S45. [DOI] [PubMed] [Google Scholar]
- 36.Mendez, B., C. Tachibana, and S. B. Levy. 1980. Heterogeneity of tetracycline-resistance determinants. Plasmid 3:99-108. [DOI] [PubMed] [Google Scholar]
- 37.Molbak, K. 2004. Spread of resistant bacteria and resistance genes from animals to humans—the public health consequences. J. Vet. Med. B 51:364-369. [DOI] [PubMed] [Google Scholar]
- 38.Moro, M. H., G. W. Beran, R. W. Griffith, and L. J. Hoffman. 2000. Effects of heat stress on the antimicrobial drug resistance of Escherichia coli of intestinal flora of swine. J. Appl. Microbiol. 88:836-844. [DOI] [PubMed] [Google Scholar]
- 39.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 2nd ed. NCCLS document M31-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 40.Ng, L.-K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline-resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 41.Nuru, S., G. W. Osbaldiston, E. C. Stowe, and D. Walker. 1972. Fecal microflora of healthy cattle and pigs. Cornell Vet. 62:242-253. [PubMed] [Google Scholar]
- 42.Orden, J. A., J. A. Ruiz-Santa-Quiteria, S. Garcia, D. Cid, and R. De La Fuente. 2000. In vitro susceptibility of Escherichia coli strains isolated from diarrhoeic dairy calves to 15 antimicrobial agents. J. Vet. Med. B 47:329-335. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, M. C. 1996. Tetracycline-resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 44.Sáenz, Y., M. Zarazaga, L. Brinas, M. Lantero, F. Ruiz-Larrea, and C. Torres. 2001. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain. Int. J. Antimicrob. Agents 18:353-358. [DOI] [PubMed] [Google Scholar]
- 45.Salmon, S. A., J. L. Watts, C. A. Case, L. J. Hoffman, H. C. Wegener, and R. J. Yancey, Jr. 1995. Comparison of MICs of ceftiofur and other antimicrobial agents against bacterial pathogens of swine from the United States, Canada, and Denmark. J. Clin. Microbiol. 33:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Sawant, A. A., L. M. Sordillo, and B. M. Jayarao. 2005. A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 88:2991-2999. [DOI] [PubMed] [Google Scholar]
- 48.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroeder, C. M., D. G. White, B. Ge, Y. Zhang, P. F. McDermott, S. Ayers, S. Zhao, and J. Meng. 2003. Isolation of antimicrobial-resistant Escherichia coli from retail meats purchased in Greater Washington, DC, USA. Int. J. Food Microbiol. 85:197-202. [DOI] [PubMed] [Google Scholar]
- 50.Shiraki, Y., N. Shibata, Y. Doi, and Y. Arakawa. 2004. Escherichia coli producing CTX-M-2 beta-lactamase in cattle, Japan. Emerg. Infect. Dis. 10:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer, R. S., S. K. Patterson, A. E. Meier, J. K. Gibson, H. L. Lee, and C. W. Maddox. 2004. Relationship between phenotypic and genotypic florfenicol resistance in Escherichia coli. Antimicrob. Agents Chemother. 48:4047-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speer, B. S., N. B. Shoemaker, and A. A. Salyers. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stockstad, E. L. R., T. H. Jukes, J. Pierce, A. C. Page, and A. L. Franklin. 1949. The multiple nature of the animal protein factor. J. Biol. Chem. 180:647-654. [PubMed] [Google Scholar]
- 54.White, D. G., C. Hudson, J. J. Maurer, S. Ayers, S. Zhao, M. D. Lee, L. Bolton, T. Foley, and J. Sherwood. 2000. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38:4593-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeruham, I., D. Elad, Y. Avidar, and T. Goshen. 2006. A herd level analysis of urinary tract infection in dairy cattle. Vet. J. 171:172-176. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase blaCMY genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]