Abstract
A new bacterial strain, displaying potent antimicrobial properties against gram-negative and gram-positive pathogenic bacteria, was isolated from food. Based on its phenotypical and biochemical properties as well as its 16S rRNA gene sequence, the bacterium was identified as Paenibacillus polymyxa and it was designated as strain OSY-DF. The antimicrobials produced by this strain were isolated from the fermentation broth and subsequently analyzed by liquid chromatography-mass spectrometry. Two antimicrobials were found: a known antibiotic, polymyxin E1, which is active against gram-negative bacteria, and an unknown 2,983-Da compound showing activity against gram-positive bacteria. The latter was purified to homogeneity, and its antimicrobial potency and proteinaceous nature were confirmed. The antimicrobial peptide, designated paenibacillin, is active against a broad range of food-borne pathogenic and spoilage bacteria, including Bacillus spp., Clostridium sporogenes, Lactobacillus spp., Lactococcus lactis, Leuconostoc mesenteroides, Listeria spp., Pediococcus cerevisiae, Staphylococcus aureus, and Streptococcus agalactiae. Furthermore, it possesses the physico-chemical properties of an ideal antimicrobial agent in terms of water solubility, thermal resistance, and stability against acid/alkali (pH 2.0 to 9.0) treatment. Edman degradation, mass spectroscopy, and nuclear magnetic resonance were used to sequence native and chemically modified paenibacillin. While details of the tentative sequence need to be elucidated in future work, the peptide was unequivocally characterized as a novel lantibiotic, with a high degree of posttranslational modifications. The coproduction of polymyxin E1 and a lantibiotic is a finding that has not been reported earlier. The new strain and associated peptide are potentially useful in food and medical applications.
The emergence of bacterial antibiotic resistance (1, 24, 27) has mobilized the search for new potent antimicrobial agents. Although much of the resistance was observed in hospital environments and related nosocomial infections, there is increasing evidence that resistant food-borne pathogens evolved due to antibiotic use in animal feed (37, 44). Antimicrobial resistance phenotypes have been recognized in many zoonotic food-transmitted pathogens, including Salmonella spp., Campylobacter spp., Listeria spp., Escherichia coli O157:H7, and Yersinia spp. (5, 7, 37, 44). Consequently, there is a strong need for new antimicrobials that have suitable pharmacokinetic properties and safety profiles, with activity against these resistant pathogens (24). Similarly, potent antimicrobials of natural sources are needed for food applications.
Lantibiotics are group I bacteriocins that undergo posttranslational modification (6). These modifications generate dehydrated amino acids, i.e., α,β-didehydroalanine (Dha) and α,β-didehydrobutyric acid (Dhb) and thioether bridges of lanthionine (Lan) and β-methyllanthionine (MeLan), as well as some other less frequently encountered modifications (6). These modified residues are believed to stabilize molecular conformations that are essential for the antimicrobial activity of lantibiotics and their resistance to proteases of the producing strains (3, 29, 41). It is noteworthy that the lantibiotics produced by lactic acid bacteria have been tested as biopreservatives in a number of food products (10-12, 26), with nisin being the most prominent member of these bacteriocins. For decades, nisin has been used worldwide as a food additive, and it is the only lantibiotic approved by the World Health Organization as a food preservative (10, 12, 19, 26). However, the solubility and efficacy of nisin are highly pH dependent (9); therefore, the bacteriocin is only useful as a preservative in acidic foods (9-11). In addition, nisin is generally inactive against gram-negative bacteria, imposing a limitation on its usage against important food-borne pathogens, such as E. coli, Salmonella spp., Campylobacter spp., and Yersinia spp. (6, 12). In fact, bacteriocins with activity against gram-negative bacteria are scarcely reported (12).
The goal of this study was to search for novel microbial strains with potent antimicrobial activity against emerging food-transmitted pathogens. In this work, we report the isolation and identification of a new food-borne bacterial strain, OSY-DF, which produces dual antimicrobial agents and exhibits activity against food-borne pathogenic and spoilage bacteria, covering both types of Gram reactions. A bacteriocin-like substance, derived from strain OSY-DF, was purified, and its tentative primary structure was determined by tandem mass spectroscopy (MS/MS) and nuclear magnetic resonance (NMR) spectroscopy.
MATERIALS AND METHODS
Strain screening.
Fermented foods, including vegetables (kimchee, a Korean-style fermented vegetable), soybean sauce, and imported cheeses made of raw or pasteurized milk (gorgonzola, le lingot, roquefort, le brovere, and roncal), were purchased from local food stores (Columbus, OH) and screened for microorganisms that produce antimicrobial agents. Briefly, food samples were suspended in 0.1% sterile peptone water and homogenized using a blender or a stomacher. The suspensions were serially diluted and passed through a hydrophobic grid-membrane filter with a pore size of 0.45 μm (ISO-GRID; Neogen Corporation, Lansing, MI). Bacteria and fungi retained on the membranes were grown into colonies by mounting the membranes onto tryptose agar and acidified potato dextrose agar (Difco, BD Diagnostic Systems, Sparks, MD), respectively, and incubating at 30°C for 48 h. To rule out any false-positive inhibition caused by acid production, the basal media were supplemented with 0.6% CaCO3. The colony-carrying membranes were then removed and held in reserve in sterile petri dishes at 4°C. The incubated agar plates, left after removing the membranes, were overlaid with a soft agar medium seeded with Escherichia coli K-12. The medium consisted of tryptic soy broth supplemented with 0.6% yeast extract (TSBYE) and 0.75% agar. The overlaid plates were incubated at 37°C for an additional 12 h to manifest inhibition areas. Isolates corresponding to inhibition areas were located on the membrane filter and streaked onto tryptose agar plates. A sample of kimchee yielded a bacterium (OSY-DF) that produces potent antimicrobial agents.
Cultures and media.
The isolated bacterial strain, OSY-DF, was propagated on tryptic soy agar supplemented with 0.6% yeast extract (TSAYE) at 30°C. For stock preparation, the culture was cultivated overnight at 30°C in TSBYE mixed with sterile glycerol (final concentration of 20%) and stored at −80°C. The indicator strains and media used in this study are listed in Table 1.
TABLE 1.
Antimicrobial activity of Paenibacillus polymyxa OSY-DF culture supernatant and purified paenibacillin
| Gram reaction and straina | Broth mediumb | Antimicrobial activity
|
|
|---|---|---|---|
| Culture supernatant | Paenibacillin | ||
| Gram-negative bacteria | |||
| Escherichia coli K-12 | TSBYE | + | − |
| E. coli O157:H7 ATCC 43889 | TSBYE | + | − |
| E. coli O157:H7 EDL-933 (mutant) | TSBYE | + | − |
| E. coli O157:H7 EDL-933 (wild type) | TSBYE | + | − |
| E. coli O157:H12 | TSBYE | + | − |
| Salmonella enterica serovar Enteritidis | TSBYE | + | − |
| Pseudomonas putida | TSBYE | + | − |
| S. enterica serovar Typhimurium OSU 228 | TSBYE | + | − |
| S. enterica serovar Typhimurium DT 109 | TSBYE | + | − |
| S. enterica serovar Typhimurium FM 12501-51 | TSBYE | + | − |
| Yersinia enterocolitica OSU 602 | TSBYE | + | − |
| Gram-positive bacteria | |||
| Bacillus cereus ATCC 14579 | TSBYE | + | + |
| Bacillus subtilis ATCC 6633 | TSBYE | + | + |
| Clostridium sporogenes OSU 392 | TSBYE | + | + |
| Lactobacillus acidophilus ATCC 19992 | MRS | + | + |
| Lactobacillus casei ATCC 7469 | MRS | + | + |
| Lactobacillus plantarum ATCC 8014 | MRS | + | + |
| Lactococcus lactis ATCC 11454 | MRS | + | + |
| Leuconostoc mesenteroides | MRS | + | + |
| Listeria innocua ATCC 33090 | TSBYE | + | + |
| L. monocytogenes OSY-8578 | TSBYE | + | + |
| L. monocytogenes Scott A | TSBYE | + | + |
| Pediococcus cerevisiae | MRS | + | + |
| Staphylococcus aureus | TSBYE | + | + |
| Streptococcus agalactiae OSU 602 | TSBYE | + | + |
Strains were obtained from the culture collection of the Ohio State University Food Safety Laboratory.
TSBYE, tryptic soy broth supplemented with yeast extract; MRS, lactobacillus MRS broth.
Phenotypic and biochemical characterizations of the OSY-DF isolate.
The morphological characteristics of OSY-DF were observed by Gram staining, spore staining, and scanning electron microscopy examination. For scanning electron microscopy observation, the strain was grown on TSAYE at 30°C for 48 h. The resulting colonies, on the plate, were fixed with 3.0% glutaraldehyde (vol/vol) in 0.1 M phosphate buffer (pH 7.4) for 3.5 h. Subsequently, the agar surface was rinsed three times (15 min each) with the same buffer to remove traces of glutaraldehyde fixative. The agar area carrying bacteria was excised, postfixed, and dehydrated following a procedure described elsewhere (18). Bacterial cells were sputter coated with gold-palladium and examined in a scanning electron microscope at 30 kV (Philips XL-30; FEI, Inc., Hillsboro, OR).
Analyses for the biochemical properties of OSY-DF included catalase, oxidase, and urease reactions, nitrate reduction, gelatin liquefaction, starch hydrolysis, glucose fermentation, esculin hydrolysis, indole production, and H2S formation. In addition, the carbohydrate fermentation pattern of the OSY-DF isolate was determined using a biochemical test kit (API 50CH strips and API CHB/E medium; BioMerieux, Inc., Durham, NC). The results were checked after incubating OSY-DF at 30°C for 24 and 48 h, and the identification was done by referring to the database provided by the kit manufacturer.
16S rRNA gene amplification, cloning, and sequencing.
Genomic DNA of the OSY-DF isolate was prepared by suspending two to three colonies from a 24-h culture on TSAYE in 100 μl double-distilled water and boiling for 20 min. A pair of high-performance liquid chromatography (HPLC)-grade universal primers specific for bacterial 16S rRNA, fD1 and rD1 (42), were used to amplify the corresponding gene. Amplification by PCR involved using a Taq DNA polymerase kit (QIAGEN, Valencia, CA) under the following conditions: after an initial 3-min incubation at 95°C, the mixture was subjected to 30 cycles, each including 1 min at 95°C, 30 s at 52°C, and 2 min at 72°C. A final extension was performed at 72°C for 10 min. The amplified 16S rRNA was purified using a commercial DNA extraction kit (QIAquick gel extraction kit; QIAGEN), ligated to pGEM-T Easy vector (Promega Corporation, Madison, WI), and transformed into E. coli DH5α cells via electroporation. The recombinant plasmid was harvested from a 5-ml overnight culture in LB medium using silica spin columns (QIAprep Spin Miniprep kit; QIAGEN) and sequenced (3730 DNA Analyzer; Applied Biosystems, Foster City, CA) using T7 terminator and SP6 promoter primers. The derived 16S rRNA gene sequence (∼1.5 kb) was compared to known bacterial sequences in the NCBI GenBank using BLAST. Only results from the highest-score queries were considered for phylotype identification, with 98% minimum similarity (33).
Isolation of antimicrobial agents from fermentation broth.
A single colony of OSY-DF was subcultured into 10 ml TSBYE and incubated at 30°C for 24 h. The resulting culture was used to inoculate a 2-liter flask containing 500 ml TSBYE. The flask was incubated at 30°C for 24 h in a rotary shaker (New Brunswick Scientific, Edison, NJ) with agitation at 195 rpm. Cells in the fermentation broth were separated by centrifugation at 12,000 × g for 20 min. The resulting cell-free supernatant was μ-filtered (0.45-μm-pore-size filter; Millipore) and mixed with Amberlite XAD-7 resin (Sigma, St. Louis, MO) at a 10% level, and the mixture was maintained at 4°C for 24 h with stirring to allow maximum adsorption. The resin, with adsorbed antimicrobials, was collected by filtration and washed sequentially with 2 liters distilled water and 1 liter 30% (vol/vol) ethanol. The resin was resuspended in 250 ml ethanol (75% [vol/vol]; pH 2.0) and maintained at 25°C for 4 h with agitation followed by filtration. The resulting ethanol fraction, which contained the antimicrobial agents, was condensed by a rotary evaporator at 35°C under vacuum, and the concentrate was freeze dried. The generated powder (approximately 0.5 g) was reconstituted in 5 ml distilled water followed by centrifugation. The resulting supernatant, herein referred to as the antimicrobial crude extract (CE), contained 2 × 105 arbitrary units (AU)/ml as determined by the bioassay method described below.
Antimicrobial activity determination.
A qualitative and quantitative bioassay for antimicrobial potency was done using the spot-on-lawn method. An indicator lawn was prepared by pouring 10 ml soft agar (seeded with 200 μl overnight indicator culture) onto tryptose agar as a basal medium, in a petri dish. Escherichia coli K-12 and Lactobacillus plantarum ATCC 8014 were generally used as the gram-negative and gram-positive sensitive indicators, respectively, but other bacteria were tested to determine the OSY-DF antimicrobial spectrum (Table 1). For qualitative tests, aliquots (10 μl) of cell-free culture supernatant (pH 6.5) were spotted on indicator lawns, and the plates were incubated overnight for inhibition area observation. A clear inhibition area of ≥3 mm in diameter was recorded as positive. For the quantitative measurements, the cell-free culture supernatant was twofold serially diluted and dilutions were spotted onto the indicator lawn as just described. Antimicrobial activity was expressed in AU/ml; these values are the reciprocal of the highest dilution displaying a clear zone of inhibition that corresponds to 1 ml of the nondiluted supernatant.
Separation and purification by HPLC.
The HPLC system consisted of a pump (model SP8800; Thermo Separation Products, Fremont, CA), UV-Vis monitor (model 1706; Bio-Rad Laboratories, Milford, MA), and an integrator (HP 3396 series III; Hewlett-Packard). Separation was achieved using an ether-linked phenyl-based reversed-phase, 250- by 2.0-mm column with 4-μm particle size (Phenomenex Synergi; Phenomenex, Torrance, CA). The mobile phase consisted of (i) methanol and (ii) HPLC-grade water containing 0.1% trifluoroacetic acid (TFA). A 30-μl aliquot of CE was loaded and separated on the column by a linear biphasic gradient of 20 to 40% methanol over 10 min (2% methanol/min), 40 to 60% over 5 min (4% methanol/min), and 60 to 70% over 10 min (1% methanol/min) at a flow rate of 0.3 ml/min. Elution was monitored at a wavelength of 220 nm, and fractions were collected manually for the antimicrobial activity bioassay. Fractions that exhibited antimicrobial activity, at a given retention time, were collected from different HPLC runs, pooled, and lyophilized. Powder from pooled anti-gram-positive fractions was reconstituted and repurified using the same HPLC conditions described earlier. Collected fractions from multiple HPLC runs were lyophilized again, and the resulting powder was checked for efficacy against gram-positive bacteria. The antimicrobial agent in this powder will be referred to as the OSY-DF peptide.
Sensitivity to heat, pH, and degradative enzymes.
Crude extracts of Paenibacillus polymyxa OSY-DF and the HPLC-purified OSY-DF peptide were readily soluble in neutral water. The purified peptide was tested for sensitivity to heat, pH changes, and degradative enzymes. The qualitative spot-on-lawn bioassay was used to monitor the changes in antimicrobial potency after these treatments. For thermal stability testing, aliquots of OSY-DF peptide solution were exposed to 25, 30, 37, 50, and 70°C for 72 h and 120°C for 5 min. For the pH stability test, solutions of OSY-DF peptide were adjusted to pH 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, and 9.0, followed by incubation at 25°C for 2 h. The residual antimicrobial activity was assessed after neutralizing the sample to pH 6.5.
Sensitivity of the OSY-DF peptide to various degradative enzymes was determined. Enzymes tested were α-chymotrypsin (48 U/mg), β-amylase (26.8 U/mg), bromelain (1.15 U/mg), ficin (0.22 U/mg), lipase (type I; 7.9 U/mg), papain (1.5 U/mg), protease (type XIII; 0.6 U/mg), and trypsin (10,700 U/mg). All enzymes were purchased from Sigma, and their solutions were prepared in 25 mM phosphate buffer, pH 7.0, each containing 1 mg/ml except lipase solution, which contained 0.1 mg/ml. Solutions of the antimicrobial peptide were prepared in the same buffer. All stock solutions were separately sterilized by filtrating through low-protein binding filter (MILLEX-GV 0.22-μm filter unit; Millipore, Carrigtwohill, County Cork, Ireland). The stock solutions of OSY-DF peptide and enzymes were mixed at a 1:1 ratio (vol/vol) and incubated at 37°C for 1 h before residual antimicrobial activity measurement.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on the OSY-DF peptide using a 16.5%, 8- by 10-cm Tris-Tricine/peptide precast gel system (Ready Gel; Bio-Rad, Laboratories, Inc., Hercules, CA) (28, 32). After electrophoresis, one-half of the gel was stained with Coomassie blue G-250, while the other was washed three times, 15 min each, with sterile distilled water (45) and then overlaid with MRS soft agar seeded with L. plantarum ATCC 8014. The latter was examined for antimicrobial activity after overnight incubation at 30°C.
LC-MS.
The antimicrobial CE fraction was analyzed by liquid chromatography-MS (LC-MS) under the same conditions as described for the HPLC purification, except that 15 μl of sample was injected. A Micromass LCT (Wythenshawe, United Kingdom) with an orthogonal electrospray source (Z-spray) was coupled to the outlet of the HPLC using a T-splitter. Samples were infused into the electrospray source at a flow rate of about 20 μl/min. For optimum electrospray ionization conditions, capillary voltage was 3 kV, source temperature was 100°C, and cone voltage was 50 V. Sodium iodide was used as an external mass calibration standard over the m/z range of 500 to 2,500. Data were acquired in continuum mode at the rate of 1 scan/s. All spectra were obtained in the positive ion mode.
N-terminal amino acid sequence determination.
The purified antimicrobial peptide was subjected to Edman degradation and analyzed by a protein sequencing system (model 494, Procise sequencing system; Applied Biosystems) at the Microchemistry and Proteomics Analysis Facility, Harvard University (Cambridge, MA) using standard protocols (8, 43).
Ni2B-based desulfurization/reduction of the antimicrobial peptide.
The peptide modification reaction was adapted from the methods of Martin et al. (25). A portion (∼0.5 mg) of the OSY-DF peptide was dissolved in a 70:30 (vol/vol) methanol-water solution containing 0.1% TFA. Ten mg NiCl2 (Sigma) was added to 1 ml peptide solution, and the suspension was stirred until the solution became clear. The resulting solution was transferred to a 1.5-ml screw-cap flask containing 10 mg NaBH4 (Sigma) and sealed rapidly. A black precipitate formed immediately (Ni2B), with evolution of hydrogen gas. The mixture was then stirred for 1 h at 50°C followed by centrifugation to separate Ni2B precipitate from the supernatant. The Ni2B precipitate was washed sequentially by (i) 0.5 ml methanol, (ii) water, and (iii) 70:30 methanol-water (all solvents contained 0.1% TFA). Each washing was followed by centrifugation and decanting the wash solution. All the decanted wash solutions were analyzed as described below.
Tryptic digestion profile of modified antimicrobial peptide.
Sequencing-grade trypsin (Promega) was added to the modified OSY-DF peptide in 100 mM NH4HCO3 buffer (pH 8.0). The mixture, with a 1:25 enzyme-substrate ratio (wt/wt), was incubated at 37°C for 16 h before quenching by adding 0.1% TFA. Samples were then desalted with a peptide desalting trap (Michrom BioResources Inc., Auburn, CA) before mass spectrometric analysis.
MALDI-TOF analysis.
The OSY-DF peptide and its chemically modified derivate were subjected to matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis. The matrix, α-cyano-4-hydroxy cinnamic acid, was prepared as a saturated solution in 50% acetonitrile with 0.1% TFA in water. Aliquots consisting of 5 μl matrix and 1 μl sample were thoroughly mixed, spotted (1.0 μl) on the target plate, and allowed to air dry. Analysis was performed on a Bruker Reflex III time-of-flight mass spectrometer (Bruker Daltonics Inc., Billerica, MA) operated in reflection positive ion mode at an accelerating voltage of 28 kV. The N2 laser was operated at the minimum threshold level required to generate signal and minimize dissociation.
Quadrupole-time of flight MS/MS.
Detailed sequence information of the native and reduced OSY-DF peptide was further investigated on a Micromass Q-Tof II apparatus (Micromass, Wythenshawe, United Kingdom) equipped with an orthogonal electrospray source (Z-spray) and operated in positive ion mode. For external mass calibration, NaI was used over the m/z range of 200 to 2,500. The antimicrobial peptide, dissolved in the mixture of H2O-CH3OH-HAc (50:50:2.5), was infused into the electrospray source at a 2-μl/min flow rate. To achieve the optimal electrospray, capillary voltage was set at 3,000 V, source temperature was 150°C, and cone voltage was 60 V. The first quadrupole, Q1, was set to pass ions between 200 and 2,500 m/z. The target ion was isolated and fragmented within the second quadrupole by adding a voltage of between 20 and 40 V. The fragment ions were then analyzed in the time-of-flight tube. Data were acquired in continuum mode until well-averaged data were obtained.
NMR analysis.
Purified and lyophilized OSY-DF peptide (∼0.3 mg) was dissolved into 600 ml 99.9% deuterium oxide (D2O; Cambridge Isotope Lab., Andover, MA). One-dimensional 1H-NMR spectroscopy and two-dimensional 1H-homonuclear total correlation spectroscopy (TOCSY) were performed at 20°C on a Bruker DMX-600 spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) equipped with a triple resonance probe as well as three-axis gradient coils. The two-dimensional TOCSY experiment employed a DIPSI2 mixing sequence with the sensitivity enhancement feature (4). The spectral width and mixing time were 6,600 Hz and 60 ms, respectively. The data were recorded with 2,048 time-domain complex points, 210 increments in the indirectly detected dimension, and 96 scans per t1 increment. Data were processed using XWIN-NMR 3.1 software (Bruker). Briefly, the appropriate window function was applied on each dimension followed by Fourier transformation and baseline correction. Chemical shifts were referenced to the external standard, 2,2-dimethyl-2-silapentane-5-sulfonate.
RESULTS
Isolation and identification of an antimicrobial-producing strain from food.
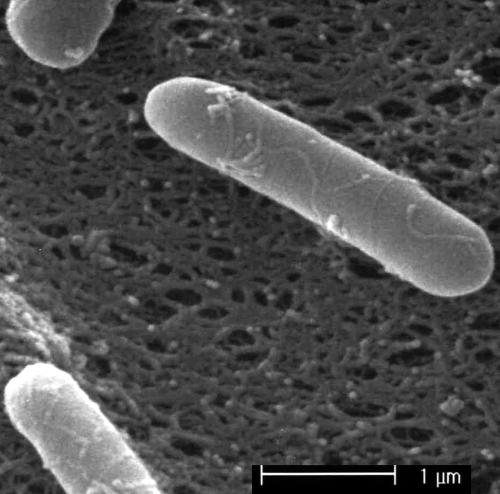
By applying a convenient hydrophobic grid-membrane-based method, a large number of food isolates were screened for antimicrobial activity against E. coli K-12. An isolate from kimchee (pH 4.05) showed a distinct inhibition area on basal tryptose agar. Culture supernatant of this isolate was active against several gram-positive and gram-negative bacteria (Table 1). This isolate formed pale colonies on TSAYE. Morphologically, the isolate was rod shaped, 0.6 by 3.0 μm (Fig. 1), gram-positive bacterium. The cell was motile with peritrichous flagella (data not shown). Upon prolonged incubation on agar medium, cells produced central endospores.
FIG. 1.
Scanning electron microscope observation of Paenibacillus polymyxa OSY-DF.
The isolate is positive for catalase, nitrate reduction, gelatin liquefaction, starch hydrolyzation, glucose fermentation, and esculin hydrolysis but negative for oxidase, urease, indole production, and H2S formation. The bacterium grew well in TSBYE and MRS broth under aerobic conditions. The isolate grew in medium supplemented with ethanol as the sole carbon source, and it further oxidized ethanol to acetic acid in a medium containing 7% ethanol (data not shown). Genomic analysis showed the 16S rRNA gene of the isolate shares >99.0% sequence similarity with that of Paenibacillus polymyxa. Carbohydrate fermentation analysis (Api 50 CH kit) confirmed the high similarity of the isolate (>99%) with P. polymyxa. Thus, it was concluded that the isolate belongs to P. polymyxa, and it was given the strain designation OSY-DF.
Antimicrobial spectrum of P. polymyxa OSY-DF culture supernatant.
When tested against a panel of gram-negative and gram-positive bacteria, OSY-DF cell-free culture supernatant (CFCS) exhibited a broad spectrum of antimicrobial activity. All pathogenic bacteria tested in this study were sensitive to the CFCS of OSY-DF; these are Escherichia coli 0157:H7 (three strains), Salmonella enterica serovar Enteritidis, S. enterica serovar Typhimurium (four strains, including the multidrug-resistant DT109 and FM 12501-51), Yersinia enterocolitica, Bacillus cereus, Listeria monocytogenes (three strains, including the processing-resistant OSY-8578), and Staphylococcus aureus (Table 1). However, the CFCS of OSY-DF had no activity against fungi (data not shown).
Isolation, purification, and characterization of antibacterial substances produced by P. polymyxa OSY-DF.
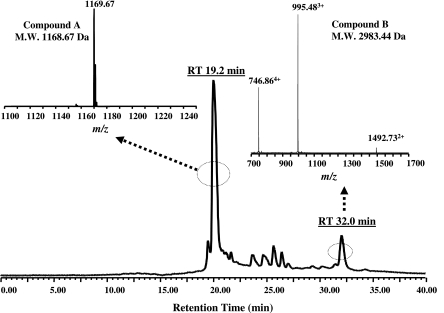
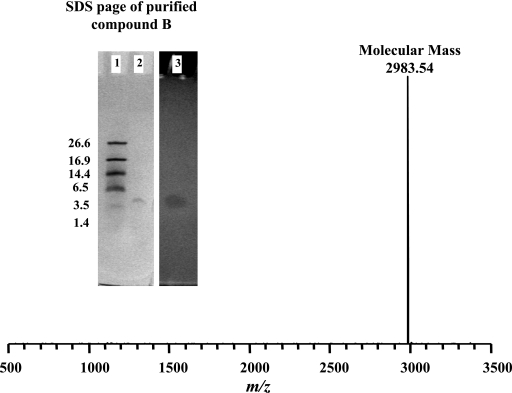
Several commercially available microbiological media were tested for supporting the growth and production of antimicrobials by the OSY-DF strain. Among these media, TSBYE supported the highest antimicrobial potency (1,600 AU/ml) when the culture supernatant was tested against E. coli K-12 or L. plantarum ATCC 8014. Extraction of the antimicrobial substances from OSY-DF fermentate was achieved using Amberlite XAD-7 adsorbent, a nonionic macro-reticular resin that adsorbs and releases ionic species through hydrophobic and polar interactions. By applying XAD-7 resin to cell-free culture supernatant, the antimicrobial substances were selectively adsorbed, whereas most other water-soluble components remained in the liquid phase. The antimicrobial substances were eluted from XAD-7 by 75% ethanol, and the resulting fraction was freeze-dried to a CE powder, which retained most of the antimicrobial activity. Components of CE were separated further by HPLC, using a specialized column. In the HPLC profile, fractions corresponding to two peaks with retention times (RT) of 20 min and 32 min were active against E. coli K-12 and L. plantarum ATCC 8014, respectively (Fig. 2). These results suggest that OSY-DF produces dual antimicrobial compounds with different antimicrobial spectra. The chemical nature of OSY-DF antimicrobial compounds was elucidated by LC-MS (Fig. 3). This analysis produced a chromatographic profile similar to that observed earlier in the HPLC results, except for minor shifts in the RT, which may have been caused by the reduced loading volume (from 30 to 15 μl) and differences in system void volumes. Data from LC-MS analysis showed that the fraction corresponding to a 19.2-min RT contains a pure compound (compound A) with a molecular mass of 1,168.67 Da. Association of this fraction with the anti-gram-negative activity of the OSY-DF cell extract was confirmed by the bioassay. Subsequent MS/MS analysis of compound A showed a fragmentation pattern identical to that of polymyxin E1 (see the supplemental material), an antibiotic that is specifically active against gram-negative bacteria. Another compound in the 32.0-min RT fraction (compound B) had a molecular mass of 2,983.44 Da, and it was active against gram-positive bacteria. Compound B was further purified to homogeneity by using the established HPLC procedures, and it was analyzed by MALDI-TOF MS for purity and molecular mass verification. Results of this analysis (Fig. 4) proved this HPLC-purified agent contained only one compound with identical molecular mass to the one obtained earlier in the LC-MS experiment. When compound B was subjected to SDS-PAGE, only a single band (∼3,000 Da) was detected (Fig. 4), confirming the high purity of the antimicrobial compound in the sample. The unstained half of the gel was overlaid with soft agar seeded with L. plantarum ATCC 8014; this produced an inhibition zone that corresponded to the band observed in the stained half (Fig. 4). This SDS-PAGE experiment and the subsequent analysis by proteolytic enzymes, as described later, confirmed the proteinaceous nature of compound B; this peptide was designated as paenibacillin.
FIG. 2.
High-performance liquid chromatography profile of a crude extract of Paenibacillus polymyxa OSY-DF culture supernatant. Fraction A has activity against Escherichia coli K-12; fraction B has activity against Lactobacillus plantarum ATCC 8014.
FIG. 3.
LC-MS profile of a crude extract of Paenibacillus polymyxa OSY-DF culture supernatant.
FIG. 4.
Verification of molecular mass and purity of compound B, as determined by MALDI-TOF MS analysis (main figure) and SDS-PAGE (figure insert). Lanes 1 and 2, molecular mass marker (in kilodaltons) and purified paenibacillin, stained with Coomassie blue G-250; lane 3, purified paenibacillin, overlaid with soft agar containing Lactobacillus plantarum ATCC 8014 after incubation.
Antimicrobial activity and stability of paenibacillin.
Purified paenibacillin was active against a panel of food-borne gram-positive pathogenic and spoilage bacteria, including Bacillus spp., Clostridium sporogenes, Lactobacillus spp., Lactococcus lactis, Lactococcus mesenteroides, Listeria spp., Pediococcus cerevisiae, S. aureus, and Streptococcus agalactiae, but it was inactive against gram negatives (Table 1). Although paenibacillin targets gram-positive organisms only, it has a considerably broad antimicrobial spectrum within this group of bacteria. The purified paenibacillin was also tested for sensitivity to changes in pH and temperature. Paenibacillin retained most of its antimicrobial activity, as judged by the results of the spot-on-lawn bioassay, when (i) held at 30, 37, 50, or 75°C for 3 days, (ii) autoclaved at 121°C for 5 min, or (iii) subjected to different pH values from 2.0 to 9.0 (data not shown). Purified paenibacillin lost its activity totally and partially when digested with ficin and trypsin, respectively (data not shown); this provides additional evidence for its proteinaceous nature. However, the antimicrobial activity of paenibacillin was not affected by β-amylase or lipase, implying that the compound is a pure peptide, without polysaccharide or lipid moieties.
Amino acid sequencing of native paenibacillin.
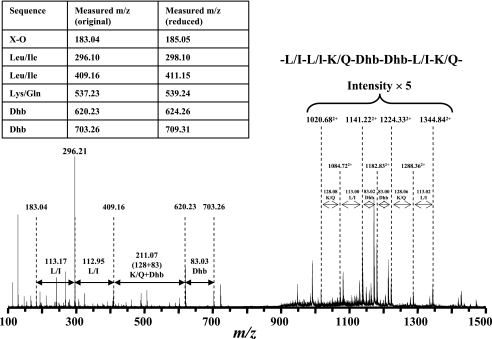
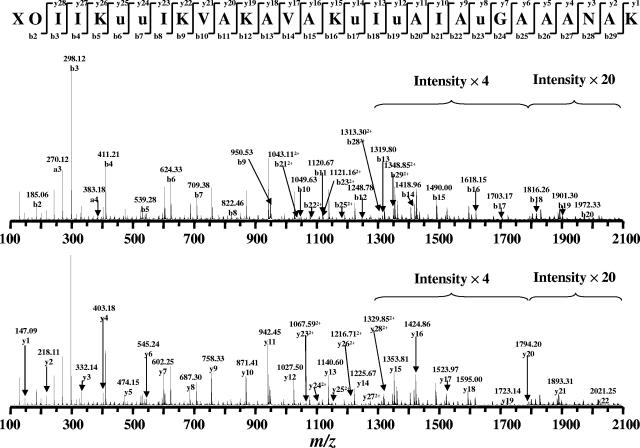
No amino acid residues were detected by direct N-terminal amino acid sequencing (Edman degradation) of paenibacillin. Consequently, it was hypothesized that the N terminal of the peptide is blocked by an unusual structure. Sequencing the native peptide using MS/MS analysis was only partially successful. As shown in Fig. 5, little cleavage was observed between 720 and 1,200 (m/z), suggesting the presence of an intramolecular thioether bridge (Lan and/or MeLan), a common feature among lantibiotics; such a bridge may have impeded the fragmentation of native paenibacillin during the MS/MS analysis. Nevertheless, a partial sequence was revealed: Leu/Ile-Leu/Ile-Lys/Gln-Dhb-Dhb-Leu/Ile-Lys/Gln or, in the reverse order, Lys/Gln-Leu/Ile-Dhb-Dhb-Lys/Gln-Leu/Ile-Leu/Ile, in which Leu and Lys could not be differentiated from Ile and Gln, respectively, due to the same (Leu/Ile) or virtually identical (Lys/Gln) masses (Fig. 5). Analysis by NMR also confirmed the presence of Dhb residues (Fig. 6), which were readily identified by the unique quartet peaks of Hβ at ∼6.7 ppm in the 1D 1H NMR spectroscopy and the through-bond cross-peaks between Hβ and Hγ (∼1.80 ppm) in the TOCSY experiment (38, 39). Although limited information was obtained on the native paenibacillin, the observation of a fragment containing Dhb tandem, together with crude information regarding the several flanking residues, is sufficiently unique to claim that the peptide is a novel lantibiotic. This conclusion is further strengthened in the following work on the chemically modified peptide.
FIG. 5.
MS/MS sequencing of native paenibacillin. The partial sequence was identified as -Leu/Ile-Leu/Ile-Lys/Gln-Dhb-Dhb-Leu/Ile-Lys/Gln-. Leu/Ile and Lys/Gln cannot be differentiated due to the identical and similar molecular masses, respectively.
FIG. 6.
NMR evidence for the existence of two Dhb residues in paenibacillin. The expansions of NMR spectra show the unique quartet peaks of Hβ at ∼6.7 ppm in one-dimensional 1H-NMR spectroscopy (top) and the through-bond cross-peaks between Hβ and Hγ (∼1.80 ppm) (left) in the TOCSY experiment (middle).
Elucidating the paenibacillin sequence after chemical modification and enzyme digestion.
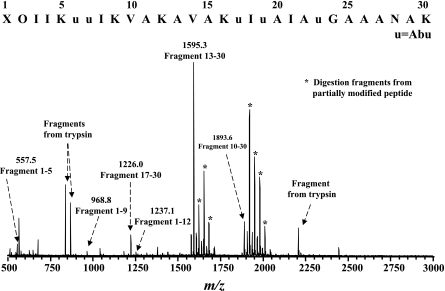
The newly reported Ni2B-based desulfurization/reduction approach (25), which converts an intralinked lantibiotic into a linear structure, significantly facilitated the sequencing of paenibacillin by MS/MS analysis. In such a treatment, a Lan is converted into two Ala and MeLan is converted into Ala and Abu, while Dha and Dhb are reduced to Ala and Abu, respectively. The method has been adapted and successfully applied to paenibacillin, and the following sequential information was obtained for this chemically modified form: X-O-Leu/Ile-Leu/Ile-Lys/Gln-Abu-Abu-Leu/Ile-Lys/Gln-Val-Ala-Lys/Gln-Ala-Val-Ala-Lys/Gln-Abu-Leu/Ieu-Abu-Ala-Leu/Ile-Ala-Abu-Gly-Ala-Ala-Ala-Asn-Ala-Lys/Gln, in which X and O are yet undetermined and ambiguities still remained regarding the Leu/Ile and Lys/Gln identification (Fig. 7; Table 2). After digestion of modified paenibacillin with trypsin, which specifically cleaves at Arg and Lys but not Gln, all Lys residues but one at the C terminus were determined (Fig. 8). The desulfurization/reduction reaction was repeated with a deuterated reducing agent, sodium borodeuteride (NaBD4), followed by MS/MS analysis. Since two deuterium atoms were added to the double bonds in both Dha and Dhb, whereas only one deuterium atom was added to either Abu in MeLan or Ala in Lan, the mass shift of the isotope effect provides a means to differentiate the sources of Ala and Abu in the derivative peptide (25). The results of this analysis are summarized in Table 2. For example, a 2-Da mass shift was observed on Abu6, Abu7, and Ala27, leading to the confirmation and identification of Dhb6, Dhb7, and Dha27 in the native form. On the other hand, only a 1-Da mass shift was observed in Abu17, Abu19, Abu23, Ala11, Ala15, Ala20, Ala22, Ala25, Ala26, and Ala29, inferring the existence of three MeLan and two Lan thioether bridges. Lastly, the Edman degradation method was repeated on this chemically modified paenibacillin, but no amino acid residues were detected during this sequencing attempt. This result implies that the N terminus of paenibacillin is blocked for an unknown reason. However, the 2-Da mass shift, observed for the X-O fragment (183.05 Da in the native form versus 185.05 Da in the reduced form), suggests the presence of Dha, Dhb, or a variant of these residues in the N-terminal XO region (Table 2). Taken together, a tentative sequence with 15 modified residues (in italics) is proposed for the native paenibacillin: (X-Dha/Dhb)-Leu/Ile-Leu/Ile-Lys-Dhb-Dhb-Leu/Ile-Lys-Val-Ala-Lys-Ala-Val-Ala-Lys-Abu-Leu/Ile-Abu-Ala-Leu/Ile-Ala-Abu-Gly-Ala-Ala-Dha-Asn-Ala-Lys/Gln, where the highlighted Abu and Ala residues are engaged in the formation of Lan or MeLan thiother bridges.
FIG. 7.
MS/MS sequencing results of the modified paenibacillin; Lys 5, 9, 12, 16, and 30 cannot be differentiated from Gln due to the similar mass, whereas Ile in 3, 4, 8, 18, and 21 cannot be distinguished from Leu due to the identical mass.
TABLE 2.
Detailed MS/MS analysis of paenibacillin reduced with NaBH4 and NaBD4
| Δm between y-n (D) and y-n (H) | Fragment ion | Measured m/z
|
Sequencea | Measured m/z
|
Fragment ion | Δm between b-n (D) and b-n (H) | ||
|---|---|---|---|---|---|---|---|---|
| NaBD4 | NaBH4 | NaBD4 | NaBH4 | |||||
| X | ||||||||
| O (2) | 187.02 | 185.06 | b-2 | 1.96 | ||||
| y-28 | 1,329.852+ | Leu | 300.13 | 298.12 | b-3 | 2.01 | ||
| 16.02 | y-27 | 1,281.292+ | 1,273.282+ | Leu | 413.21 | 411.21 | b-4 | 2.00 |
| 16.06 | y-26 | 1,224.742+ | 1,216.712+ | Lys | 541.27 | 539.28 | b-5 | 1.99 |
| 15.98 | y-25 | 1,160.652+ | 1,152.662+ | Abu (2) | 628.33 | 624.33 | b-6 | 4.00 |
| 14.00 | y-24 | 1,117.122+ | 1,110.122+ | Abu (2) | 715.41 | 709.38 | b-7 | 6.03 |
| 12.06 | y-23 | 1,073.622+ | 1,067.592+ | Leu | 828.48 | 822.46 | b-8 | 6.02 |
| 12.01 | y-22 | 1,017.132+ | 2,021.25 | Lys | 956.53 | 950.53 | b-9 | 6.00 |
| y-21 | 1,893.31 | Val | 1,055.61 | 1,049.63 | b-10 | 5.98 | ||
| 11.98 | y-20 | 903.592+ | 1,794.20 | Ala (1) | 1,127.69 | 1,120.67 | b-11 | 7.02 |
| 11.00 | y-19 | 867.572+ | 1,723.14 | Lys | 1,255.77 | 1,248.78 | b-12 | 6.99 |
| 10.98 | y-18 | 803.492+ | 1,595.00 | Ala (0) | 663.892+ | 1,319.80 | b-13 | 6.98 |
| 10.96 | y-17 | 1,534.93 | 1,523.97 | Val | 713.462+ | 1,418.96 | b-14 | 6.96 |
| 11.02 | y-16 | 1,435.88 | 1,424.86 | Ala (1) | 749.492+ | 1,490.00 | b-15 | 7.98 |
| 10.95 | y-15 | 682.382+ | 1,353.81 | Lys | 813.562+ | 1,618.15 | b-16 | 7.97 |
| 10.01 | y-14 | 1,235.68 | 1,225.67 | Abu (1) | 856.572+ | 1,703.17 | b-17 | 8.97 |
| 9.01 | y-13 | 1,149.61 | 1,140.60 | Leu | 913.162+ | 1,816.26 | b-18 | 9.06 |
| 9.01 | y-12 | 1,036.52 | 1,027.50 | Abu (1) | 1,901.30 | b-19 | ||
| 8.01 | y-11 | 950.46 | 942.45 | Ala (1) | 992.142+ | 1,972.33 | b-20 | 10.95 |
| 7.01 | y-10 | 878.42 | 871.41 | Leu | 1,048.592+ | 1,043.112+ | b-21 | 10.96 |
| 7.01 | y-9 | 765.34 | 758.33 | Ala (1) | 1,078.642+ | b-22 | ||
| 6.00 | y-8 | 693.30 | 687.30 | Abu (1) | 1,127.692+ | 1,121.162+ | b-23 | 13.06 |
| 5.02 | y-7 | 607.27 | 602.25 | Gly | 1,149.682+ | b-24 | ||
| 5.01 | y-6 | 550.25 | 545.24 | Ala (1) | 1,192.202+ | 1,185.212+ | b-25 | 13.98 |
| 4.00 | y-5 | 478.21 | 474.21 | Ala (1) | 1,220.742+ | b-26 | ||
| 3.00 | y-4 | 406.18 | 403.18 | Ala (2) | 1,264.802+ | 1,256.272+ | b-27 | 17.06 |
| 0.99 | y-3 | 333.13 | 332.14 | Asn | 1,321.782+ | 1,313.302+ | b-28 | 16.96 |
| 1.00 | y-2 | 219.11 | 218.11 | Ala (1) | 1,348.852+ | b-29 | ||
| −0.01 | y-1 | 147.08 | 147.09 | Lys | ||||
The number in parentheses in the sequence column represents the mass shift caused by deuterium labeling on that specific residue: 1, a mass shift of 1 Da, suggesting this residue was from Lan or MeLan; 2, a mass shift of 2 Da, suggesting this residue is from Dha or Dhb.
FIG. 8.
MALDI-TOF MS analysis of the tryptic digestion product of modified paenibacillin, confirming the presence of Lys5, 9, 12, and 16, whereas Lys/Gln30 remains undetermined; Leu and Ile also remain undistinguishable.
DISCUSSION
A new bacterial strain, P. polymyxa OSY-DF, was isolated during a screening of fermented foods for microorganisms with potent antimicrobial activity. The isolate has antimicrobial efficacy against both gram-positive and gram-negative food-borne pathogenic bacteria. The identity of the isolate was determined after morphological, biochemical, and genetic analyses. The new strain uniquely produces two antimicrobial compounds concurrently: an anti-gram-negative antibiotic (polymyxin E1) and a new lantibiotic (group I bacteriocin) that is active against gram-positive bacteria.
Several strains of P. polymyxa are known to produce polymyxin antibiotics, a group of cyclic peptides with linear side chains. This group consists of five chemically different compounds, polymyxins A to E (14, 15, 17). Polymyxin E (colistin) has been used in clinical practice as a topical otic and ophthalmic solution for decades (15). In contrast, production of bacteriocins by P. polymyxa has scarcely been documented. A strain, P13, was found to produce polyxin, a bacteriocin-like peptide with activity against a wide range of gram-positive and gram-negative bacterial species, but no structural information was documented except that it has a molecular mass of 10 kDa (30). Another example is the recent discovery of a class IIa bacteriocin (molecular mass, 3,864 Da) from P. polymyxa NRRL-B-30509, which has been used for the control of Campylobacter spp. in poultry (34, 35). However, the coproduction of polymyxin and a lantibiotic by natural isolates has never been reported.
In this work, isolation and enrichment of two antimicrobial agents from P. polymyxa OSY-DF fermentate was accomplished by a one-step procedure involving adsorption to XAD resin. The coexistence of two compounds with complementary antimicrobial spectra in one preparation is potentially valuable in topical treatment, bio-control, feed additives, and other applications that aim at eradicating gram-positive and gram-negative pathogens or nonpathogenic contaminants in the targeted environment.
Polymyxin E1 is known to be active against Pseudomonas aeruginosa and Acinetobacter baumannii (15, 36). The compound was commercially released as early as 1959 but was subsequently relegated to use as a second-line antibiotic because of early reports about its potential toxicity (15). However, new evidence shows that polymyxins, particularly polymyxin E1, have less toxicity than previously thought. Recent emergence of multidrug-resistant gram-negative pathogens, such as P. aeruginosa, Salmonella spp., and Acinetobacter spp., has become a major clinical problem (2, 21-23). Some of these pathogens (e.g., A. baumannii, a pathogen causing bloodstream infections in military medical facilities) have developed substantial antimicrobial resistance, and they are only susceptible to polymyxins (36). The scarcity of newly introduced antibiotics against resistant gram-negative bacteria and the recent confirmation of polymyxin safety have favored the use of this antibiotic in the therapy of multidrug-resistant gram-negative bacterial infections (21). Paenibacillus polymyxa OSY-DF copiously produced only one type of polymyxin; thus, the strain may be used industrially to synthesize a high-purity polymyxin E1.
The newly discovered antimicrobial agent, paenibacillin, belongs to the group I bacteriocins (lantibiotics). At the present, about 40 different lantibiotics have been reported; among these, nisin, subtilin, and ericin S undergo the most posttranslational modifications, each having 13 modified residues (6). Despite a lack of detailed sequencing in the current work, 15 modified residues are suggested, based on the deduced primary sequence of paenibacillin; thus, the antimicrobial peptide could be equally distinct in the extent of chain modification.
Paenibacillin exhibited a relatively broad antimicrobial spectrum, showing activity against a panel of gram-positive bacteria including spore and nonspore formers and pathogenic and spoilage bacteria. This peptide is quite stable at all temperatures tested in this study; it even retained its activity after a short autoclaving. These characteristics suggest the feasibility of using this peptide in preserving a wide range of foods or in pharmaceutical compositions that require heating during preparation. Unlike nisin, which is stable at low pH (pH 2.0) but loses activity sharply in the neutral pH region (9, 10, 31), paenibacillin is quite stable over a wide pH range, from 2.0 to 9.0. The new peptide is also easy to dissolve in water, and it has multiple positively charged residues (no less than four Lys). These features grant the OSY-DF peptide the potency and flexibility for formulations in food production or pharmaceutical applications.
Tandem mass spectrometry has been used extensively for elucidating primary structures of proteins and peptides. In lantibiotics, however, complete fragmentation is impeded by the presence of modified residues in general, and Lan/MeLan thioether bridges specifically. The Ni2B-based modification developed by Martin et al. (25) proved effective in this work to resolve the blockage and to generate a linear structure suitable for MS/MS sequencing. Furthermore, use of deuterium-labeled Ni2B-based desulfurization/reduction reagents helped in inferring the structural information and locations of Dha, Dhb, Lan, and MeLan in the native peptide. The successful application of this approach has generated supportive information about this peptide and facilitated proposing a tentative primary sequence using submilligram quantities of the peptide. However, this method does not reveal how the thioether bridges are linked in paenibacillin, since the sequencing is based on the fragmentation analysis rather than direct pairing analysis or correlation of the two moieties of each thioether bridge (25). In addition, two issues remain to be addressed: (i) the mass ambiguities, i.e., Leu/Ile isomers and Lys/Gln; and (ii) the N-terminal capping.
Interpretation of MS/MS data is sometimes challenging without ultrahigh mass accuracy. For example, a mass difference of 128 Da between two fragments can be interpreted as Lys (128.09496), Gln (128.05858), or Gly-Ala/Ala-Gly (128.05857). Although trypsin digestion can be used to single out the existence of Lys, as successfully applied in this study, it remains difficult to draw a conclusion when the lysine residue is followed by a proline or located at the C terminus. Currently, the last residue in the OSY-DF peptide could be Lys or Gln, but there is also a slight possibility that it is an Ala-Gly or Gly-Ala pair. Finally, when the peptide is bigger than 10 amino acids, the b1 ion sometimes cannot be observed, making the interpretation of the N terminus difficult, particularly when a yn-1 ion is not available.
In this study, we provided evidence for the novelty of paenbacillin and proposed a tentative sequence, but more efforts are needed to fully elucidate its primary structure. It is well known that NMR spectroscopy is an excellent alternative method for lantibiotic sequencing (13, 16, 20, 25, 38, 39, 40). Compared with MS/MS and Edman degradation, however, NMR spectroscopy requires a relatively larger amount of pure sample, especially when performing naturally abundant heteronuclear NMR experiments. Additionally, the method is time consuming in terms of data collection and analysis. Lack of a sufficient amount of pure sample has prevented us from applying a comprehensive NMR analysis to paenibacillin. All the sequence puzzles as well as the assignment of the thioether bridges should be resolved by more NMR experiments when a larger amount pure paenibacillin is available in the future.
In summary, the present study identifies a food isolate, P. polymyxa OSY-DF, with a promising broad antimicrobial spectrum. The strain coproduces polymyxin E1 and a lantibiotic, paenibacillin, a phenomenon that has never been reported. Analysis of the purified paenibacillin revealed a novel lantibiotic with attractive physico-chemical properties and considerable antimicrobial efficacy against gram-positive bacteria. Based on these findings, paenibacillin is potentially useful for food or medicinal applications.
Supplementary Material
Acknowledgments
In the experiment, the scanning electron microscopy observation was conducted at The Ohio State University Campus Microscopy and Imaging Facility; sequencing of plasmid DNA was accomplished at The Ohio State University Plant-Microbe Genomics Facility; and the Edman degradation sequencing was done in Harvard's Microchemistry & Proteomics Analysis facility. We thank Polly D. Courtney for assistance in 16S rRNA gene sequencing and Ben Jones (Mass Spectrometry and Proteomics Facility, Campus Chemical Instrument Center, Ohio State University) for performing tandem MS experiments.
Footnotes
Published ahead of print on 27 October 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amabile-Cuevas, C. F. 2003. New antibiotics and new resistance. Am. Sci. 91:138-149. [Google Scholar]
- 2.Berlana, D., J. M. Llop, E. Fort, M. B. Badia, and R. Jodar. 2005. Use of colistin in the treatment of multiple-drug-resistant gram-negative infections. Am. J. Health Syst. Pharm. 62:39-47. [DOI] [PubMed] [Google Scholar]
- 3.Bierbaum, G., C. Szekat, M. Joste, C. Heidrich, C. Kempter, G. Jung, and H. G. Sahl. 1996. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Appl. Environ. Microbiol. 62:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh, J., and M. Rance. 1990. Sensitivity improvement in isotropic mixing (TOCSY) experiments. J. Magn. Reson. 88:72-85. [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food: 10 States, United States, 2005. Morbid. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 6.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633-684. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert, A. 2006. Introduction: emerging antimicrobial resistance mechanisms in the zoonotic foodborne pathogens Salmonella and Campylobacter. Microbes Infect. [Epub ahead of print.] [DOI] [PubMed]
- 8.Cornwell, G. G., K. Sletten, B. Johansson, and P. Westermark. 1988. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem. Biophys. Res. Commun. 154:648-653. [DOI] [PubMed] [Google Scholar]
- 9.Davies, E. A., H. E. Bevis, R. Potter, J. Harris, G. C. Williams, and J. Delves-Broughton. 1998. Research note: the effect of pH on the stability of nisin solution during autoclaving. Lett. Appl. Microbiol. 27:186-187. [Google Scholar]
- 10.Delves-Broughton, J. 1990. Nisin and its application as a food preservative. J. Soc. Dairy Technol. 43:73-76. [Google Scholar]
- 11.Delves-Broughton, J. 1990. Nisin and its use as food preservative. Food Technol. 40:100-117. [Google Scholar]
- 12.Diep, D. B., and I. F. Nes. 2002. Ribosomally synthesized antibacterial peptides in gram positive bacteria. Curr. Drug Targets 3:107-122. [DOI] [PubMed] [Google Scholar]
- 13.Ekkelenkamp, M. B., M. Hanssen, S. T. Danny Hsu, A. de Jong, D. Milatovic, J. Verhoef, and N. A. van Nuland. 2005. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 579:1917-1922. [DOI] [PubMed] [Google Scholar]
- 14.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333-1341. [DOI] [PubMed] [Google Scholar]
- 15.Falagas, M. E., and S. K. Kasiakou. 2006. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit. Care 10:R27 [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freund, S., G. Jung, O. Gutbrod, G. Folkers, W. A. Gibbons, H. Allgaier, and R. Werner. 1991. The solution structure of the lantibiotic gallidermin. Biopolymers 31:803-811. [DOI] [PubMed] [Google Scholar]
- 17.Govaerts, C., J. Orwa, A. Van Schepdael, E. Roets, and J. Hoogmartens. 2002. Characterization of polypeptide antibiotics of the polymyxin series by liquid chromatography electrospray ionization ion trap tandem mass spectrometry. J. Pept. Sci. 8:45-55. [DOI] [PubMed] [Google Scholar]
- 18.Kaletunc, G., J. Lee, H. Alpas, and F. Bozoglu. 2004. Evaluation of structural changes induced by high hydrostatic pressure in Leuconostoc mesenteroides. Appl. Environ. Microbiol. 70:1116-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaenhammer, T. R. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337-349. [DOI] [PubMed] [Google Scholar]
- 20.Krull, R. E., P. Chen, J. Novak, M. Kirk, S. Barnes, J. Baker, N. R. Krishna, and P. W. Caufield. 2000. Biochemical structural analysis of the lantibiotic mutacin II. J. Biol. Chem. 275:15845-15850. [DOI] [PubMed] [Google Scholar]
- 21.Li, J., R. L. Nation, R. W. Milne, J. D. Turnidge, and K. Coulthard. 2005. Evaluation of colistin as an agent against multi-resistant gram-negative bacteria. Int. J. Antimicrob. Agents 25:11-25. [DOI] [PubMed] [Google Scholar]
- 22.Linden, P. K., S. Kusne, K. Coley, P. Fontes, D. J. Kramer, and D. Paterson. 2003. Use of parenteral colistin for the treatment of serious infection due to antimicrobial-resistant Pseudomonas aeruginosa. Clin. Infect. Dis. 37:154-160. [DOI] [PubMed] [Google Scholar]
- 23.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 24.Livermore, D. M. 2004. The need for new antibiotics. Clin. Microbiol. Infect. 10(Suppl. 4):1-9. [DOI] [PubMed] [Google Scholar]
- 25.Martin, N. I., T. Sprules, M. R. Carpenter, P. D. Cotter, C. Hill, R. P. Ross, and J. C. Vederas. 2004. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry 43:3049-3056. [DOI] [PubMed] [Google Scholar]
- 26.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 27.Menichetti, F. 2005. Current and emerging serious gram-positive infections. Clin. Microbiol. Infect. 11(Suppl. 3):22-28. [DOI] [PubMed] [Google Scholar]
- 28.Navaratna, M. A., H. G. Sahl, and J. R. Tagg. 1998. Two-component anti-Staphylococcus aureus lantibiotic activity produced by Staphylococcus aureus C55. Appl. Environ. Microbiol. 64:4803-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ösapay, G., L. Prokai, H. S. Kim, M. F. Medzihradszky, D. H. Coy, G. Liapakis, T. Reisine, G. Malacini, Q. Zhu, S. H. H. Wang, R. H. Mattern, and M. Goodman. 1997. Lanthionine-somatostatin analogs: synthesis, characterization, biological activity and enzymatic studies. J. Med. Chem. 40:2241-2251. [DOI] [PubMed] [Google Scholar]
- 30.Piuri, M., C. Sanchez-Rivas, and S. M. Ruzal. 1998. A novel antimicrobial activity of a Paenibacillus polymyxa strain isolated from regional fermented sausages. Lett. Appl. Microbiol. 27:9-13. [DOI] [PubMed] [Google Scholar]
- 31.Rollema, H. S., O. P. Kuipers, P. Both, W. M. de Vos, and R. J. Siezen. 1995. Improvement of solubility and stability of the antimicrobial peptide nisin by protein engineering. Appl. Environ. Microbiol. 61:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 33.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Stern, N. J., E. A. Svetoch, B. V. Eruslanov, Y. N. Kovalev, L. I. Volodina, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, and V. P. Levchuk. 2005. Paenibacillus polymyxa purified bacteriocin to control Campylobacter jejuni in chickens. J. Food Prot. 68:1450-1453. [DOI] [PubMed] [Google Scholar]
- 35.Svetoch, E. A., N. J. Stern, B. V. Eruslanov, Y. N. Kovalev, L. I. Volodina, V. V. Perelygin, E. V. Mitsevich, I. P. Mitsevich, V. D. Pokhilenko, V. N. Borzenkov, V. P. Levchuk, O. E. Svetoch, and T. Y. Kudriavtseva. 2005. Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associated bacteriocins. J. Food Prot. 68:11-17. [DOI] [PubMed] [Google Scholar]
- 36.Tankovic, J., P. Legrand, G. De Gatines, V. Chemineau, C. Brun-Buisson, and J. Duval. 1994. Characterization of a hospital outbreak of imipenem-resistant Acinetobacter baumannii by phenotypic and genotypic typing methods. J. Clin. Microbiol. 32:2677-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tauxe, R. V. 2002. Emerging foodborne pathogens. Int. J. Food Microbiol. 78:31-41. [DOI] [PubMed] [Google Scholar]
- 38.van de Kamp, M., H. W. Van den Hooven, R. N. H. Konings, G. Bierbaum, H.-G. Sahl, O. P. Kuipers, R. J. Siezen, W. M. De Vos, C. W. Hilbers, and F. J. M. Van de Ven. 1995. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Cloning and characterization of the epilancin K7 encoding gene and NMR analysis of mature epilancin K7. Eur. J. Biochem. 230:587-600. [DOI] [PubMed] [Google Scholar]
- 39.van de Kamp, M., L. M. Horstink, H. W. Van den Hooven, R. N. H. Konings, C. W. Hilbers, A. Frey, H.-G. Sahl, J. W. Metzger, and F. J. M. van de Ven. 1995. Sequence analysis by NMR spectroscopy of the peptide lantibiotic epilancin K7 from Staphylococcus epidermidis K7. Eur. J. Biochem. 227:757-771. [DOI] [PubMed] [Google Scholar]
- 40.van den Hooven, H. W., F. M. Lagerwerf, W. Heerma, J. Haverkamp, J. C. Piard, C. W. Hilbers, R. J. Siezen, O. P. Kuipers, and H. S. Rollema. 1996. The structure of the lantibiotic lacticin 481 produced by Lactococcus lactis: location of the thioether bridges. FEBS Lett. 391:317-322. [DOI] [PubMed] [Google Scholar]
- 41.van Kraaij, C., E. Breukink, H. S. Rollema, R. S. Bongers, H. A. Kosters, B. de Kruijff, and O. P. Kuipers. 2000. Engineering a disulfide bond and free thiols in the lantibiotic nisin Z. Eur. J. Biochem. 267:901-909. [DOI] [PubMed] [Google Scholar]
- 42.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wescombe, P. A., and J. R. Tagg. 2003. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 69:2737-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, D. G., S. Zhao, R. Singh, and P. F. McDermott. 2004. Antimicrobial resistance among gram-negative foodborne bacterial pathogens associated with foods of animal origin. Foodborne Pathog. Dis. 1:137-152. [DOI] [PubMed] [Google Scholar]
- 45.Whitford, M. F., M. A. McPherson. R. J. Forster, and R. M. Teather. 2001. Identification of bacteriocin-like inhibitors from rumen Streptococcus spp. and isolation and characterization of bovicin 255. Appl. Environ. Microbiol. 67:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.