Abstract
A new mutant of the industrial enzyme penicillin G acylase (PGA) from Escherichia coli has been designed to improve its reversible immobilization on anionic exchangers (DEAE- or polyethyleneimine [PEI]-coated agarose) by assembling eight new glutamic residues distributed homogeneously through the enzyme surface via site-directed mutagenesis. The mutant PGA is produced and processed in vivo as is the native enzyme. Moreover, it has a similar specific activity to and shows the same pH activity profile as native PGA; however, its isoelectric point decreased from 6.4 to 4.3. Although the new enzyme is adsorbed on both supports, the adsorption was even stronger when supports were coated with PEI, allowing us to improve the enzyme stability in organic cosolvents. The use of restrictive conditions during the enzyme adsorption on anionic exchangers (pH 5 and high ionic strength) permitted us to still further increase the strength of adsorption and the enzyme stability in the presence of organic solvents, suggesting that these conditions allow the penetration of the enzyme inside the polymeric beds, thus becoming fully covered with the polymer. After the enzyme inactivation, it can be desorbed to reuse the support. The possibility to improve the immobilization properties on an enzyme by site-directed mutagenesis of its surface opens a promising new scenario for enzyme engineering.
The use of enzymes as industrial biocatalysts usually requires their previous immobilization to simplify the control of the reactor, to avoid product contaminations, and mainly to recover and reuse the enzyme for many reaction cycles (5, 6, 8, 10, 37). However, enzyme immobilization may be expensive (considering support and process costs) and time-consuming. In this sense, immobilization of proteins on ionic exchangers presents several advantages because it does not involve complex protocols for enzyme immobilization; it only requires putting the enzyme solution in contact with the support under very mild conditions. The most important advantage of this method is that it is possible to reutilize the supports after enzyme inactivation (8, 31, 35, 37, 46). Therefore, ionic exchangers provide a simple solution for some immobilization problems, and in fact they were used as the first strategy to produce an industrial biocatalyst (10, 24).
Nevertheless, the main drawback of the ionic immobilization strategy is the desorption of the proteins during operation, promoting biocatalyst inactivation and product contamination (25, 44). The use of ionic polymers coating the surface of solid supports (e.g., polyethyleneimine [PEI] or dextran sulfate) not only provides a stronger adsorption but also prevents the alteration of the three-dimensional protein structure since the polymer becomes adapted to the enzyme instead of forcing the protein to become adapted to the support (13, 31). The immobilization of proteins on this polymer-coated support may have some additional advantages, preventing subunit dissociation of multimeric enzymes (2, 14, 31) or generating hydrophilic nanoenvironments that stabilize the proteins against the presence of organic solvents (12). However, in some cases a protein may not be adsorbed strongly enough even on these polymeric ionic beds for a particular application. When improving the properties of the support is no longer possible, to further increase the strength of protein adsorption we propose to remodel the enzyme surface by site-directed mutagenesis in order to improve its complementarity with the ionic exchanger, increasing the number of ionic groups in the enzyme surface capable of interacting with the support (that is, exposed to the medium). Therefore, we propose to carry out genetic modifications of the proteins, not to directly improve the enzyme properties (18, 28, 29, 53) but to produce an enzyme with better characteristics to facilitate its immobilization by a very simple technique as its adsorption on anionic exchangers.
This strategy has been used scarcely, and most trials have been directed to control the enzyme orientation on the support (20, 30, 39). Nevertheless, we have recently described the use of Lys-enriched penicillin G acylase (PGA) to establish an intense multipoint covalent attachment to glyoxyl agarose supports (1). However, this technique has not been proposed to be used to improve protein immobilization on ionic exchangers so far. This approach does not require a knowledge of protein structure as deep as that required to directly improve the stability of soluble enzymes, since in this case we only have to be able to predict the groups that will be exposed to the medium. Moreover, it is assumed that site-directed mutagenesis on these surface groups should not cause dramatic negative changes on the enzyme properties (1).
To demonstrate the feasibility of this strategy, we have chosen as a model enzyme the PGA from Escherichia coli (34, 48), one of the most important industrial enzymes because it is currently used by pharmaceutical companies to produce beta-lactam antibiotics (3, 33, 42). Moreover, PGA can be utilized in many other reactions, like enzymatic synthesis of antibiotics, resolution of racemic mixtures, synthesis of amides, and selective deprotections (16, 22, 38, 43, 47, 49-51). PGA suffers a complex posttranslational processing to become active, since it is synthesized in E. coli as an inactive monomer that is autoprocessed during its secretion to the periplasmic space to form an active alpha-beta heterodimer form (23, 52).
In this work, we have increased the number of carboxylic groups in the PGA surface to improve its adsorption on anionic exchangers, where the native enzyme was not significantly adsorbed (M. Fuentes, P. Batalla, V. Grazú, C. C. Benevides, C. Mateo, T. Montes, J. Hermoso, J. M. Guisán, and R. Fernández-Lafuente, submitted for publication). Taking into account that support adsorption may involve a large percentage of the protein surface and even more in the case of polymeric-coated supports where the enzyme may penetrate into the polymeric bed, we have created eight glutamic residues distributed all around the protein surface, trying to reduce the putative negative effects of these mutations on PGA activity or stability.
MATERIALS AND METHODS
Materials.
A High Pure plasmid isolation kit was purchased from Roche Diagnostics SL (Barcelona, Spain). A QIAquick gel extraction kit for DNA fragment purification was purchased from QIAGEN GmbH (Hilden, Germany). Pfu DNA polymerase (Stratagene, La Jolla, CA) was used for site-directed mutagenesis. Oligonucleotides were synthesized by Genotek (Madrid, Spain). Penicillin G was kindly provided by Antibióticos S.A. (León, Spain). PEI (25, 60, and 600 kDa), streptomycin sulfate, and isopropyl-β-d-thiogalactopyranoside (IPTG) were supplied by Sigma-Aldrich (St. Louis, MO). DEAE-Sepharose and cyanogen bromide (CNBr)-activated Sepharose 4B were purchased from Amersham Biosciences (Uppsala, Sweden). Cross-linked 4% agarose beads (4BCL) were donated by Hispanagar S.A. (Burgos, Spain). All other reagents were of analytical grade. PEI supports were prepared from glyoxyl agarose (15, 32) as previously described (31)
Molecular dynamics on PGA enzyme structure.
Models of the different mutants were built on the basis of the crystal structure of the PGA named “OLE_LINK1” from E. coli ATCC 11105 (19). Amino acid changes were introduced using the O graphic program (21) running on a Silicon Graphics workstation. Side-chain rotamers were chosen from a database of more-common conformers (41). First models were energy minimized using the Powell minimizer algorithm implemented in X-PLOR (9), version 3.851. The Engh and Huber (11) force field was used in all energy minimization and molecular dynamic simulations. Subsequently, a slow-cooling molecular dynamic protocol (9) was carried out over a period of 1.5 ps and applied by using a weak temperature coupling method (4). The target temperature of 2,500 K was decreased by 25 K every 100 steps to reach the final temperature of 300 K. The time step was set to 0.005 fs. Finally, the conformation of the different mutants trapped at 300 K was subjected to 500 additional steps of energy minimization.
Bacterial strains, plasmids, and culture media.
Escherichia coli strains DH5α (laboratory stock) and One Shot Top 10 (Invitrogen, Paisley, United Kingdom) were used for routine cloning procedures. Overproduction of PGA was performed on E. coli strain BL21 Star(DE3) (Invitrogen). The E. coli strains were routinely cultured at 37°C in Luria-Bertani (LB) broth, in either liquid or solid media containing ampicillin (150 μg/ml). For overproduction experiments, cells were cultured at lower temperatures to avoid the formation of inclusion bodies (see below). Plasmid pET101/D-TOPO (Invitrogen, Paisley, United Kingdom) was used for cloning purposes according to the supplier's procedures (Invitrogen, Paisley, United Kingdom). Plasmid pOAF, a pET101/D-TOPO plasmid derivative containing the wild-type pac gene from E. coli ATCC 11105, was previously described (1).
Site-directed mutagenesis of PGA.
DNA manipulations were performed as described by Sambrook and Russell (45). PGA mutants were constructed by a site-directed mutagenesis PCR technique (17) using plasmid pOAF as the template and the mutagenic primers shown in Table 1. The eight amino acid changes (Asn or Gln replaced by Glu) were introduced by three PCR steps. First, nine DNA fragments covering the complete sequence of the pac gene and carrying the proposed nucleotide mutations were independently amplified by PCR: fragment A (pOAF as the template and primers P1 and P2R), fragment B (pOAF as the template and primers P2F and P3R), fragment C (pOAF as the template and primers P3F and P4R), fragment D (pOAF as the template and primers P4F and P5R), fragment E (pOAF as the template and primers P5F and P6R), fragment F (pOAF as the template and primers P6F and P7R), fragment G (pOAF as the template and primers P7F and P8R), fragment H (pOAF as the template and primers P8F and P9R), and fragment I (pOAF as the template and primers P9F and P10). Second, these amplified DNA fragments were purified by gel electrophoresis and connected in pairs or triplets by PCR to create four new fragments: fragment AB (fragments A and B as templates and primers P1 and P3R), fragment CDE (fragments C, D, and E as templates and primers P3F and P6R), fragment FG (fragments F and G as templates and primers P6F and P8R), and fragment HI (fragments H and I as templates and primers P8F and P10). Finally, the four DNA fragments (AB, CDE, FG, and HI) were purified by gel electrophoresis and connected all together in a single PCR to reconstruct the mutated pac gene using these fragments as templates and primers P1 and P10. The resulting DNA fragment (ABCDEFGHI), carrying the complete pac gene containing the eight desired mutations, was purified by gel electrophoresis and cloned into plasmid pET101/D-TOPO according to the suppliers (Invitrogen, Paisley, United Kingdom). The resulting plasmid (named pPGA8glu), expressing the mutated pac gene, was completely sequenced to verify the absence of unwanted mutations.
TABLE 1.
PCR primers used for site-directed mutagenesis of the pac genea
| Primer | Oligonucleotide sequence |
|---|---|
| (P1) TOPO 5′ | (−5) 5′-CACCAATGAAAAATAGAAATCGTATGA-3′ |
| (P2F) asn A108 glu | (+387) 5′-GACTGATAAGGTAGAAACCAATCCAGAG-3′ |
| (P2R) asn A108 glu | 3′-CTGACTATTCCATCTTTGGTTAGGTCTC-5′ |
| (P3F) gln B112 glu | (+1189) 5′-GTGAAAAATGGTGAGGCAGAGACCTTTAC-3′ |
| (P3R) gln B112 glu | 3′-CACTTTTTACCACTCCGTCTCTGGAAATC-5′ |
| (P4F) gln B131 glu | (+1243) 5′-ATTCTCCAAACTGACGAGACGACACAAACG-3′ |
| (P4R) gln B131 glu | 3′-TAAGAGGTTTGACTGCTCTGCTGTGTTTGC-5′ |
| (P5F) gln B164 glu | (+1344) 5′-GGCCAAAAATTGGGAGGAGTGGACACAG-3′ |
| (P5R) gln B164 glu | 3′-CCGGTTTTTAACCCTCCTCACCTGTGTC-5′ |
| (P6F) gln B233 glu | (+1552) 5′-GTGTATAACCCCGAGTCGGGATATATTG-3′ |
| (P6R) gln B233 glu | 3′-CACATATTGGGGCTCAGCCCTATATAAC-5′ |
| (P7F) gln B312 glu | (+1786) 5′-ACATCTGGTTTGACAGAGAGCGATCCGCGT-3′ |
| (P7R) gln B312 glu | 3′-TGTAGACCAAACTGTCTCTCGATAGGGGCA-5′ |
| (P8F) gln B380 glu | (+1993) 5′-TACGAAACAACCGAGGACGGCCCAACT-3′ |
| (P8R) gln B380 glu | 3′-ATGCTTTGTTGGCTCCTGCCGGGTTGT-5′ |
| (P9F) gln B420 glu | (+2112) 5′-GGGAAACCACAGGAGGAGGTTGTGTTG-3′ |
| (P9R) gln B420 glu | 3′-CCCTTTGGTGTCCTCCTCCAACACAAC-5′ |
| (P10) TOPO 3′ | (+2594) 3′-TACAACCTCCCGACCAATGAAA-5′ |
The amino acid changed and its position on the α or β chain of mature PGA are indicated. The mutated Glu codons are presented in bold in the primer sequences. F and R indicate forward and reverse sequences regarding the coding sequence of the pac gene. Numbers in brackets indicate the position of the first nucleotide of the mutagenic primer on the pac sequence, considering position +1 the first nucleotide of the start codon. The TOPO 5′ primer (P1) provides the start codon of the pac gene (boldfaced and underlined). The TOPO 3′ primer (P10) hybridizes with the downstream 3′ noncoding region of the pac gene present in plasmid pOAF to allow the amplification of the C-terminal coding region of the pac gene.
Protein expression and purification.
Cells of E. coli BL21(DE3) were transformed with pOAF or pPGA8glu plasmid to facilitate the overexpression of wild-type or mutant pac genes, respectively, under the control of the T7 promoter. In brief, E. coli BL21(DE3) recombinant cells were cultured at 22°C and 200 rpm in 100 ml to 2 liters LB medium containing ampicillin (150 μg/ml) up to an optical density at 600 nm of 0.6, and then 0.1 mM IPTG was added to the culture. After 48 h of incubation at the same temperature, cells were harvested by centrifugation (10,000 × g, 5 min), resuspended in 20 mM phosphate buffer, pH 7.0, and disrupted by sonication. The insoluble fraction was separated by centrifugation (20,000 × g, 20 min) at 4°C. The supernatant containing the overexpressed wild-type or mutant PGA was used as crude extract for the first step of enzyme purification. DNA contained in crude extracts was precipitated with 2% (wt/vol) streptomycin sulfate with mild shaking for 20 min at 4°C. Precipitated extract was then centrifuged (20,000 × g, 15 min), and the supernatant was dialyzed against 50 mM Tris-HCl buffer, pH 8.5. The dialyzed extract of wild-type PGA was then incubated in the presence of a PEI (25 kDa) agarose-ion exchanger (31) at pH 8.5 for 4 h at 4°C on a batch reactor. The PGA activity remained in the supernatant; thus, after filtration, the extract containing the enzyme PGA was further purified by fast-performance liquid chromatography with a Q-Sepharose high-performance column (Pharmacia) using a gradient of 0 to 100 mM NaCl in 50 mM Tris-HCl, pH 8.5, as the elution buffer.
On the other hand, the mutant PGA was purified by fast-performance liquid chromatography by directly loading the dialyzed extract prepared after DNA precipitation on a Q-Sepharose high-performance column (Pharmacia) and eluting the proteins with a gradient of 0 to 200 mM NaCl in 50 mM Tris-HCl buffer, pH 8.5.
Protein concentration was determined by the method of Bradford (7). The purity of proteins was checked on 12% (wt/vol) sodium dodecyl sulfate-polyacrylamide gels as described by Laemmli (27).
Isoelectric point determination.
Analytical isoelectric focusing was performed with a Pharmacia Phast system in a PhastGel IEF, pH 3 to 9 (0.35 mm thin, 5 by 4 cm size, 5% T and 3% C). Running conditions were 2,000 V, 2.5 mA, and 3.5 W at 15°C until 410 V/h was reached. Sample loading was 1 μl per sample (0.150 mg/ml). The gel was stained with Coomassie dye following the supplier's indications, and the isoelectric points of the purified mutant and the wild type were estimated from the positions relative to those of standard proteins (36).
Determination of PGA activity.
Enzyme activity was determined using an automatic titrator (DL50 Mettler Toledo) to titrate the release of phenylacetic acid produced by the hydrolysis of 10 mM penicillin G in 0.1 M sodium phosphate-0.5 M NaCl at pH 8.0 and 25°C. A 100 mM NaOH solution was used as the titrating reagent. One international unit of PGA activity was defined as the amount of enzyme that hydrolyzes 1 μmol of penicillin G per minute at pH 8 and 25°C. All experiments were performed at least in triplicate, and the results are presented as mean values. Experimental error was never over 5%.
In some cases, PGA activity was followed spectrophotometrically using 6-nitro-3-phenylacetamido benzoic acid.
Immobilization on ionic exchangers.
Immobilization was carried out by adding 1 volume of the corresponding supports to 4 volumes of a solution containing 2.5 IU of native or mutant PGA in 5 mM sodium phosphate at pH 7.0 or pH 5.0 at 25°C (standard ratio was 1 g of wet support to 4 ml of enzyme solution). In some cases, immobilizations were carried out in the presence of different concentrations of NaCl.
During adsorption, samples were withdrawn from the supernatant and the suspension to assay enzyme activity as described above. After immobilization, the derivatives were washed with distilled water and stored at 4°C.
Reference suspensions were prepared with exactly the same enzyme concentration and medium conditions (pH, temperature, and ionic strength) but with addition of the corresponding amount of inert agarose instead of the active support. The PGA activity in the supernatant of this reference suspension was fully preserved in all cases; therefore, the decrease in PGA activity observed in the supernatant of the “immobilization suspension” can be directly correlated to the amount of enzyme adsorbed on the ionic exchangers. Experimental error was never over 7%.
Desorption of the proteins adsorbed on the ionic exchangers.
The different immobilized PGA preparations (10 IU/g of support) were incubated at increasing concentrations of NaCl at 25°C and pH 7.0, and after 30 min (longer incubation times did not reveal any change in the results) the PGA activity of the suspension and supernatant was assayed as described above. Experimental error was never over 9%.
Thermal stability assays.
Enzyme preparations (soluble or immobilized preparations) were incubated in 25 mM sodium phosphate buffer, pH 7.0, at 55°C. Samples of the suspension were withdrawn periodically, and enzyme activity was analyzed as described above. PGA residual activity was expressed as a percentage of initial activity at the given incubation time. Experimental error was never over 5%.
Inactivation in the presence of dioxane.
Enzyme preparations were incubated in different percentages of dioxane (vol/vol) in 25 mM sodium phosphate buffer, pH 6.5, at 4°C. Samples of the suspension were withdrawn periodically, and enzyme activity was analyzed as described above. PGA residual activity was expressed as a percentage of initial activity at the given incubation time. Dioxane was used for its deleterious effect on PGA stability and high hydrophobicity, which permit its partitioning when generating hydrophilic nanoenvironments (12). Experimental error was never over 10%.
RESULTS
Selection of the amino acid residues to be mutated.
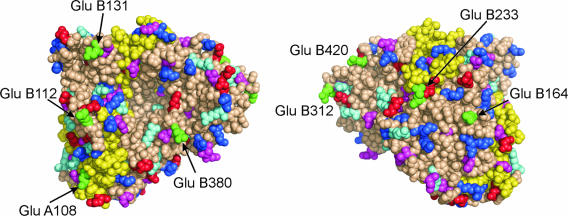
As stated in the introduction, the aim of this work was to demonstrate the utility of increasing the number of acidic residues in the PGA surface by using a site-directed mutagenesis approach to improve its adsorption on anionic exchangers. To create the new PGA, we decided to distribute the acidic mutations homogenously through the PGA surface to decrease the likely impact in the enzyme properties. To fulfill this goal, we theoretically divided the enzyme surface in eight sectors and then selected the asparagines or glutamines exposed to the medium (one per sector) according to the positions shown in Fig. 1. By using the procedures described above, we first checked whether the predicted structure of the mutated enzyme would remain almost unaltered compared to that of the native enzyme (root mean square deviation was 0.164). Once demonstrated by this theoretical analysis that the structure of the mutant PGA would most probably remain unaltered, we decided to produce the new protein by using the site-directed mutagenesis procedure described above.
FIG. 1.
Space-filled model of the three-dimensional structure of the mutated PGA containing eight new Glu residues. Mutated residues are in green. Lys, Arg, Glu, and Asp residues are in dark blue, light blue, red, and pink, respectively. The α-subunit of PGA is shown in yellow.
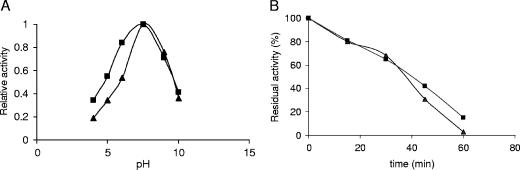
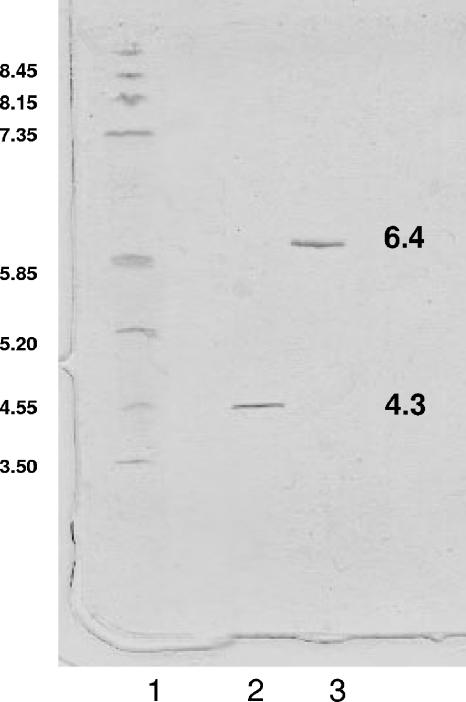
Remarkably, the amount of mutant PGA produced in the recombinant E. coli BL21(DE3)(pPGA8glu) cells was very similar to that of the wild-type enzyme produced by E. coli BL21(DE3)(pOAF) (around 200 IU per liter of culture), suggesting that the introduced mutations did not affect the in vivo complex posttranslational processing of this enzyme (23, 52). More importantly, the mutant enzyme showed a specific activity nearly identical to that of wild-type PGA (at pH 8 and 25°C, it was 25 ± 3 IU). In addition, the stabilities of both enzymes were very similar (Fig. 2). The pH/activity profiles also remained quite similar, with the mutant enzyme being slightly more active at an acidic pH value. All of these results taken together demonstrate that the predictions of in silico analysis correspond with the actual structure of the mutant enzyme and that we have created a new form of PGA with activity and stability very similar to those of the wild type but with a very different external surface. By analyzing the structure of the native enzyme and the model of the mutant enzymes, we studied the number of charged groups exposed to the medium and therefore useful to interact with a support. Thus, the newly designed mutant PGA contains 85 exposed Glu-plus-Asp residues, while the native enzyme exposed in the surface only 77 acidic residues (Table 2) , and such an increase in the number of negative charges reduces the isoelectric point of the mutant PGA from 6.4 to 4.3 (Fig. 3).
FIG. 2.
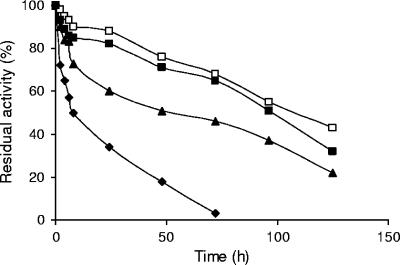
pH activity profiles and thermal inactivation courses of both native and mutant PGA. (A) pH activity profiles of enzymes, determined by using penicillin G as the substrate. Experiments were performed at 25°C. (B) Thermal inactivation courses of enzymes. Inactivation experiments were carried out at 55°C in 25 mM sodium phosphate buffer, pH 7.0. Enzyme activity was determined as described in Materials and Methods. Symbols: ▴, native PGA; ▪, mutant PGA.
TABLE 2.
Analysis of ionizable groups of native and mutant PGA accessible to the medium
| Type of PGA | No. of residues
|
|||
|---|---|---|---|---|
| Lys | Arg | Glu | Asp | |
| Native | 36 | 19 | 41 | 36 |
| Mutant | 36 | 19 | 49 | 36 |
FIG. 3.
Isoelectric focusing electrophoresis of PGA. Electrophoresis was performed with a Pharmacia Phast system in a PhastGel IEF, pH 3 to 9. Lane 1, pI calibration kit, pH 3 to 9; lane 2, mutant PGA; lane 3, native PGA. The isoelectric points of the purified mutant and wild-type enzymes were estimated from their positions relative to those of standard proteins. See details in Materials and Methods.
Adsorption of native and mutant enzymes on anionic exchangers.
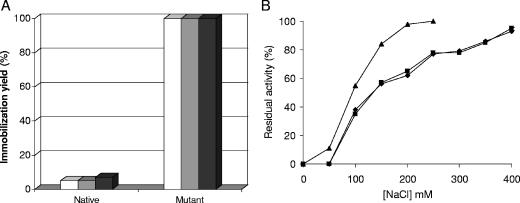
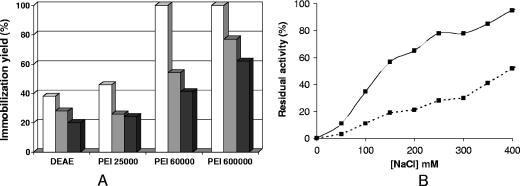
Both mutant and native PGA enzymes in 5 mM sodium phosphate, pH 7.0, were offered to DEAE and two supports coated with PEI of different sizes (Fig. 4). Interestingly, the native PGA did not become significantly immobilized on any of the three supports, while the mutant enzyme became fully immobilized on them. In all cases, the enzyme activity was unaltered during adsorption, due to the very mild immobilization conditions used in these experiments.
FIG. 4.
Immobilization and desorption of PGA on different anionic exchangers. (A) Immobilization courses of native and mutant PGA on different anionic exchangers. White columns, DEAE; gray columns, PEI (25 kDa); black columns, PEI (600 kDa). (B) Desorption of mutant PGA from different ionic exchangers. Immobilizations were performed in 5 mM sodium phosphate buffer, pH 7.0, at 25°C using 10 IU of enzyme/ml support (see Materials and Methods for details). Symbols: ▴, DEAE; ▪, PEI (25 kDa); ⧫, PEI (600 kDa).
Desorption of mutant PGA from anionic exchangers.
The mutant PGA adsorbed on DEAE was desorbed at moderate ionic strength, since full desorption was achieved at 200 mM NaCl (Fig. 4), while the enzyme adsorbed on PEI could not be fully desorbed even with 400 mM NaCl. However, both PEI-coated supports gave the same desorption profile, suggesting very similar adsorption strengths.
Enzyme stability in the presence of dioxane.
Adsorption of the enzymes on polymeric beds has been reported to stabilize the enzymes against the inactivation caused by organic solvents. Figure 5 shows the stabilities of different immobilized preparations of the enzyme in the presence of dioxane. The mutant PGA adsorbed on DEAE was more stable than the enzyme covalently immobilized on CNBr agarose, but the most stable preparations were those where the enzyme was adsorbed on PEI. No differences between PEI-coated supports could be found.
FIG. 5.
Inactivation courses of immobilized mutant PGA in the presence of dioxane. Experiments were carried out in the presence of 60% (vol/vol) dioxane in 25 mM sodium phosphate, pH 6.5, at 4°C. Symbols: ⧫, mutant PGA immobilized onto CNBr-Sepharose; ▴, mutant PGA immobilized onto DEAE-agarose; ▪, mutant PGA immobilized onto PEI (25 kDa)-agarose; □, mutant PGA immobilized onto PEI (600 kDa).
Optimization of the adsorption of mutant PGA on PEI-coated supports.
The moderate stabilization of the PGA immobilized on PEI-coated supports in the presence of dioxane together with the absence of differences between the properties of the mutant PGA adsorbed on both supports coated with PEI in spite of their very different sizes suggested that the enzyme does not penetrate the bed formed by the polymer and that it interacts only with the superficial layer of the film formed by the ionic groups. This possibility is reinforced by the fact that the enzyme may be fully and rapidly adsorbed on conventional supports covered with cationic groups under these conditions.
On the other hand, the use of a high ionic strength during adsorption to increase the adsorption strength has been reported previously (40), suggesting that these more restrictive conditions make the multipoint adsorption necessary to fix a protein on an ionic exchanger more difficult (26), allowing a better penetration of the enzyme on the polymeric bed.
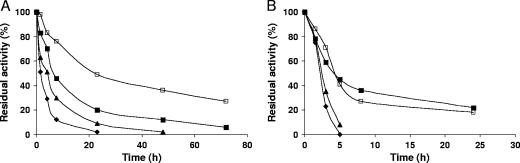
Thus, to improve the properties of the immobilized PGA on PEI-coated supports (i.e., adsorption strength and stability against dioxane), the adsorption conditions were changed towards apparently less favorable conditions, taking advantage of the new properties of the engineered PGA and the use of a very strong anionic exchanger like PEI. Therefore, enzyme adsorptions under lower pH and higher ionic strength conditions were studied (Fig. 6). At pH 5.0, adsorption of the mutant enzyme was greatly reduced on DEAE (adsorption accounted for less than 40% of the enzyme) and the use of a higher ionic strength further reduced the percentage of adsorbed enzyme. The adsorption on supports coated with 25-kDa PEI under similar conditions produced similar results, although when this support was used the adsorption was slightly higher. Remarkably, the use of PEI of larger sizes (60 kDa or 600 kDa) progressively increases the percentage of immobilized enzyme; in fact, with the largest polymer more than 60% of the enzyme was adsorbed at pH 5.0 and 150 mM NaCl. Figure 6 shows that the mutant PGA adsorbed under these conditions required much higher concentrations of NaCl to be desorbed from the PEI-coated support than when adsorption was performed under standard conditions. The concentration of NaCl required to release 50% of the enzyme from the support increased from 150 mM to 400 mM NaCl. More interestingly, the enzyme adsorbed under the new conditions became much more stable in the presence of dioxane (Fig. 7A), although the thermal stabilities of both preparations were quite similar (Fig. 7B). Therefore, under these conditions, it seems that PGA is able to penetrate deeper inside the polymeric bed.
FIG. 6.
(A) Immobilization at pH 5.0 of mutant PGA on supports coated with PEI (600 kDa). Experiments were carried out in 25 mM acetate buffer, pH 5.0, at 25°C using 10 IU of enzyme/ml support. White columns, 25 mM NaCl; gray columns, 100 mM NaCl; black columns, 150 mM NaCl. (B) Desorption of mutant PGA immobilized at pH 5 on support coated with PEI (600 kDa). Desorption was performed at pH 7 and 25°C as described in Materials and Methods. Activity released from PEI (600 kDa) is depicted as follows: —▪—, mutant PGA adsorbed in 5 mM sodium phosphate, pH 7.0, and - - - - ▪- - - - , mutant PGA adsorbed in the presence of 150 mM NaCl and pH 5.0.
FIG. 7.
Inactivation courses of immobilized mutant PGA. (A) Inactivation by dioxane. Experiments were carried out in the presence of 75% (vol/vol) dioxane in 25 mM sodium acetate buffer (pH 5.5) at 4°C. Symbols: ⧫, mutant and native PGA immobilized onto CNBr-Sepharose; ▴, mutant PGA immobilized onto DEAE at pH 7.0; ▪, mutant PGA immobilized onto PEI (600 kDa) at pH 7.0; □, mutant PGA immobilized onto PEI (600 kDa) in the presence of 150 mM NaCl at pH 5.0. (B) Thermal inactivation. Experiments were carried out at 55°C in 25 mM phosphate buffer, pH 5.0. Symbols: ⧫, mutant and native PGA immobilized onto CNBr-Sepharose; ▴, mutant PGA immobilized onto DEAE at pH 7.0; ▪, mutant PGA immobilized onto PEI (600 kDa) at pH 7.0; □, mutant PGA immobilized onto PEI (600 kDa) in the presence of 150 mM NaCl at pH 5.0.
In this way, a greater surface area of the protein contacts the polymer, generating a stronger adsorption and a higher stabilization against the action of dioxane. The loading capacity of the support was around 20 mg of PGA/g of support.
An enzyme preparation bearing 10 IU (5 g of biocatalyst in 50 ml of reaction mixture) was used at pH 8 and 25°C in the hydrolysis of 5% of penicillin G for 10 consecutive cycles without detecting any change in the enzyme activity, confirming that there is no enzyme desorption under these conditions.
DISCUSSION
The remodeling of enzyme surfaces by site-directed mutagenesis in order to improve enzyme immobilization on tailor-made supports has proven to be a very powerful tool to prepare industrial biocatalysts. In this paper, the increase in the number of glutamic residues on the surface of PGA via site-directed mutagenesis allowed immobilization of the enzyme on anionic exchangers, a process that was not possible with the native enzyme. We were able to introduce eight mutations which did not produce any relevant effect on its enzymatic properties but completely modified its capacity to become adsorbed on these supports. The adsorption is performed by a multipoint process that is achieved only by enzymes able to interchange several amino acid residues with the support.
However, to take full advantage of the immobilization of proteins via ionic exchange, it has been necessary to use highly improved supports (i.e., supports coated with polymeric cationic beds) and, moreover, to use stringent optimized immobilization conditions. In fact, the best results for enzyme stabilization on dioxane and adsorption strength were achieved if the adsorption was performed under apparently unfavorable conditions, i.e., low pH and high ionic strength. The adsorption of the enzyme under more favorable conditions permits the enzyme immobilization just by interaction with the external groups of the polymeric bed, avoiding the possibility that the enzyme could be fully covered by the polymer (40). This may explain the lack of effect of the size of the polymer coating the support on the enzyme stability and adsorption strength (when the enzyme was adsorbed at pH 7.0 and low ionic strength), conditions under which the mutant enzyme becomes fully and rapidly adsorbed even on DEAE-coated supports. However, the use of more restrictive conditions requires the interaction of the polymer with a larger surface area of the protein and involves a high number of groups of the support. In this way, the enzyme may be more extensively covered by the polymer, increasing the protection against inactivation by dioxane and the adsorption strength. The fact that the eight new Glu residues were distributed homogenously throughout the entire enzyme surface contributed to fully cover PGA with the polymer. Thus, in our best optimal preparation the enzyme activity remains fully unaltered after immobilization, and it may be used at a relatively high ionic strength and under a wide range of pH values and present a significant stabilization against the deleterious action of dioxane. Moreover, in spite of this strong adsorption, PGA can be desorbed when it becomes inactivated during operation by incubating the biocatalyst under different conditions (e.g., 100 mM HCl), allowing the reuse of the support for at least five cycles (results not shown).
Thus, reversible immobilization of PGA on supports coated with polymers may permit increase of the stability of the enzyme in the presence of dioxane. Multipoint covalent attachment on proper supports may permit a higher stabilization rigidification of the enzyme, with higher stabilization in thermal inactivations (1, 15, 32). However, the generation of hydrophilic nanoenvironments by this new strategy yields stabilization factors in the presence of dioxane that are very similar to those obtained by multipoint covalent attachment, permitting full use of the advantages of the reversible immobilization.
Acknowledgments
We gratefully recognize support from the Spanish CICYT (projects BIO-2005-8576 and BIO-2003-05309-C04-02). We gratefully recognize Spanish MEC for the fellowships for T. Montes and F. López-Gallego.
We kindly appreciate the help of C. Talavera with the isoelectric focusing technique. The help and suggestions of Ángel Berenguer (MCMA, Universidad de Alicante) are gratefully recognized.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Abián, O., V. Grazú, J. Hermoso, R. González, J. L. García, R. Fernández-Lafuente, and J. M. Guisán. 2004. Stabilization of penicillin G acylase from Escherichia coli: site-directed mutagenesis of the protein surface to increase multipoint covalent attachment. Appl. Environ. Microbiol. 70:1249-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arica, M. Y., and G. Bayramoglu. 2006. Invertase reversibly immobilized onto polyethylenimine-grafted poly(GMA-MMA) beads for sucrose hydrolysis. J. Mol. Catal. B 38:131-138. [Google Scholar]
- 3.Arroyo, M., I. De La Mata, C. Acebal, and M. P. Castillón. 2003. Biotechnological applications of penicillin acylases: state-of-the-art. Appl. Microbiol. Biotechnol. 60:507-514. [DOI] [PubMed] [Google Scholar]
- 4.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684-3690. [Google Scholar]
- 5.Bickerstaff, G. F. 1997. Methods in biotechnology, vol. 1. Humana Press, Inc., Totowa, NJ.
- 6.Bornscheuer, U. T. 2003. Immobilizing enzymes: how to create more suitable catalysts. Angew. Chem. Int. Ed. Engl. 42:3336-3337. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brena, B. M., L. G. Ryden, and J. Porath. 1994. Immobilization of beta-galactosidase on metal-chelate-substituted gels. Biotechnol. Appl. Biochem. 19:217-231. [PubMed] [Google Scholar]
- 9.Brünger, A. T., A. Krukowski, and J. Erickson. 1990. Slow cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. A 46:585-593. [DOI] [PubMed] [Google Scholar]
- 10.Chibata, I., and T. Tosa. 1976. Industrial applications of immobilized enzymes and immobilized microbial cells, p. 239-360. In L. B. Wingard, Jr., E. Katchalski-Katzir, and L. Goldstein (ed.), Series applied biochemistry and bioengineering, vol. 1. Academic Press, London, United Kingdom. [Google Scholar]
- 11.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A 47:392-400. [Google Scholar]
- 12.Fernandez-Lafuente, R., C. M. Rosell, L. Caanan-Haden, L. Rodes, and J. M. Guisán. 1999. Facile synthesis of artificial enzyme nano-environments via solid-phase chemistry of immobilized derivatives: dramatic stabilization of penicillin acylase versus organic solvents. Enzyme Microb. Technol. 24:96-103. [Google Scholar]
- 13.Fuentes, M., B. C. C. Pessela, J. V. Maquiese, C. Ortiz, R. L. Segura, J. M. Palomo, O. Abián, R. Torres, C. Mateo, R. Fernández-Lafuente, and J. M. Guisán. 2004. Reversible and strong immobilization of proteins by ionic exchange on supports coated with sulfate-dextran. Biotechnol. Prog. 20:1134-1139. [DOI] [PubMed] [Google Scholar]
- 14.González, P., F. Batista-Viera, and B. M. Brena. 2004. Polyethylenimine coated agarose supports, for the reversible immobilisation of beta-galactosidase from Aspergillus oryzae. Int. J. Biotechnol. 6:338-345. [Google Scholar]
- 15.Guisán, J. M. 1988. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzyme Microb. Technol. 10:375-382. [Google Scholar]
- 16.Guranda, D. T., L. M. Van Langen, F. Van Rantwijk, R. A. Sheldon, and V. K. Svedas. 2001. Highly efficient and enantioselective enzymatic acylation of amines in aqueous medium. Tetrahedron Asymmetry 12:1645-1650. [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Hult, K., and P. Berglund. 2003. Engineered enzymes for improved organic synthesis. Curr. Opin. Biotechnol. 14:395-400. [DOI] [PubMed] [Google Scholar]
- 19.Hunt, P. D., S. P. Tolley, R. J. Ward, C. P. Hill, and G. G. Dodson. 1990. Expression, purification and crystallization of penicillin G acylase from Escherichia coli ATCC 11105. Protein Eng. 3:635-639. [DOI] [PubMed] [Google Scholar]
- 20.Iwakura, M., and T. Kokubu. 1993. Immobilization of dihydrofolate reductase by engineered cysteine residue attached to its C-terminal end. J. Biochem. 114:339-343. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 22.Kadereit, D., and H. Waldmann. 2001. Enzymatic protecting group techniques. Chem. Rev. 101:3367-3396. [DOI] [PubMed] [Google Scholar]
- 23.Kasche, V., Z. Ignatova, H. Markl, W. Plate, N. Punckt, D. Schmidt, K. Wiegandt, and B. Ernst. 2005. Ca2+ is a cofactor required for membrane transport and maturation and is a yield-determining factor in high cell density penicillin amidase production. Biotechnol. Prog. 21:432-438. [DOI] [PubMed] [Google Scholar]
- 24.Katchalski-Katzir, E. 1993. Immobilized enzymes—learning from past successes and failures. Trends Biotechnol. 11:471-478. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, J. F., E. H. M. Melo, and K. Jumel. 1990. Immobilized enzymes and cells. Chem. Eng. Prog. 86:81-89. [Google Scholar]
- 26.Kumar, A., I. Y. Galaev, and B. Mattiasson. 2000. Polymer displacement/shielding in protein chromatography. J. Chromatogr. B 741:103-113. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Liu, H. L., Y. Doleyres, P. M. Coutinho, C. Ford, and P. J. Reilly. 2000. Replacement and deletion mutations in the catalytic domain and belt region of Aspergillus awamori glucoamylase to enhance thermostability. Protein Eng. 13:655-659. [DOI] [PubMed] [Google Scholar]
- 29.Luo, S., G. Kim, and R. L. Levine. 2005. Mutation of the adenylylated tyrosine of glutamine synthetase alters its catalytic properties. Biochemistry 44:9441-9446. [DOI] [PubMed] [Google Scholar]
- 30.Mansfeld, J., G. Vriend, B. Van Den Burg, V. G. H. Eijsink, and R. Ulbrich-Hofmann. 1999. Probing the unfolding region in a thermolysin-like protease by site-specific immobilization. Biochemistry 38:8240-8245. [DOI] [PubMed] [Google Scholar]
- 31.Mateo, C., O. Abian, R. Fernández-Lafuente, and J. M. Guisán. 2000. Reversible enzyme immobilization via a very strong and nondistorting ionic adsorption on support-polyethylenimine composites. Biotechnol. Bioeng. 68:98-105. [DOI] [PubMed] [Google Scholar]
- 32.Mateo, C., J. M. Palomo, M. Fuentes, L. Betancor, V. Grazú, F. Lopez-Gallego, C. C. Pessela, A. Hidalgo, G. Fernández-Lorente, R. Fernández-Lafuente, and J. M. Guisán. 2006. Glyoxyl-agarose: a fully inert hydrophilic support for immobilization and high stabilization of proteins. Enzyme Microb. Technol. 39:274-280. [Google Scholar]
- 33.Matsumoto, K. 1993. Production of 6-APA, 7-ACA, and 7-ADCA by immobilized penicillin and cephalosporin amidases. Bioprocess Technol. 16:67-88. [PubMed] [Google Scholar]
- 34.Merino, E., P. Balbas, F. Recillas, B. Becerril, F. Valle, and F. Bolivar. 1992. Carbon regulation and the role in nature of the Escherichia coli penicillin acylase (pac) gene. Mol. Microbiol. 6:2175-2182. [DOI] [PubMed] [Google Scholar]
- 35.Nahalka, J., and P. Gemeiner. 2006. Thermoswitched immobilization—a novel approach in reversible immobilization. J. Biotechnol. 123:478-482. [DOI] [PubMed] [Google Scholar]
- 36.Olsson, I., U. B. Axiö-Fredriksson, M. Degerman, and B. Olsson. 1988. Fast horizontal electrophoresis. I. Isoelectric focusing and polyacrylamide gel electrophoresis using PhastSystem. Electrophoresis 9:16-22. [DOI] [PubMed] [Google Scholar]
- 37.Ovsejevi, K., C. Manta, and J. A. Carlsson. 1991. A new method for the reversible immobilization of thiol biomolecules based on solid phase bound thiosulfonates groups. Appl. Biochem. Biotechnol. 31:175-195. [Google Scholar]
- 38.Parmar, A., H. Kumar, S. S. Marwaha, and J. F. Kennedy. 2000. Advances in enzymatic transformation of penicillins to 6-aminopenicillanic acid (6-APA). Biotechnol. Adv. 18:289-301. [DOI] [PubMed] [Google Scholar]
- 39.Persson, M., L. Bulow, and K. Mosbach. 1990. Purification and site-specific immobilization of genetically engineered glucose dehydrogenase on thiopropyl-Sepharose. FEBS Lett. 270:41-44. [DOI] [PubMed] [Google Scholar]
- 40.Pessela, B. C. C., L. Betancor, F. Lopez-Gallego, R. Torres, G. M. Dellamora-Ortiz, N. Alonso-Morales, M. Fuentes, R. Fernández-Lafuente, J. M. Guisán, and C. Mateo. 2005. Increasing the binding strength of proteins to PEI coated supports by immobilizing at high ionic strength. Enzyme Microb. Technol. 37:295-299. [Google Scholar]
- 41.Ponder, J. W., and F. M. Richards. 1987. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 193:775-791. [DOI] [PubMed] [Google Scholar]
- 42.Rajendhran, J., and P. Gunasekaran. 2004. Recent biotechnological interventions for developing improved penicillin G acylases. J. Biosci. Bioeng. 97:1-13. [DOI] [PubMed] [Google Scholar]
- 43.Rocchietti, S., A. S. V. Urrutia, M. Pregnolato, A. Tagliani, J. M. Guisán, R. Fernández-Lafuente, and M. Terreni. 2002. Influence of the enzyme derivative preparation and substrate structure on the enantioselectivity of penicillin G acylase. Enzyme Microb. Technol. 31:88-93. [Google Scholar]
- 44.Rosevear, A. 1984. Immobilized biocatalysts—a critical review. J. Chem. Technol. Biotechnol. 34B:127-150. [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 47.Sio, C. F., and W. J. Quax. 2004. Improved beta-lactam acylases and their use as industrial biocatalysts. Curr. Opin. Biotechnol. 15:349-355. [DOI] [PubMed] [Google Scholar]
- 48.Valle, F., P. Balbas, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 16:36-40. [DOI] [PubMed] [Google Scholar]
- 49.Van De Sandt, E. J. A. X., and E. De Vroom. 2000. Innovations in cephalosporin and penicillin production: painting the antibiotics industry green. Chim. Oggi 18:72-75. [Google Scholar]
- 50.Van Langen, L. M., N. H. P. Oosthoek, D. T. Guranda, F. Van Rantwijk, V. K. Svedas, and R. A. Sheldon. 2000. Penicillin acylase-catalyzed resolution of amines in aqueous organic solvents. Tetrahedron Asymmetry 11:4593-4600. [Google Scholar]
- 51.Waldmann, H., A. Heuser, and S. Schulze. 1996. Selective enzymatic removal of protecting groups: the phenylacetamide as amino protecting group in phosphopeptide synthesis. Tetrahedron Lett. 37:8725-8728. [Google Scholar]
- 52.Xu, Y., M. Y. Hsieh, N. Narayanan, W. A. Anderson, J. M. Scharer, M. Moo-Young, and C. P. Chou. 2005. Cytoplasmic overexpression, folding, and processing of penicillin acylase precursor in Escherichia coli. Biotechnol. Prog. 21:1357-1365. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, W., Y. Liu, H. Zheng, S. Yang, and W. Jiang. 2005. Improving the activity and stability of GL-7-ACA acylase CA130 by site-directed mutagenesis. Appl. Environ. Microbiol. 71:5290-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]