Abstract
Lactobacillus plantarum is a common inhabitant of mammalian gastrointestinal tracts, and L. plantarum strain WCFS1 is a human isolate with a known genome sequence. L. plantarum WCFS1 survives intestinal passage in an active form, and its transit time and transcriptional activities were monitored in 15 BALB/c mice at 2, 4, 6, 8, and 24 h after being fed a single intragastric dose of this organism. Enumeration of viable cells isolated from fecal material revealed that the majority of the L. plantarum inoculum transited the mouse intestine within 4 h after ingestion. Three mice were sacrificed at each time point, and total RNA was isolated from the mouse intestinal compartments (stomach through colon). Quantification of L. plantarum 16S rRNA by quantitative real-time reverse-transcription-PCR revealed that L. plantarum was present at elevated levels in the stomach and small intestine for at least 4 h following ingestion and for over 8 h in the cecum and colon. We also examined the expression of 9 L. plantarum housekeeping genes and 15 L. plantarum in vivo-inducible (ivi) genes previously identified by recombination-based in vivo expression technology to be induced in the mouse gastrointestinal tract. The relative expression levels of the ivi genes increased up to 350-fold in the mouse intestine compared to levels observed for L. plantarum WCFS1 cells grown in a rich laboratory medium. Moreover, several genes displayed intestinal compartment-specific (small intestine versus colon) activities. These results confirm that L. plantarum displays specific and differential responses at various sites along the mammalian intestine.
The human gastrointestinal tract contains over 1013 bacterial cells, comprising more than 500 different species (13; for a review, see reference 40). These microorganisms perform many critical functions, including digestion and assimilation of nutrients, protection against pathogen colonization, activation of immunological surveillance signals, regulation of fat storage, and stimulation of intestinal angiogenesis (3, 39).
Members of the genus Lactobacillus are commonly found in human and animal gastrointestinal tracts and are considered to be among the most dominant organisms colonizing the small intestine (SI). Lactobacilli belong to the lactic acid bacteria (LAB), which comprise a variety of microorganisms applied to a variety of industrial and artisanal dairy, meat, and plant fermentations. Some selected strains of Lactobacillus are believed to be beneficial to human and animal health and are marketed as probiotics (28). While consumer interest in probiotics is growing (1), the activities of probiotics in the gastrointestinal tract and the mechanisms by which they exert their health-modulating effects remain largely unknown (16).
The genome of Lactobacillus plantarum WCFS1 was the first Lactobacillus genome to be sequenced (15). This strain originates from human saliva and displays good survival and persistence properties in the human gastrointestinal tract compared to other LAB (36). In-depth genome annotation has provided molecular maps and detailed catalogues of metabolic pathways for this organism (14, 31, 34). Additionally, whole-genome genotyping of different L. plantarum strains by using L. plantarum WCFS1-based DNA microarrays revealed variable genomic regions that are probably involved in strain-specific adaptation to different habitats (20). Such studies also enabled genotype-phenotype matching, leading to the identification of the L. plantarum gene encoding a mannose-specific adhesion (25). This L. plantarum adhesion might protect the host from infection by competing with pathogenic bacteria for attachment at mannose-receptor binding sites on the intestinal epithelial surface.
The specific activities of Lactobacillus in the gastrointestinal tract are being investigated through the application of in vitro- and in vivo-based approaches. In vitro genetic screening strategies, including promoter trapping (6, 7) and transcriptome analysis (9), were used to identify bile- and salt-inducible genes of L. plantarum WCFS1 and acid tolerance genes of Lactobacillus acidophilus (2). Because low pH, bile acids, and increasing osmolarity are encountered by bacteria during gastrointestinal tract transit, the genes identified in these studies are likely to encode functions that enable Lactobacillus to compete and survive better in this harsh environment. Several strains of Lactobacillus also appear to be metabolically active in vivo in the intestine. Lactobacillus casei was shown to actively transcribe genes involved in central metabolism and initiate de novo protein synthesis at various sites along the mouse intestine (21, 23). Moreover, application of in vivo expression technology (IVET) resulted in the identification of gut-inducible genes of Lactobacillus reuteri and L. plantarum WCFS1 in mouse model systems (5, 38). Three L. reuteri genes with in vivo-inducible (ivi) gene activities were identified, one of which (encoding methionine sulfoxide reductase) was found to be required for competitive intestinal colonization of mice (37, 38). In total, 72 L. plantarum WCFS1 ivi genes were identified using a recombination-based IVET (R-IVET) approach (5). These genes encode various classes of proteins, including those involved in nutrient acquisition and synthesis, transcription regulation, and adaptation to environmental stresses (5).
The R-IVET approach identified L. plantarum WCFS1 genes that are expressed at a low level in culture medium but are induced in the mouse intestine. However, ivi gene expression levels and the intestinal locations at which these genes were most highly expressed remained to be determined. Here, we address this issue by using real-time reverse-transcription (RT)-PCR to measure the transcript abundances of 15 L. plantarum WCFS1 ivi genes during mouse intestinal transit. This analysis confirmed that most ivi genes, and not L. plantarum housekeeping genes, are up-regulated in vivo. Moreover, the expression levels of several ivi genes differed considerably between the mouse intestinal compartments, thereby illustrating the dynamic activities of L. plantarum in the intestinal tract.
MATERIALS AND METHODS
Preparation of bacterial strain and administration to mice.
Wild-type L. plantarum WCFS1 (15) was grown at 37°C in 200 ml Mann Rogosa Sharpe (MRS) broth (Difco, Surrey, United Kingdom) (12) without aeration (14.5 h). The cells were collected by centrifugation for 10 min at 600 × g and then resuspended in MRS broth to a final volume of 2.5 ml. This cell suspension was divided into separate aliquots for RNA isolation, viable-cell-number determination, and inoculation into mice. Fifteen 7-week-old female BALB/c mice (Wageningen University, Harlan, Horst, The Netherlands) were fed directly into the stomach by gavage with 100 μl of the L. plantarum cell suspension and placed in groups of three into separate cages (time zero). The mice were housed in standard cages with free access to tap water and mouse chow throughout the duration of the experiment. The mice were sacrificed by cervical dislocation in groups of three at 2, 4, 6, 8, and 24 h after inoculation. An additional group of three mice were sacrificed 4 h after being fed sterile MRS broth (100 μl). The mouse digestive tracts were immediately excised, sectioned, frozen in liquid nitrogen, and stored at −80°C until RNA isolation. The animal welfare committee of Wageningen University (Wageningen, The Netherlands) approved the experimental protocol used in this study.
Enumeration of viable lactobacilli in mouse feces.
Mouse fecal material was collected before and after intragastric gavage. To ensure an absence of fecal contamination from earlier time points, the mice were transferred into cages containing fresh bedding at the time point prior to sacrifice. Approximately 0.1 g of mouse fecal material was placed into tubes containing 3 ml sterile phosphate-buffered salt solution (30) and 1.0 g glass beads (2 mm) for homogenization by vortexing. Numbers of viable cells in fecal material were determined by plating serial dilutions of the suspension on MRS agar, followed by aerobic incubation for 3 days at 30°C. For species identification, the V1-to-V3 regions of the 16S rRNA genes of individual colonies were amplified by PCR according to standard procedures (33). Purified PCR products were then sequenced, and these sequences were compared to 16S rRNA sequences from the Ribosomal Database Project (11).
RNA isolation.
Total RNA was isolated from L. plantarum and mouse tissues (weighing between 0.1 and 0.7 g) by using a bead-beating procedure as described previously, with the exception that the frozen samples were placed directly in ice-cold tubes containing 0.5 g of 0.1 mm zirconia/silica beads (Biospec, Bartlesville, OK), 0.5 ml phenol, and 0.5 ml TE (Tris-EDTA) buffer containing 1% sodium dodecyl sulfate and 0.01% macaloid clay (Kronos Titan GmbH, Germany) (7). RNA was further purified and treated with DNase I on QIAGEN RNeasy Midi columns (QIAGEN, Venlo, The Netherlands) according to the manufacturer's protocol. The concentration and quality of isolated RNA were assessed by spectrophotometry at absorbance levels of 260 and 280 nm (Ultrospec 2000; GE Healthcare, Diegem, Belgium) and by analysis with a 2100 Bioanalyzer (Agilent Technologies, Amstelveen, The Netherlands). All bioanalyzer profiles showed distinct peaks for mouse 28S and 18S rRNA, whereas measurable levels of 23S and 16S rRNA (typical 23S-versus-16S rRNA ratio of 1.7) were observed only for the cecum and colon samples.
Primer design.
All primers used in this study are listed in Table 1. Primers were designed using Primer 3 (29) and the software package Primer Express (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). The presence of secondary structures, including possible primer-dimers, was evaluated using NetPrimer (Premier Biosoft International, Palo Alto, CA). All primers were designed to have melting temperatures of 58 to 60°C according to Primer Express and amplicon sizes between 70 and 130 bp. The specificities of the primers to L. plantarum WCFS1 were evaluated by nucleotide similarity searches with the BLAST algorithm for short, nearly exact matches at the NCBI website (http://www.ncbi.nlm.nih.gov) (19). In silico comparisons and PCR amplification products confirmed that the 16S rRNA gene primer set was specific for both L. plantarum and the closely related species Lactobacillus pentosus but not other Lactobacillus species (data not shown).
TABLE 1.
Gene targets and primers
| Locus taga | Gene | Function | Forward primer | Reverse primer | E value | Source or reference |
|---|---|---|---|---|---|---|
| General | ||||||
| 16S rRNA | SSU ribosome (5 copies) | TGATCCTGGCTCAGGACGAA | TGCAAGCACCAATCAATACCA | 1.90 | 7 | |
| lp_0006 | gyrB | DNA gyrase B | GGAATTGATGAAGCCCTAGCAG | GAATCCCACGACCGTTATCA | 1.90 | This study |
| lp_0537 | ldhL | l-Lactate dehydrogenase | TGATCCTCGTTCCGTTGATG | CCGATGGTTGCAGTTGAGTAAG | 1.92 | This study |
| lp_0727 | groES | Chaperone | CCCAAAGCGGTAAGGTTGTT | CTTCACGCTGGGGTCAACTT | 1.97 | This study |
| lp_1021 | rpoB | DNA-directed RNA polymerase | CACCGTACCCGTAGAAGTTATGC | GGAGACCTTGATCCAAGAACCA | 1.91 | This study |
| lp_1027 | fusA2 | Elongation factor G | CCCATGATGGTGCTTCACAA | TCGTGGCAGCAGAGGTAATG | 1.89 | 7 |
| lp_1144 | pcrA | ATP-dependent DNA helicase | AGGAGGTCTGGGTCTCAACG | AAGGTCCGTTGCTCGCTAGT | 1.95 | This study |
| lp_1898 | pfk | 6-Phosphofructokinase | GTGGCGACGGTTCTTACCAT | CCCTGGAAGACCAATCGTGT | 1.93 | 7 |
| lp_2187 | ileS | Isoleucine-tRNA ligase | GGCACCTTACGTTCCTGGTT | CGTCATCTTCTTGCGGTCAT | 1.90 | This study |
| lp_2301 | recA | Recombinase A | GGCAGAACAGATCAAGGAAGG | TATCCACTTCGGCACGCTTA | 1.91 | This study |
| ivi gene | ||||||
| lp_0017 | proA | Gulamate-5-semialdehyde dehydrogenase | CGTGAGTTTGCCAGATCCAA | CATGCCGATAACACCTAATGG | 2.00 | This study |
| lp_0237 | Integral membrane protein (hypothetical) | GTACTGATATGGTTGTCGGGAATTA | ACGGGTGCGTAGAAGAAGC | 1.92 | 7 | |
| lp_0419 | plnI | Membrane-bound protease, immunity protein | CGCTTCGATGATGACTGCTT | CTGACCATACGGGCTGTGAT | 1.91 | This study |
| lp_0775 | argG | Argininosuccinate synthase | GCTCTTGCACCGGATATCAA | TTTCTTCTTCCCGTGACCAGT | 2.00 | 7 |
| lp_0800 | Cell surface protein precursor | CGATTAATGCGGCAACAACA | CCGGTTGTTCAGCCTTTGAG | 1.90 | This study | |
| lp_1019 | clpC | clpC (mecB gene) | TTCGCAAGCTAGGTGTCAGTG | AGGTTGGCGTTCCTTCAGTC | 2.00 | This study |
| lp_1164 | celB | Cellobiose PTS, EIIC | GGGCATCTTCCTCGCACTAT | TCGATCTCCTGGTGGATGTGT | 1.91 | This study |
| lp_1403 | Cell surface protein | AGTCCCAGTCGATGCTAACG | CGTCAGGCGAATACAACCAT | 1.99 | This study | |
| lp_1603 | hem | Hemolysin homologue | TGGTTCAGTCGTTGCCCTAA | AACAGCAGGATCACGGACAA | 1.82 | This study |
| lp_2940 | Cell surface protein precursor | ATGGCACGGTCAGTTTAGCA | TTGCACCGCTTGTGTTACCT | 1.88 | This study | |
| lp_3055 | copA | Copper-transporting ATPase | CGCACTTGTGACCACTTTCG | TTCCGCTTCCTTGGCTTGTA | 1.95 | This study |
| lp_3176 | pkn2 | Serine-threonine protein kinase | CAACGGAGCGATCTATATTCGT | GCCCATGTTGAATCCTGTGT | 1.94 | This study |
| lp_3473 | ram2 | Rhamnosidase | CAACCACGCTGACGTTACCA | CCGTGACCACTGGATTGCTA | 1.91 | This study |
| lp_3660 | rbsK3 | Ribokinase | TTATTGGTGCGGTTGGTGAC | TCTTTGTGATCCCTGCCAAG | 1.93 | This study |
| lp_3662 | adhE | Alcohol and acetaldehyde dehydrogenase | GCCGCACTGGACAATCATA | TCGCTGGCGTAGATGTTCTT | 1.99 | This study |
Designated gene number for the annotated L. plantarum WCFS1 chromosome.
cDNA construction and real-time PCR assays.
To eliminate remaining genomic DNA contamination, a second DNase treatment (DNase I; Invitrogen, Breda, The Netherlands) was included for all RNA samples prior to first-strand cDNA synthesis. For every 5 μg of total RNA, 20-μl reaction mixtures containing 200 U Superscript III reverse transcriptase (Invitrogen), 40 U RNaseOUT RNase inhibitor (Invitrogen), 1 mM of each deoxynucleoside triphosphate, and 2 pmol of each gene-specific primer complementary to the mRNA of specified genes (Table 1) were prepared. RT reactions were performed in duplicate and contained 3 ng or 60 ng total RNA collected from L. plantarum WCFS1 cells grown in laboratory culture and either 80 ng or 12 μg mouse digestive tract total RNA depending on whether subsequent PCRs would be performed for detection of 16S rRNA or protein-encoding gene transcript analysis, respectively. RT was carried out according to the manufacturer's protocol. The resulting cDNA samples were stored at −20°C until use.
Real-time PCR amplification was performed in 96-well plates on an ABI Prism 7700 sequence detection system (Applied Biosystems), using the double-stranded DNA intercalating fluorescent agent SYBR green for product detection. Each well contained SYBR green Master Mix (Applied Biosystems), 200 nM of each primer, and a template. Templates consisted of cDNA products equivalent to either 0.01 or 1 ng RNA from the L. plantarum WCFS1 laboratory culture and either 2 or 200 ng mouse digestive tract total RNA. PCR amplification was initiated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 55°C for 60 s. Control PCRs were included to detect background contamination (no-template control) and remaining chromosomal DNA (RT reactions in which Superscript III was omitted). Spiking experiments in which L. plantarum WCFS1 RNA was added to the mouse-derived RNA samples confirmed that real-time RT-PCR amplification was not inhibited, even when these reactions were initiated with large amounts of mouse-derived RNA.
PCR specificity and product detection were checked postamplification by examining the dissociation curves of the PCR products. These melting curve profiles were generated by first heating the samples to 95°C and then cooling them to 55°C and slowly heating them at 2°C/min to 95°C for detection of SYBR green fluorescence. Melting curve profiles were analyzed and compared using Dissociation Curve software 1.0 (Applied Biosystems).
Schema for analysis of L. plantarum gene transcripts in the mouse intestine.
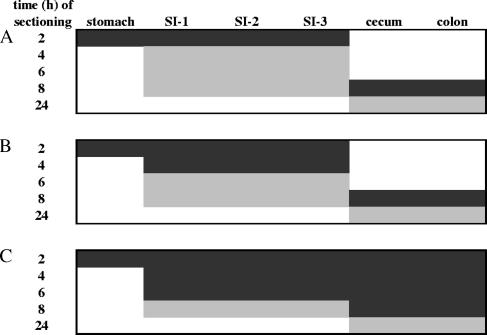
Real-time RT-PCR was performed for the quantification of 15 L. plantarum ivi genes and 9 housekeeping gene transcripts in the stomach and small intestine (regions SI-1 to SI-3) at 2 h and in the cecum and colon at 8 h after the ingestion of L. plantarum WCFS1 (Fig. 1A). To determine whether L. plantarum gene transcripts were measurable in the small intestine at other time points, equivalent amounts of compartment-specific, total RNA from individual mouse samples were combined for real-time RT-PCR transcript analysis (Fig. 1A). Transcripts of some genes were detected in the small intestine samples at 4 h but not at 6 and 8 h, and therefore, the analysis was expanded to quantify these gene transcripts among individual mice at this time point (Fig. 1B). Finally, a smaller set of highly abundant housekeeping and ivi gene transcripts were also examined in the ceca and colons of individual mice at all possible time points to obtain a comprehensive view of their expression through time and location in the digestive tract (Fig. 1C).
FIG. 1.
Mouse intestinal tract samples selected for gene transcript analysis. Real-time RT-PCR was performed on RNA from individual mice (n = 3) (black panels) or pooled samples (gray panels). (A) The minimal number of samples in which a housekeeping or ivi gene was examined. (B) argG, clpC, pkn2, plnI, proA, pts14C(celB), rbsK3, ram2, lp_0237, lp_0800, lp_1403, and lp_2940 were also monitored in the small intestines of individual mice at 4 h. (C) groES, fusA, ldhL, pfk, rpoB, copA, and adhE were monitored as described for panels A and B, with the addition of the small intestine at 6 h and the cecum and colon at 2, 4, and 6 h.
Data analysis.
Real-time PCR quantification is based on the number of cycles required for amplification-associated fluorescence to reach the detection threshold (as expressed by the cycle threshold [CT] value). CT values were obtained for each reaction by manually setting the baseline to a level at which fluorescence was noticeably above the background level and exponential amplification was under way. Two independent RT reactions and real-time PCR amplifications were performed for each gene in every sample, and the average of the observed CT values was used for further analysis. Real-time PCR amplification efficiencies were determined for each primer pair by standard curves generated by plotting the starting RNA concentration against the observed CT values over a 1 × 104-fold dilution range of cDNA input. The slope of the calibration curve was used to determine the reaction efficiency (E) as E = 10−1/slope (Table 1) (26).
To estimate the number of L. plantarum cells contained in the mouse digestive tract compartments, the CT values obtained for L. plantarum 16S rRNA in these samples were first compared to those for samples containing known amounts of L. plantarum WCFS1 total RNA. The estimated L. plantarum total RNA in the mouse samples was then correlated to the corresponding number of L. plantarum cells based on the observation that we typically isolated approximately 0.02 pg total RNA per laboratory culture-grown L. plantarum cell.
To identify endogenous reference genes, a panel of nine L. plantarum housekeeping genes (Table 1) was evaluated for gene expression stability according to pair-wise variation procedures outlined for the geNorm approach (35). Following this method, we calculated a normalization factor based on the geometric mean of the linear-transformed expression values for the three most stable housekeeping genes. These values were then used to express the relative amounts of the ivi and housekeeping gene transcripts in the mouse-derived RNA samples compared to those in L. plantarum WCFS1 grown in MRS broth.
RESULTS
Enumeration of viable Lactobacillus in mouse feces.
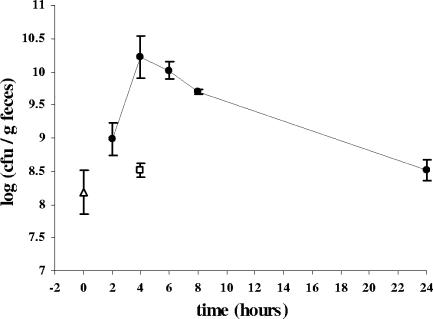
A single dose of wild-type L. plantarum WCFS1 (2 × 1010 cells grown in MRS broth) (12) was fed by gavage directly into the stomachs of 15 BALB/c mice (time zero). During the subsequent 24 h, mouse fecal material was collected periodically to determine the time required for L. plantarum WCFS1 to transit through the digestive tract. Fecal samples collected before inoculation of L. plantarum and after control inoculation with sterile culture medium contained on average 108 cells/g feces able to form colonies on MRS agar (Fig. 2). 16S rRNA gene sequence analysis of seven colonies with distinct colony morphologies indicated that three fecal isolates were L. plantarum, while the others were very similar (>97% nucleotide identity) to either Lactobacillus murinus (two of seven) or Enterococcus hirae (two of seven). Hence, colony enumeration on MRS agar enabled the monitoring of the endogenous Lactobacillus and related LAB populations contained in the feces as well as the L. plantarum WCFS1 fed to the mice.
FIG. 2.
Enumeration of viable Lactobacillus cells and related LAB isolated from feces of mice immediately prior to the start of the experiment (open triangle) and inoculated with either L. plantarum WCFS1 (closed circles) or sterile MRS broth (open square). Shown are the averages ± standard deviations for feces excreted by three mice during the time period prior to sacrifice.
The number of fecal bacteria able to grow on MRS agar increased within 2 h after ingestion of L. plantarum WCFS1 (Fig. 2). Over the next 2 hours, this population reached its highest level at 1010 cells/g feces. Subsequently, after 6 h, the population started to decline and continued to decline in time until colony counts reached preinoculation levels. 16S rRNA gene sequence analysis confirmed that the observed population dynamics were the result of the excretion of L. plantarum WCFS1 into the mouse feces (data not shown).
Transit of L. plantarum through intestinal compartments.
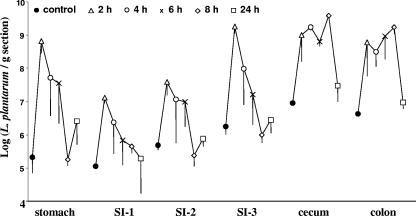
At 2, 4, 6, 8, and 24 h after ingestion of L. plantarum WCFS1, three mice were sacrificed and the stomach, small intestine (in three parts approximately representing the duodenum [SI-1], jejunum [SI-2], and ileum [SI-3]), cecum, and colon of each mouse were collected for RNA isolation. In order to monitor the dynamics of L. plantarum transit through the different intestinal compartments, we applied real-time RT-PCR to measure the levels L. plantarum 16S rRNA present in the samples, using primers described previously (Table 1) (7). Estimates of the number of L. plantarum cells were obtained by correlating the real-time RT-PCR signals for 16S rRNA in the mouse samples to those obtained for reactions in which the number of L. plantarum WCFS1 cells and the RNA template were known. This approach revealed that the control mice harbored approximately 2 × 105 L. plantarum cells in the stomach and over 5 × 106 cells in the cecum and colon (per gram tissue) (Fig. 3).
FIG. 3.
Population dynamics of L. plantarum cells contained in mouse digestive tract sections as determined by 16S rRNA-targeted real-time RT-PCR. Each point represents the average ± standard error of the mean values for three mice at each time point.
Upon inoculation of L. plantarum WCFS1, the number of L. plantarum cells in the different intestinal compartments increased by 2 to 3 orders of magnitude compared to what was found for the control mice (Fig. 3). The amounts of L. plantarum in the stomach and small intestine remained large for at least 4 h but then declined to background levels, indicating that the majority of L. plantarum WCFS1 cells were no longer present in these compartments. In contrast, the cecum and colon contained approximately 109 L. plantarum cells per gram tissue for at least 8 h (Fig. 3). Remarkably, in one mouse sacrificed at 24 h, the stomach and the upper duodenum (SI-1) contained elevated levels of L. plantarum. This result could be explained by the fact that mice are coprophilic (consume their own feces), and hence, their fecal material likely served as a secondary source of L. plantarum inoculum.
L. plantarum housekeeping gene expression levels in the mouse intestine.
The transcript abundances of the ivi genes were compared between the intestinal samples and the L. plantarum WCFS1 cells that were used to gavage the mice (time zero). To control for errors introduced by differences in the concentrations of L. plantarum total RNA isolated from the mouse intestine and laboratory-grown cultures, the observed gene expression values were first normalized to those for endogenous L. plantarum reference housekeeping gene transcripts. Housekeeping genes were evaluated on the basis that the gene transcript was detected in mouse samples collected from mice fed L. plantarum WCFS1 and that it was stably expressed relative to all other housekeeping genes examined. Real-time RT-PCR of RNA isolated from the intestinal compartments of the control mice did not result in a detectable product for L. plantarum housekeeping or ivi genes. In contrast, transcripts for five of the nine housekeeping genes were present in measurable levels in the intestinal compartments of mice fed L. plantarum WCFS1 (Fig. 1A). Transcripts for ileS, pcrA, gyrB, and recA were not consistently detected (see Fig. S2 in the supplemental material) and were therefore not included in subsequent analyses. Pair-wise comparisons (35) of the expression values obtained for the five remaining housekeeping genes indicated that fusA, rpoB, and pfk were the most stably expressed housekeeping genes among L. plantarum cells contained in the mouse intestine and grown in MRS medium. Sample-specific normalization factors comprising the geometric mean of their expression values were used to quantify L. plantarum WCFS1 gene transcript abundance.
Analysis of the real-time RT-PCR data revealed that most L. plantarum housekeeping genes were not differentially regulated (see Fig. S1 and S2 in the supplemental material). Only the transcription of gyrB and ldhL was induced more than twofold, on average, in the stomach and SI-1 (gyrB) or cecum and colon (ldhL). Conversely, groES was down-regulated at the distal end (SI-3) of the small intestine, cecum, and colon of most mice.
In vivo-inducible (ivi) gene expression levels in the mouse intestine.
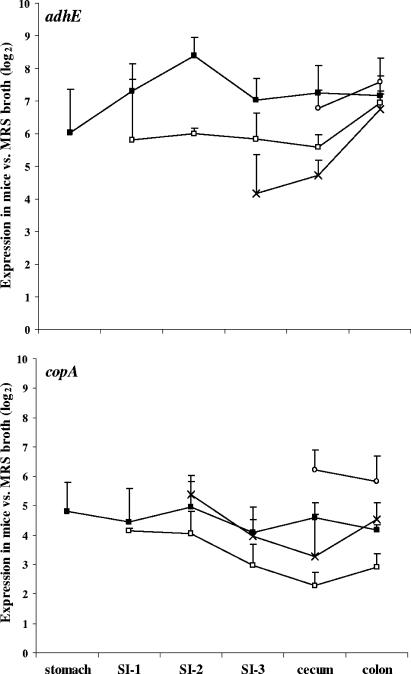
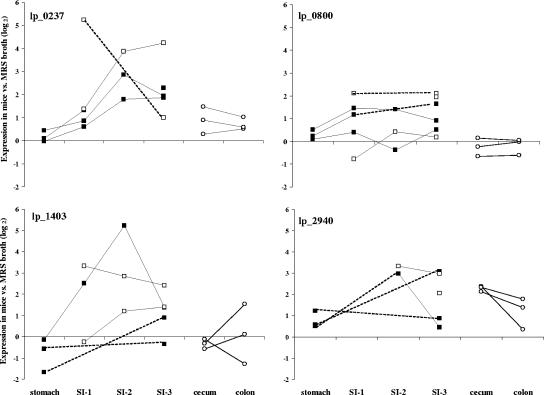
A total of 15 ivi genes, selected to represent the diversity of cellular activities identified in the R-IVET screen, were analyzed for their in vivo expression levels by real-time RT-PCR (Table 2). Strikingly, copA, encoding a putative copper-transporting ATPase, and adhE, encoding bifunctional alcohol dehydrogenase, were highly induced in all intestinal compartments and at all time points examined (Table 2 and Fig. 4). These loci were up-regulated in the mouse stomach, and their expression remained induced at similar levels throughout the digestive tract. The gene encoding a lytic enzyme, annotated as a putative hemolysin, hem, was the only ivi gene that was consistently down-regulated in all intestinal samples examined (Table 2). All other ivi genes examined exhibited at least modest levels of induction in the mouse intestine, and in some cases, this activity was dependent upon the intestinal tract compartment. For example, L. plantarum genes encoding cell surface and membrane-bound proteins, the lp_0237, lp_0800, lp_1403, and lp_2940 proteins, were induced primarily in the small intestine (Table 2 and Fig. 5). Although there was considerable mouse-to-mouse variation and transcripts were not detected in all samples, three of these genes (lp_0237, lp_0800, and lp_1403) were induced along the length of the small intestine, whereas lp_2940 was most active at the distal end of this compartment and in the cecum. Similar to the cell surface proteins, genes encoding protein kinase Pkn2 and argininosuccinate synthetase ArgG (Table 2) (see Fig. S3 in the supplemental material) were typically induced in the small intestine. Genes that appeared to be up-regulated but for which transcripts were detected in only a few intestinal samples included plnI and rbsK, encoding a plantaricin immunity protein and ribokinase, respectively (Table 2) (see Fig. S3 in the supplemental material). Finally, transcripts for clpC and proA were detected in the intestine but were not present in significantly larger amounts than those found in L. plantarum cells contained in MRS broth (Table 2) (see Fig. S3 in the supplemental material).
TABLE 2.
ivi gene expression in micea
| Gene |
ivi expression level at indicated site
|
|||||
|---|---|---|---|---|---|---|
| Stomach | SI-1 | SI-2 | SI-3 | Cecum | Colon | |
| adhE | 6.03 | 6.10 | 6.85 | 5.68 | 6.43 | 7.12 |
| argG | 0.52 | 1.76 | 1.88 | 1.24 | 0.58 | 0.25 |
| celB | −0.09 | BD | 1.73 | 0.76 | −0.08 | 0.29 |
| clpC | 0.16 | 0.99 | 0.94 | 0.37 | −0.36 | −0.06 |
| copA | 4.80 | 4.51 | 4.81 | 3.67 | 4.22 | 4.38 |
| hem | −0.61 | −0.44 | −0.56 | −0.71 | −1.00 | −1.38 |
| pkn2 | 0.23 | 1.93 | 1.76 | 1.81 | −0.18 | −0.02 |
| plnI | 1.06 | 1.82 | 4.31 | 4.28 | 2.23 | 2.60 |
| proA | 0.52 | 0.39 | 0.69 | 0.34 | −0.25 | −0.50 |
| rbsK3 | 2.70 | BD | 4.86 | 3.51 | 2.27 | 2.04 |
| ram2 | 0.79 | BD | 4.15 | 3.53 | BD | 0.11 |
| lp_0237 | 0.18 | 1.88 | 2.84 | 2.26 | 0.88 | 0.70 |
| lp_0800 | 0.27 | 0.86 | 0.49 | 1.22 | −0.27 | −0.20 |
| lp_1403 | −0.79 | 1.86 | 3.08 | 1.14 | −0.34 | 0.11 |
| lp_2940 | 0.77 | BD | 3.15 | 1.89 | 2.28 | 1.17 |
Average change (n-fold) in expression levels (log2) for L. plantarum ivi genes in mouse intestinal compartments compared to those in MRS culture medium (time zero). The average includes all mice and time points examined. The values in bold indicate gene transcripts that could be detected in three or more individual mice and were, on average, more than twofold induced according to the one-tailed t test (P < 0.05). BD, below detection.
FIG. 4.
Relative expression levels of the copA and adhE genes in the mouse intestine at 2 h (▪), 4 h (□), 6 h (×), and 8 h (○) after inoculation of L. plantarum WCFS1. The normalized gene transcript amounts in the intestinal sections were compared to those found for L. plantarum in the mouse inoculum (time zero). Expression values are reported as the averages ± standard errors of the means for three mice at each time point.
FIG. 5.
Relative expression levels of lp_0237, lp_0800, lp_1403, and lp_2940 in the stomachs and small intestines of individual mice at 2 h (▪) and 4 h (□) and in the ceca and colons at 8 h (○) after inoculation of L. plantarum WCFS1. The normalized gene transcript amounts in the intestinal sections were compared to those found for L. plantarum in the mouse inoculum (time zero). Expression values for individual mice are connected by solid or dotted lines, depending on whether transcripts were detected in consecutive or nonconsecutive intestinal sections, respectively.
DISCUSSION
This study revealed that L. plantarum WCFS1 undergoes dynamic changes in ivi gene expression in the digestive tracts of mice. To aid the quantitative detection of L. plantarum gene transcripts, the transit of L. plantarum through the mouse intestinal compartments was also measured. The number of viable Lactobacillus cells in the mouse feces increased within 2 h after inoculation of L. plantarum WCFS1, reached its highest level in the subsequent 2-h period, and returned to preinoculation levels within 24 h (Fig. 2). These transit dynamics are similar to those reported for L. casei and Bacillus subtilis spores fed to human flora-associated mice (22). Because L. plantarum WCFS1 was not selectively monitored on the MRS medium used here, it is not possible to conclude whether this organism was still present once the number of Lactobacillus cells in the mouse feces returned to the initial level. A previous study found that an antibiotic-resistant variant of L. plantarum WCFS1 could still be detected in the feces of mice for up to 7 days after inoculation (24). Therefore, it remains likely that while the majority of the L. plantarum WCFS1 cells transited the digestive tract rather rapidly in the present study, a small but persistent population of this organism was retained in the mice.
Consistent with the number of Lactobacillus cells present in the mouse feces, real-time RT-PCR of L. plantarum 16S rRNA isolated from the intestinal compartments confirmed substantial increases in the number of L. plantarum cells 2 h after inoculation of L. plantarum WCFS1 (Fig. 3). Transit of this organism was most rapid through the mouse stomach and small intestine (regions SI-1 through SI-3), and within 6 h, the numbers of L. plantarum returned to preinoculation levels at these sites. In contrast, the cecum and colon retained large amounts of L. plantarum for at least 8 h. These transit dynamics are in agreement with analyses of L. plantarum WCFS1 housekeeping and ivi gene transcripts such that they could be detected only in mouse compartments containing elevated numbers of L. plantarum cells.
Based on the L. plantarum intestinal tract population sizes estimated according 16S rRNA-based real-time RT-PCR analysis and the signals obtained for individual gene transcripts in the different samples, the detection limit for L. plantarum mRNA was in the range of 107 cells per intestinal compartment. This range is similar to what was found for other DNA-based real-time PCR detection applied to fecal samples (18). Because there was considerable mouse-to-mouse variation in the intestinal transit of L. plantarum WCFS1, particularly through the small intestine, the approach taken here of collecting mouse intestinal compartments at multiple time points proved to be essential for determining which samples were likely to contain detectable levels of L. plantarum gene transcripts.
Because different quantities of L. plantarum total RNA were isolated from the mice, it was necessary to normalize the real-time RT-PCR data collected for the L. plantarum housekeeping and ivi gene transcripts. 16S rRNA is among the most commonly used cellular products used for transcript normalization (10). However, 16S rRNA was not a suitable normalizer for the mouse intestinal samples because it is not specific for L. plantarum WCFS1 and is much more abundant than mRNA. In comparison, the detection limit for L. plantarum WCFS1 housekeeping gene transcripts was similar to that found for the ivi gene transcripts. We previously compared the expression levels of L. plantarum WCFS1 housekeeping genes in actively dividing, stationary-phase, and nutrient-starved L. plantarum cells in vitro (M. L. Marco and M. Kleerebezem, submitted for publication). Real-time RT-PCR of the housekeeping gene transcripts revealed considerable variation in their abundance levels according to several data normalization methods. Only internal pair-wise comparisons made according to the geNorm approach accounted for variations in cell viability, fluctuating rRNA levels, and total extracted L. plantarum RNA. Therefore, we used this approach to analyze L. plantarum transcript levels in mice.
copA and adhE were the most highly induced ivi genes examined in this study, exhibiting between 5- and 350-fold-higher levels of expression throughout the mouse digestive tract. Interestingly, the expression levels of these genes were similar within the intestinal compartments at the different time points examined, suggesting that these ivi genes are consistently up-regulated in the mouse intestinal tract over time. While copA is annotated as a putative copper-transporting ATPase, its cation specificity and direction of transport are not known. Remarkably, genes involved in cation transport were frequently identified in other IVET screens in other bacteria (27). In addition, the importance of the copA gene for L. plantarum during its residency in the intestine was recently confirmed in studies whereby L. plantarum copA deletion mutants showed reduced levels of persistence and survival in mice (8). AdhE is predicted to function as a bifunctional alcohol and acetaldehyde dehydrogenase involved in metabolism during fermentation. Although the relevance of this gene for L. plantarum intestinal activities remains to be established, adhE was also found to be up-regulated in pathogenic Escherichia coli K1 during intestinal colonization (17) and therefore is likely involved in bacterial adaptation to the changing environment and nutritional resources available in the gastrointestinal tract.
While copA and adhE were up-regulated throughout the entire digestive tract, most other ivi genes were preferentially induced in the small intestine. This result is in agreement with the commonly held assumption that L. plantarum is among the most predominant and active microorganisms in the small intestine. Several ivi genes encoding membrane-bound proteins were among those that were most highly up-regulated at this location. Both lp_0800 and lp_2940 encode proteins containing LPXTG-like motifs (LPQTNE) involved in anchoring them extracellularly to the L. plantarum cell wall (4). The protein product of lp_1403 is also likely exported outside the cell, but it lacks any known binding domains. Finally, lp_0237 is annotated as an integral membrane protein of an unknown function. Both this gene and argG were previously confirmed to be up-regulated in the mouse small intestine in independent real-time RT-PCR experiments (7). These genes were also found to be induced in the presence of bile salts and hence may be responding to these compounds in the small intestine (7).
Most L. plantarum ivi genes appeared to be only moderately induced in the intestinal tract. Although this observation might be due to uniform and relatively small changes in L. plantarum WCFS1 ivi gene expression in the intestine, it is also possible that only a fraction of the total L. plantarum WCFS1 cells was responding to the stimulus required for individual ivi gene expression. As real-time RT-PCR provides an average of gene activities for all cells from which RNA was isolated, it is likely that some L. plantarum cells contained large amounts of the ivi gene transcripts, while many others were either not sensing the stimulus necessary for ivi gene expression or were metabolically inactive and unable to initiate a response. Such genes would be identified by R-IVET because this screening technique requires only single or small groups of L. plantarum cells to be activated at these loci in order to be identified as being in vivo induced. Therefore, although the hemolysin homolog appeared to be down-regulated in the mouse digestive tract according to the real-time RT-PCR studies performed here, it remains plausible that this gene was induced in L. plantarum cells located in specific intestinal microsites. A second factor that might have influenced the observed ivi gene expression levels is that the relative gene transcript abundance was dependent on the in vitro condition to which they were compared. Comparisons of transcript levels found in stationary- and exponential-phase L. plantarum cells revealed that many ivi genes were more highly expressed in the stationary-phase cells fed to the mice (see Table S1 in the supplemental material). However, independent of the growth phase to which the in vivo data were compared, the conclusion remains that the L. plantarum ivi genes examined here were more highly expressed in the mouse gastrointestinal tract than in culture medium.
Real-time RT-PCR has yet to be commonly applied to confirm and quantify the expression levels of genes identified in an IVET-based approach (27). However, this technique proved to be valuable for the study of L. plantarum in the mouse intestine, whereas other, less-sensitive methods may not have detected specific L. plantarum gene transcripts. Genome-wide transcript analyses using DNA microarrays also provide opportunities for even more comprehensive and integrative views of bacterial activities occurring within the intestinal tract. The potential of this approach was exemplified by studies reporting full-genome transcriptome profiles of Bacteroides thetaiotaomicron residing in the ceca of germfree mice (32). However, only real-time RT-PCR permits direct measurements of bacterial gene expression in densely populated, species-rich digestive tracts. Application of such complementary techniques will improve our understanding of bacterial gene expression in situ in the gastrointestinal tract and aid in the development of molecular models for describing bacterial activities in vivo and the corresponding host responses. Such models will be required for our understanding of the mechanisms by which probiotic strains exert their functional benefits in the consumer and will ultimately enable the selection or construction of improved strains with predestined health benefits.
Supplementary Material
Footnotes
Published ahead of print on 27 October 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arvanitoyannis, I. S., and M. Van Houwelingen-Koukaliaroglou. 2005. Functional foods: a survey of health claims, pros and cons, and current legislation. Crit. Rev. Food Sci. Nutr. 45:385-404. [DOI] [PubMed] [Google Scholar]
- 2.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bron, P. A., S. M. Hoffer, I. I. Van Swam, W. M. de Vos, and M. Kleerebezem. 2004. Selection and characterization of conditionally active promoters in Lactobacillus plantarum using alanine racemase as a promoter probe. Appl. Environ. Microbiol. 70:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bron, P. A., M. Meijer, R. S. Bongers, W. M. De Vos, and M. Kleerebezem. 2004. The molecular response of Lactobacillus plantarum to intestinal passage and conditions, p. 129-153. Thesis. Wageningen University, The Netherlands.
- 9.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100:728-738. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of Lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkhoven, R., F. H. van Enckevort, J. Boekhorst, D. Molenaar, and R. J. Siezen. 2004. Visualization for genomics: the Microbial Genome Viewer. Bioinformatics 20:1812-1814. [DOI] [PubMed] [Google Scholar]
- 15.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17:204-210. [DOI] [PubMed] [Google Scholar]
- 17.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 18.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGinnis, S., and T. L. Madden. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20-W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oozeer, R., J. P. Furet, N. Goupil-Feuillerat, J. Anba, J. Mengaud, and G. Corthier. 2005. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 71:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oozeer, R., N. Goupil-Feuillerat, C. A. Alpert, M. van de Guchte, J. Anba, J. Mengaud, and G. Corthier. 2002. Lactobacillus casei is able to survive and initiate protein synthesis during its transit in the digestive tract of human flora-associated mice. Appl. Environ. Microbiol. 68:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oozeer, R., D. D. G. Mater, N. Goupil-Feuillerat, and G. Corthier. 2004. Initiation of protein synthesis by a labeled derivative of the Lactobacillus casei DN-114 001 strain during transit from the stomach to the cecum in mice harboring human microbiota. Appl. Environ. Microbiol. 70:6992-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavan, S., P. Desreumaux, and A. Mercenier. 2003. Use of mouse models to evaluate the persistence, safety, and immune modulation capacities of lactic acid bacteria. Clin. Diagn. Lab. Immunol. 10:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pretzer, G., J. Snel, D. Molenaar, A. Wiersma, P. A. Bron, J. Lambert, W. M. de Vos, R. van der Meer, M. A. Smits, and M. Kleerebezem. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187:6128-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen, R. 2001. Quantification on the LightCycler, p. 21-34. In S. Meuer, C. Wittwer, and K. Nakagawara (ed.), Rapid cycle real-time PCR, methods and applications. Springer Press, Heidelberg, Germany.
- 27.Rediers, H., P. B. Rainey, J. Vanderleyden, and R. De Mot. 2005. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 69:217-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid, G. 1999. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 65:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rozen, S., and H. Skaletsky. 2000. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Smid, E. J., F. J. van Enckevort, A. Wegkamp, J. Boekhorst, D. Molenaar, J. Hugenholtz, R. J. Siezen, and B. Teusink. 2005. Metabolic models for rational improvement of lactic acid bacteria as cell factories. J. Appl. Microbiol. 98:1326-1331. [DOI] [PubMed] [Google Scholar]
- 32.Sonnenburg, J. L., J. Xu, D. D. Leip, C.-H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 33.te Giffel, M. C., R. R. Beumer, N. Klijn, A. Wagendorp, and F. M. Rombouts. 1997. Discrimination between Bacillus cereus and Bacillus thuringiensis using specific DNA probes based on variable regions of 16S rRNA. FEMS Microbiol. Lett. 146:47-51. [DOI] [PubMed] [Google Scholar]
- 34.Teusink, B., F. H. J. van Enckevort, C. Francke, A. Wiersma, A. Wegkamp, E. J. Smid, and R. J. Siezen. 2005. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 71:7253-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:34.1-34.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]
- 37.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, J., N. C. K. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoetendal, E. G., E. E. Vaughan, and W. M. de Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.