Abstract
A sensitive rRNA-targeted reverse transcription-quantitative PCR (RT-qPCR) method was developed for exact and sensitive enumeration of subdominant bacterial populations. Using group- or species-specific primers for 16S or 23S rRNA, analytical curves were constructed for Escherichia coli, Enterococcus faecalis, Staphylococcus aureus, Clostridium perfringens, and Pseudomonas aeruginosa, and the threshold cycle value was found to be linear up to an RNA amount of 10−3 cell per RT-PCR. The number of bacteria in culture was determined by RT-qPCR, and the results correlated well with the CFU count over the range from 100 to 105 CFU. The bacterial counts obtained by RT-qPCR were the same as the CFU counts irrespective of the growth phase in vitro, except for C. perfringens during starvation periods; the viable cell counts obtained by using a combination of 4′,6-diamidino-2-phenylindole (DAPI) staining and SYTO9-propidium iodide double staining were in good agreement with the RT-qPCR counts rather than with the CFU counts. The RT-qPCR method could detect endogenous Enterobacteriaceae and P. aeruginosa in feces of hospitalized patients (n = 38) at a level of 103 cells per g of feces, and for enumeration of S. aureus or P. aeruginosa spiked into human peripheral blood, the lower detection limit for RT-qPCR quantification of the bacteria was 2 cells per ml of blood, suggesting that this method was equivalent to the conventional culture method. As only 5 h was needed for RT-qPCR quantification, we suggest that rRNA-targeted RT-qPCR assays provide a sensitive and convenient system for quantification of commensal bacteria and for examining their possible invasion of a host.
For almost a century, culture techniques have been recognized as the “gold standards” for determining viable bacterial counts. As the human fecal flora has been reported to consist of approximately 400 bacterial species (12, 35) and these species are present at a concentration of 1011 viable microorganisms per g of contents (42), conventional culture techniques for enumeration of different populations involve the use of selective microbiological media, followed by isolation of pure cultures and the use of confirmatory biochemical tests. Recently, a number of molecular methods based on immunological and genotypic techniques have been developed (41, 48). In analyses of the gut microflora, a number of molecular methods have been used in place of cultivation-based techniques. Techniques such as the clone library method (42, 46), denaturing gradient gel electrophoresis (13), and terminal restriction fragment length polymorphism (31, 36) allow analysis of predominant bacteria that are difficult to culture. The fluorescent in situ hybridization method (18, 43) and the quantitative PCR (qPCR) method with rRNA-targeted oligonucleotide probes or primers have also been used as culture-independent methods. Among these, PCR methods targeting mainly well-conserved 16S rRNA genes have prevailed for rapid quantification of bacteria and are recognized as having two advantages, specificity and convenience. To determine the bacterial population in the human gastrointestinal tract, the applications of qPCR have been expanded (5, 16, 29, 30). The new techniques enable accurate and convenient quantification of targeted predominant anaerobic species in the microflora, such as members of Bifidobacterium and the Bacteroideaceae, that are present at levels of more than 109 cells per g of feces. However, it has been demonstrated that the sensitivity of PCR is around 105 to 106 cells per g of feces, which does not seem to be sufficient for accurate quantification of minor but important commensal species, such as members of the Enterobacteriaceae, Enterococcus, Staphylococcus, and Clostridium perfringens that have been implicated as potential pathogens in immunocompromised hosts. Because of the lower levels of these subdominant bacterial species in healthy intestines, it is difficult to detect them accurately in the huge total bacterial population by existing techniques. In clinical examinations, it has been demonstrated that qPCR can detect bacterial contaminants with a sensitivity of 101 to 102 CFU per ml of blood (23, 38) but usually is not able to detect contamination with only a small number of bacteria (less than 10 cells per ml of blood).
We have focused on rRNAs as the target for precise and sensitive quantification of commensal subdominant bacterial populations, since rRNA is a universal constituent of bacterial ribosomes and high copy numbers (103 to 104 molecules per actively growing cell) are present as housekeeping genes (1, 17). Targeting these molecules has the potential to increase the detection sensitivity compared to the sensitivity of assays based on detection of a single copy or even multiple copies of genomic sequences. Here we describe sensitive quantification of bacterial populations with lower detection limits of 103 cells per g of feces and 100 cells per ml of peripheral blood using reverse transcription-quantitative PCR (RT-qPCR) targeting rRNA, which has almost the same sensitivity as the conventional culture method but improved performance time.
MATERIALS AND METHODS
Reference strains and culture conditions.
The strains listed in Table 1 were used. Escherichia coli ATCC 11775T, Enterococcus faecalis ATCC 19433T, Staphylococcus aureus ATCC 12600T, and Pseudomonas aeruginosa ATCC 10145T were grown aerobically in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, Md.) at 37°C, and the bacterial counts were expressed in CFU after culturing on BHI agar. C. perfringens JCM 1290T was grown anaerobically in MRS broth (Becton Dickinson) at 37°C, and the CFU counts were determined by culturing the organism on GAM agar (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) in an anaerobic chamber (Coy Laboratory Products Inc., Grass Lake, MI).
TABLE 1.
Specificity tests with newly developed primers
| Taxon | Strain | Reactions with the following primersa:
|
||||
|---|---|---|---|---|---|---|
| En-lsu3F/ En-lsu3′R | Ec-ssu1′F/ Ec-ssu1R | STPYF/ STPYR2 | PSD7F/ PSD7R | ClPER-F/ ClPER-R | ||
| Escherichia coli | ATCC 11775T | + | − | − | − | − |
| Citrobacter freundii | ATCC 13316T | + | − | − | − | − |
| Citrobacter koseri | ATCC 27028T | + | − | − | − | − |
| Enterobacter cloacae | ATCC 13047T | + | − | − | − | − |
| Enterobacter aerogenes | ATCC 13048T | + | − | − | − | − |
| Enterobacter sakazakii | JCM 1233T | + | − | − | − | − |
| Klebsiella pneumoniae | ATCC 13883T | + | − | − | − | − |
| Klebsiella oxytoca | ATCC 13182T | + | − | − | − | − |
| Serratia marcescens | ATCC 13880T | + | − | − | − | − |
| Proteus mirabilis | ATCC 29906T | + | − | − | − | − |
| Proteus vulgaris | JCM 1668 | + | − | − | − | − |
| Proteus penneri | JCM 3948T | + | − | − | − | − |
| Hafnia alvei | JCM 1666T | + | − | − | − | − |
| Edwardsiella tarda | ATCC 15947T | + | − | − | − | − |
| Providencia alcalifaciens | ATCC 9886T | + | − | − | − | − |
| Providencia rettgerii | DSM 4542T | + | − | − | − | − |
| Morganella morganii | JCM 1672T | + | − | − | − | − |
| Salmonella choleraesuis subsp. choleraesuis serotype enteritidis | DSM 9898 | + | − | − | − | − |
| Yersinia enterocolitica | DSM 4780T | + | − | − | − | − |
| Enterococcus faecalis | ATCC 19433T | − | + | − | − | − |
| Enterococcus faecium | ATCC 19434T | − | + | − | − | − |
| Enterococcus hirae | ATCC 8043T | − | + | − | − | − |
| Enterococcus avium | JCM 8722T | − | + | − | − | − |
| Enterococcus gallinarum | JCM 8728T | − | + | − | − | − |
| Enterococcus casseliflavus | JCM 8723T | − | + | − | − | − |
| Enterococcus flavescens | DSM 7370T | − | + | − | − | − |
| Staphylococcus aureus | ATCC 12600T | − | − | + | − | − |
| Staphylococcus epidermidis | ATCC 14990T | − | − | + | − | − |
| Staphylococcus schleiferi subsp. coagulans | JCM 7470T | − | − | + | − | − |
| Pseudomonas aeruginosa | ATCC 10145T | − | − | − | + | − |
| Pseudomonas putida | DSM 291T | − | − | − | + | − |
| Clostridium perfringens | JCM 1290T | − | − | − | − | + |
| Ruminococcus productus | ATCC 27340T | − | − | − | − | − |
| Ruminococcus obeum | ATCC 29174T | − | − | − | − | − |
| Clostridium orbiscindens | DSM 6740T | − | − | − | − | − |
| Bacteriodes fragilis | DSM 2151T | − | − | − | − | − |
| Bacteroides vulgatus | JCM 5826T | − | − | − | − | − |
| Bifidobacterium adolescentis | ATCC 15703T | − | − | − | − | − |
| Bifidobacterium longum | ATCC 15707T | − | − | − | − | − |
| Collinsella aerofaciens | ATCC 25986T | − | − | − | − | − |
| Eggerthella lenta | ATCC 25559T | − | − | − | − | − |
| Prevotella melaninogenica | ATCC 25845T | − | − | − | − | − |
| Veillonella parvula | ATCC 10790T | − | − | − | − | − |
| Lactobacillus acidophilus | ATCC 4356T | − | − | − | − | − |
| Lactobacillus casei | ATCC 334T | − | − | − | − | − |
| Streptococcus intermedius | ATCC 27335 | − | − | − | − | − |
| Campylobacter jejuni | ATCC 33560T | − | − | − | − | − |
| Candida albicans | ATCC 18804T | − | − | − | − | − |
| Bacillus cereus | JCM 2152T | − | − | − | − | − |
| Bacillus subtilis | ATCC 14579T | − | − | − | − | − |
The specificity of the RT-qPCR assay for target bacteria with each primer was investigated using RNA extracts corresponding to 105 cells of each strain. Specificity was judged using the criteria described in Materials and Methods.
Development of rRNA-targeted primers.
By using 16S and 23S rRNA gene sequences obtained from the DDBJ/GenBank/EMBL databases for bacteria detected in the human intestinal tract, we constructed a multiple alignment of the target groups and reference organisms with the Clustal X program (44). After comparing the sequences, we identified potential primer target sites for group-specific detection for Enterobacteriaceae, Enterococcus, Staphylococcus, and Pseudomonas. We then designed the primers for Enterobacteriaceae, Enterococcus, Staphylococcus, and Pseudomonas listed in Table 2 and checked their specificities with the database by submitting the sequences to the Probe Match program of the Ribosomal Database Project (RDP-II) (http://rdp.cme.msu.edu/) (28).
TABLE 2.
Primers based on 16S or 23S rRNA sequences
| Target | Primer | Sequence | Product size (bp) | Reference |
|---|---|---|---|---|
| Enterobacteriaceae 23S rRNA | En-lsu3F | TGCCGTAACTTCGGGAGAAGGCA | 428 | This study |
| En-lsu3′R | TCAAGGCTCAATGTTCAGTGTC | |||
| Enterococcus 16S rRNA | Ec-ssu1′F | GGATAACACTTGGAAACAGG | 115 | This study |
| Ec-ssu1R | TCCTTGTTCTTCTCTAACAA | |||
| Staphylococcus 16S rRNA | STPYF | ACGGTCTTGCTGTCACTTATA | 257 | This study |
| STPYR2 | TACACATATGTTCTTCCCTAATAA | |||
| Pseudomonas 16S rRNA | PSD7F | CAAAACTACTGAGCTAGAGTACG | 215 | This study |
| PSD7R | TAAGATCTCAAGGATCCCAACGGCT | |||
| Clostridium perfringens 16S rRNA | ClPER-F | AGATGGCATCATCATTCAAC | 793 | 21 |
| ClPER-R | GCAAGGGATGTCAAGTGT |
Fecal sampling.
Fecal samples provided by 19 hospitalized patients were weighed and then suspended in 9 volumes of sterilized anaerobic transfer medium, which contained KH2PO4 (0.0225%, wt/vol), K2HPO4 (0.0225%, wt/vol), NaCl (0.045%, wt/vol), (NH4)2SO4 (0.0225%, wt/vol), CaCl2 (0.00225%, wt/vol), MgSO4 (0.00225%, wt/vol), Na2CO3 (0.3%, wt/vol), l-cysteine hydrochloride (0.05%, wt/vol), resazurin (0.0001%, wt/vol), Lab lemco powder (1.0%, wt/vol; Oxoid Co., Ltd., Basingstoke, United Kingdom), and glycerol (10%, wt/vol; Wako Pure Chemical Industries, Ltd., Osaka, Japan). After serial dilution of the fecal suspensions with a buffer solution containing KH2PO4 (0.0225%, wt/vol), K2HPO4 (0.0225%, wt/vol), NaCl (0.045%, wt/vol), (NH4)2SO4 (0.0225%, wt/vol), CaCl2 (0.00225%, wt/vol), MgSO4 (0.00225%, wt/vol), Na2CO3 (0.3%, wt/vol), l-cysteine hydrochloride (0.05%, wt/vol), and resazurin (0.0001%, wt/vol), 50-μl portions of the appropriate dilutions were spread onto the following culture media: DHL agar (Nikken Bio Medical Laboratory Inc., Kyoto, Japan) for Enterobacteriaceae and NAC agar (Eikenkizai Co., Ltd., Tokyo, Japan) for P. aeruginosa. DHL agar and NAC agar were incubated aerobically at 37°C for 24 h. The colonies on the agar plates were then counted, and the numbers of CFU of target bacteria per g (wet weight) of feces were calculated. The lower limit of bacterial detection with this procedure was 200 CFU per g of feces.
Blood sampling.
Human peripheral blood was collected from three healthy adult volunteers, and then sodium citrate (0.38%, wt/vol) was added and the preparations were mixed immediately. Tenfold serial dilutions of S. aureus or P. aeruginosa were added to the peripheral blood. After serial dilution of the samples with physiological saline, 500-μl samples of the appropriate dilutions were spread onto BHI agar and then incubated aerobically at 37°C for 24 h. The colonies on the agar plates were then counted, and the numbers of CFU of target bacteria per ml of blood were calculated. The lower limit of bacterial detection with this procedure was 2 CFU per ml of blood.
Isolation of total RNA.
For RNA stabilization, fresh cultures of each bacterial strain (50 μl), fecal homogenate samples (200 μl), or blood samples (500 μl) were added to 2 volumes of RNAprotect bacterial reagent (QIAGEN GmbH, Hilden, Germany), and then the preparations were incubated for 5 min at room temperature. After centrifugation of each mixture at 5,000 × g for 10 min, the supernatant was discarded, and the pellet was stored at −80°C until it was used for extraction of RNA. RNA was isolated using a modified acid guanidinium thiocyanate-phenol-chloroform extraction method (8). Briefly, a thawed sample was resuspended in a solution containing 346.5 μl RLT lysis buffer (catalog no. 79216; QIAGEN Sciences, Germantown, MD), 3.5 μl β-mercaptoethanol (Sigma-Aldrich Co., St. Louis, MO), and 100 μl Tris-EDTA buffer (pH 8.0). Then 300 mg of glass beads (diameter, 0.1 mm; BioSpec Products, Inc., Bartlesville, OK) was added to the suspension, and the mixture was vortexed vigorously for 60 s using a FastPrep FP 120 (BIO 101, Vista, CA) at a power level of 5.0. Then 500 μl acid phenol (Wako Pure Chemical Industries, Ltd.) was added, and the mixture was incubated for 10 min at 60°C. After incubation, the mixture was cooled on ice for 5 min and added to 100 μl chloroform-isoamyl alcohol. After centrifugation at 12,000 × g for 10 min at 4°C, 450 μl of the supernatant was collected and added to an equal volume of chloroform-isoamyl alcohol. After centrifugation at 12,000 × g for 5 min, 400 μl of the supernatant was collected and subjected to isopropanol precipitation. Finally, the nucleic acid fraction was suspended in 50 μl nuclease-free water. To remove contaminating genomic DNA from the RNA fraction, 0.5 U RNase-free DNase I (TaKaRa Bio Inc., Shiga, Japan) per μg RNA was added to each sample in a solution containing 1× DNase I buffer (TaKaRa Bio Inc.), which was then incubated at 37°C for 20 min. After incubation, the DNase was inactivated and removed twice by acid-phenol and chloroform-isoamyl alcohol extraction as described above, and the RNA in the resultant supernatant was collected by isopropanol precipitation. Finally, the RNA was suspended in 50 μl nuclease-free water. The quantity of RNA was confirmed spectrophotometrically.
RT-qPCR.
The RT-qPCR analysis was conducted with one-step reactions using a QIAGEN OneStep RT-PCR kit (QIAGEN GmbH). Each reaction mixture (20 μl) was composed of 1× QIAGEN OneStep RT-PCR buffer, each deoxynucleoside triphosphate at a concentration of 400 μM, a 1:100,000 dilution of SYBR green I (catalog no. 50513; BioWhittaker Molecular Applications, Rockland, ME), 1 μl QIAGEN OneStep RT-PCR enzyme mixture, each of the specific primers at a concentration of 0.6 μM, and 2 μl template RNA. The reaction mixture was incubated at 50°C for 30 min for reverse transcription. The continuous amplification program consisted of one cycle at 95°C for 15 min, followed by 40 cycles at 94°C for 20 s, 60°C for 20 s, and 72°C for 50 s and finally one cycle at 94°C for 15 s. The fluorescent products were detected in the last step of each cycle. A melting curve analysis was performed after amplification to distinguish the target from the nontargeted PCR products. The melting curve was obtained by slow heating at temperatures from 60 to 95°C at a rate of 0.2°C/s with continuous fluorescence collection. Amplification and detection were performed in 96-well optical plates with an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA).
DNA extraction and qPCR.
DNA extraction was performed by using the method described by Matsuki et al., with minor modifications (29). Briefly, DNA was extracted from a fresh culture of each bacterium (50 μl) and suspended in 50 μl Tris-EDTA buffer. qPCR were conducted using a QIAGEN OneStep RT-PCR kit (QIAGEN GmbH). Each qPCR was performed in a 20-μl reaction mixture containing DNA and SYBR green I (Molecular Probes) by using the same conditions that were used for RT-qPCR except for the reverse transcription step. qPCR amplification and detection were performed in 96-well optical plates with an ABI PRISM 7900HT sequence detection system (Applied Biosystems).
Determination of bacterial number by RT-qPCR.
A standard curve was generated with the RT-qPCR data (using the threshold cycle [CT] value, the cycle number when the threshold fluorescence was reached) and the corresponding cell count, which was determined microscopically with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc., Burlingame, CA) staining using the method of Jansen et al. (20), for dilution series of the following standard strains: E. coli ATCC 11775T (for Enterobacteriaceae), E. faecalis ATCC 19433T (for Enterococcus), S. aureus ATCC 12600T (for Staphylococcus), C. perfringens JCM 1290T (for C. perfringens), and P. aeruginosa ATCC 10145T (for Pseudomonas). For determination of the bacteria present in samples, three serial dilutions of an extracted RNA sample were used for RT-qPCR, and the CT values in the linear range of the assay were applied to the standard curve generated in the same experiment to obtain the corresponding number of bacteria in each nucleic acid sample and then converted to the number of bacteria per sample.
The specificity of the RT-qPCR assay was determined as follows. Total RNA fractions extracted from the cells of 50 bacterial strains corresponding to 105 cells were assessed for the RT-qPCR using the group- or species-specific primers shown in Table 2. Using the standard curve for the representative strain of each group obtained as described above, the amplified signal was considered positive when it was greater than the signal for 104 standard cells and negative when it was less than the signal for 10−1 standard cell.
In situ viability staining.
The viability of the bacteria was assessed using a LIVE/DEAD BacLight bacterial viability kit (catalog no. L7012; Molecular Probes, Eugene, OR). Fresh bacterial cultures were incubated with 5 μM SYTO9 (Molecular Probes) and 30 μM propidium iodide (PI) (Molecular Probes) at 30°C for 10 min in the dark. SYTO9 and PI bind to DNA, and the complexes have an excitation maximum of 480 nm and an emission maximum of 500 nm for SYTO9 and an excitation maximum of 490 nm and an emission maximum of 635 nm for PI. SYTO9 is a green fluorescent dye that penetrates both viable and nonviable cells, while PI penetrates only bacteria with damaged plasma membranes (such as heat-treated or chemically treated, nonviable cells), quenching the green SYTO9 fluorescence (40). Thus, bacterial cells with compromised membranes fluoresce red, and bacterial cells with intact membranes fluoresce green. After incubation, cell suspensions were mixed with VECTASHIELD mounting medium (Vector Laboratories, Inc.) and then trapped between a glass slide and a square coverslip. The cells were imaged with a fluorescence microscope (Olympus BX-50; Olympus, Napa, CA) with a BX-FLA reflected-light fluorescence attachment using a combined fluorescein isothiocyanate-tetrarhodamine isothiocyanate filter set (catalog no. 51004v2; Chroma Technologies Corp, Brattleboro, VT). Images were then produced by using the image analysis software Image-Pro Plus, version 4 (Media-Cybernetics, Silver Spring, MD), and the ratio of the number of cells with green fluorescence (viable cells) to the total number of cells detected in each field (with both green and red fluorescence) was calculated. At the same time, the DAPI staining method was used to determine the total cell count in the bacterial culture. By multiplying the ratio for the viable cells by the total bacterial count obtained by DAPI staining, the number of viable cells in the culture was calculated with the following equation: number of viable bacteria = (number of cells labeled with SYTO9/number of cells labeled with both SYTO9 and PI) × (number of cells stained with DAPI).
Statistical analysis.
We employed the SPSS14.0 software (SPSS Japan Inc., Tokyo, Japan). A regression analysis was performed to determine the statistical correlation of the results, and Pearson's product-moment correlation coefficient was calculated. A P value of <0.05 was considered significant.
RESULTS
Quantitative detection of bacteria by RT-qPCR compared with detection by qPCR.
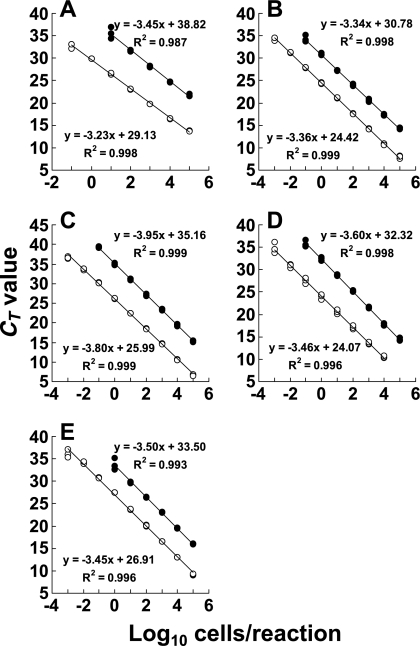
As shown in Fig. 1, the bacterial count obtained by direct staining (x axis) and the RT-qPCR value (CT value, y axis) were found to correlate well over the range of RNA dilutions corresponding to bacterial counts ranging from 105 to 10−3 cell per reaction for E. faecalis, S. aureus, C. perfringens, and P. aeruginosa (Fig. 1B to E) and ranging from 105 to 10−1 cell per reaction for E. coli (Fig. 1A) (R2, >0.99). Although there was nonspecific amplification of E. coli DNA or RNA that may have resulted from RT-qPCR reagents such as Taq DNA polymerase (data not shown), E. coli at a concentration of 10−1 cell per reaction was distinguishable (Fig. 1A). A comparison of the analytical curves for RT-qPCR with those for qPCR revealed no significant differences in slopes for the same target bacteria, indicating that the amplification efficiencies of RT-qPCR and qPCR were nearly equal, while the y-axis intercepts (CT values) of the RT-qPCR analytical curve were 6 to 10 cycles less than those of the qPCR curve, indicating that the RT-qPCR assay was 64- to 1,024-fold more sensitive than the qPCR assay.
FIG. 1.
Quantitative detection of bacteria by RT-qPCR and by qPCR. E. coli ATCC 11775T (A), E. faecalis ATCC 19433T (B), S. aureus ATCC 12600T (C), C. perfringens JCM 1290T (D), and P. aeruginosa ATCC 10145T (E) were cultivated separately in BHI or MRS broth. RNA and DNA fractions were extracted from culture samples in the early stationary phase (18 h), and the bacterial counts were determined microscopically with DAPI staining. Based on the bacterial counts, 10-fold serial dilutions of RNA or DNA from 105 to 10−3 bacteria were assessed by RT-qPCR (○) and qPCR (•) assays. The CT values for triplicate samples obtained were plotted against the log10 number of bacterial cells subjected to each reaction.
Next, total RNA extracts corresponding to 105 cells of 50 strains belonging to 50 species (Table 1) were assessed for specific detection of the target bacteria by RT-qPCR with the group-specific primers En-lsu3F and En-lsu3′R (for Enterobacteriaceae), Ec-ssu1′F and Ec-ssu1R (for Enterococcus), STPYF and STPYR2 (for Staphylococcus), and PSD7F and PSD7R (for Pseudomonas). As shown in Table 1, the primers gave positive RT-qPCR results only for the corresponding target bacterial species and did not cross-react with any of the nontarget microorganisms tested. The specificity of primers ClPER-F and ClPER-R for C. perfringens reported previously (21) was also confirmed.
Comparison of the bacterial counts in culture determined by RT-qPCR and the culture method.
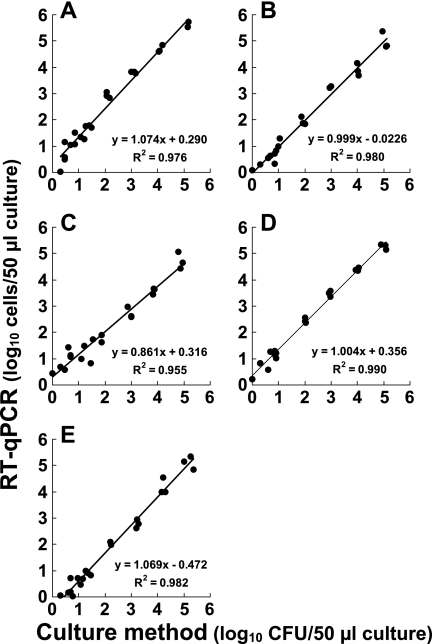
The bacterial counts in the serial dilutions of in vitro cultures were determined by RT-qPCR and compared with the corresponding CFU counts. As shown in Fig. 2, specific amplification was detected for all the samples of five species at levels less than 101 CFU, and the CT values and CFU counts were found to correlate well in the range from 100 to 105 CFU (R2, >0.90) (Fig. 2). Based on these results, we suggest that rRNA-targeted RT-qPCR can determine the number of bacteria sensitively with a detection limit of 100 CFU.
FIG. 2.
Comparison of bacterial counts in cultures determined by RT-qPCR and by the culture method. E. coli ATCC 11775T (A), E. faecalis ATCC 19433T (B), S. aureus ATCC 12600T (C), C. perfringens JCM 1290T (D), and P. aeruginosa ATCC 10145T (E) were cultivated in BHI or MRS broth. RNA fractions were extracted from 10-fold serial dilutions of each bacterial culture (50 μl) in the range from 100 to 105 CFU. The number of bacteria in each sample was determined by RT-qPCR and then plotted against the CFU count for the same sample determined on BHI (for E. coli, E. faecalis, S. aureus,and P. aeruginosa) or GAM (for C. perfringens) agar plates; data for single samples from each of the three different cultures are shown for each dilution. For RT-qPCR, an analytical curve generated with the RNA dilution series for each target strain (Fig. 1) was used.
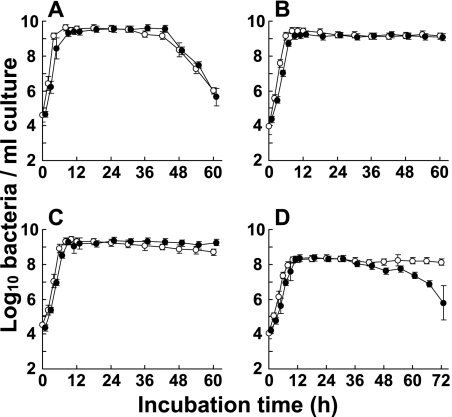
Effect of growth phase on bacterial counts determined by RT-qPCR.
The numbers of E. coli, E. faecalis, S. aureus, and C. perfringens cells in in vitro cultures were evaluated periodically throughout the growth phases until 60 h (72 h for C. perfringens) both by RT-qPCR and by the culture method using a starting concentration of around 104 CFU per ml (Fig. 3). The RT-qPCR counts were calculated using the analytical curve for each standard strain at the early stationary phase (18 h) obtained in the experiment described above (Fig. 1). Throughout the growth phase until the stationary phase, the bacterial counts obtained by RT-qPCR were in good agreement with the counts obtained by the culture method for all the bacterial species tested (Fig. 3). For E. coli, the RT-qPCR counts decreased rapidly from 42 to 60 h during incubation, showing much the same pattern as the CFU counts (Fig. 3A). For E. faecalis and S. aureus, the population levels remained unchanged for 60 h during the stationary phase without any dissociation between the RT-qPCR counts and the CFU counts (Fig. 3B and C). On the other hand, for C. perfringens, although no significant difference between the two methods was detected until 42 h, dissociation was observed during the starvation period from 42 to 72 h (Fig. 3D); the CFU counts were found to be clearly lower than the RT-qPCR counts.
FIG. 3.
Effect of growth phase on bacterial counts determined by RT-qPCR. Throughout the growth phase in broth culture, the numbers of E. coli ATCC 11775T (A), E. faecalis ATCC 19433T (B), S. aureus ATCC 12600T (C), and C. perfringens JCM 1290T (D) cells were determined by RT-qPCR (○) and the culture method (•). The analytical curves generated with the RNA dilution series for each target strain in the stationary phase (18 h) (Fig. 1) were used to quantify the bacteria. The CFU counts were determined on BHI (for E. coli, E. faecalis, and S. aureus) or GAM (for C. perfringens) agar plates. The results are the means and standard deviations of triplicate samples.
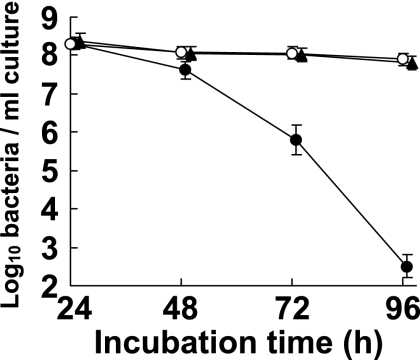
Comparison of RT-qPCR counts and viable bacterial cell counts by using a combination of DAPI staining and SYTO9-PI double staining of cultured bacteria.
To further investigate the dissociation of the RT-qPCR counts and CFU counts for C. perfringens at the later stages of culture as described above, we determined the viable cell counts under conditions in which starved C. perfringens cultures were unable to form colonies. We used the SYTO9-PI double staining method, which has been reported to be able to differentiate live and dead bacteria based on differences in plasma membrane permeability (3, 15). The number of live cells stained only with SYTO9 remained 108 throughout the test period, while the CFU counts decreased markedly, demonstrating that most of the bacteria that lost the ability to form colonies on an agar plate were still alive and maintained their cell membrane integrity and that the numbers of cells in the population that could be detected were nearly equal to those detected by RT-qPCR (Fig. 4).
FIG. 4.
Comparison of RT-qPCR counts and viable bacterial cell counts determined by a combination of DAPI staining and SYTO9-PI double staining of cultured bacteria. C. perfringens JCM 1290T was incubated in MRS broth for 4 days and examined by RT-qPCR (○), SYTO9-PI double staining (▴), and the culture method (•) at 24, 48, 72, and 96 h. The viable bacterial count was calculated with the following equation: viable bacterial count = (number of cells labeled with SYTO9/number of cells labeled with both SYTO9 and PI) × (number of cells stained with DAPI). For RT-qPCR, the analytical curve generated with the dilution series of RNA extracted from C. perfringens cells at 18 h (Fig. 1D) was used to determine the bacterial number. The CFU count was determined by culturing samples on GAM agar plates for 24 h. The results are the means and standard deviations of triplicate samples.
Comparison of the bacterial counts in human feces and blood determined by RT-qPCR and the culture method.
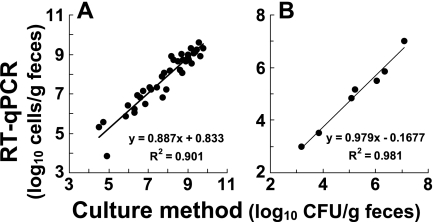
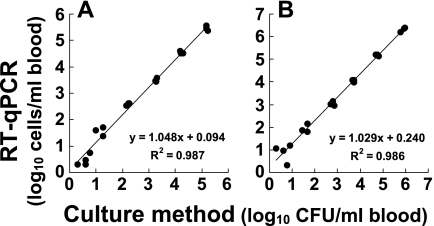
In the next series of experiments, the applicability of the RT-qPCR method for enumeration of limited bacterial populations in the fecal flora or peripheral blood was examined. As shown in Fig. 5, members of the Enterobacteriaceae were detected in 38 samples from 19 hospitalized patients and P. aeruginosa was detected in seven samples, and linear regression was performed for the number of bacteria obtained by RT-qPCR and the number of bacteria obtained by the culture method. The experimental curve obtained for the Enterobacteriaceae had a slope of 0.887 and a correlation coefficient of 0.901 (Fig. 5A), and the curve for P. aeruginosa had a slope of 0.979 and a correlation coefficient of 0.981 (Fig. 5B), suggesting that there was a good correlation between the two methods. On the other hand, when S. aureus and P. aeruginosa were spiked into human peripheral blood, 2 CFU of S. aureus and 1 CFU of P. aeruginosa in 500 μl of human peripheral blood could be detected by RT-qPCR (Fig. 6). No false-positive results were obtained for the bacterium-free controls, showing that there was neither reagent contamination nor a cross-reaction with human nucleic acids in the determination (data not shown).
FIG. 5.
Correlation between RT-qPCR counts and cultural counts in human feces. Total RNA fractions extracted from 38 human fecal homogenates were assessed by the RT-qPCR assay to determine the indigenous population levels of Enterobacteriaceae (A) and P. aeruginosa (B). The CT values obtained were applied to the analytical curves for E. coli ATCC 11775T and P. aeruginosa ATCC 10145T (Fig. 1A and E) to determine the RT-qPCR counts. The CFU counts were determined by culturing the same fecal samples on DHL (A) or NAC (B) agar plates and then were plotted against the RT-qPCR counts.
FIG. 6.
Comparison of bacterial counts in human peripheral blood determined by RT-qPCR and the culture method. Human peripheral blood samples (0.5 ml) from three individuals were spiked with various amounts of live S. aureus ATCC 12600T (A) or P. aeruginosa ATCC 10145T (B) to obtain final concentrations ranging from 100 to 106 CFU per ml. RNA fractions extracted from each sample were then assessed by the RT-qPCR assay. The CT values obtained were applied to the analytical curves for S. aureus ATCC 12600T and P. aeruginosa ATCC 10145T (Fig. 1C and E) to determine the RT-qPCR counts. The CFU counts were determined by culturing the same samples on BHI agar plates and then were plotted against the RT-qPCR counts; data for single samples from the three different donors are shown.
DISCUSSION
To develop a sensitive, specific, and convenient quantitative RT-PCR method to detect commensal subdominant bacteria, we focused on rRNA as the target. The sensitivity of the rRNA-targeted RT-qPCR method was approximately 100- to 1,000-fold higher than the sensitivity of the DNA-targeted qPCR (Fig. 1). rRNA is a universal constituent of bacterial ribosomes, and 5S, 16S, and 23S rRNAs are the components of small (30S) and large (50S) subunits that comprise the complete active ribosome (70S). In E. coli, the total number of ribosomal particles (30S, 50S, and 70S particles) per cell is known to reach a peak of more than 20,000 (1), with approximately 103 copies of each rRNA species per cell, while only seven copies of rRNA operons are present in a cell. Moreover, rRNA constitutes the largest fraction of RNA in the cell, and the proportion of rRNA in the total RNA is more than 80% (17). These aspects of rRNA, the high copy number and the high proportion of molecules, seem to contribute to the higher sensitivity of detection by RT-PCR than by PCR (Fig. 1). Although the expression of the rRNA gene has been considered more constant than the expression of other genes, which has been frequently used in quantitative studies and as an internal reference to analyze other gene expression (45, 47), the control of rRNA synthesis in bacteria has been found to be dependent on the growth rate. In rapidly dividing bacteria, the ribosome content per bacterium is much greater than that in slowly dividing cells (9), which is known as growth rate-dependent control. In the case of E. coli, the ribosome content has been reported to vary by more than 10-fold when the growth rate increased from a doubling time of 100 min to a doubling time of 24 min (10), and the rRNA synthesis is repressed by feedback mechanisms that prevent excessive production of more ribosomes than are needed for protein synthesis during balanced or steady-state growth (2, 9). These aspects of rRNA affect whether this molecule can be used as a target for standardizing bacterial populations by RT-qPCR. In this study, although comparisons of RT-qPCR counts and CFU counts during the logarithmic phase showed that the bacterial counts obtained by RT-qPCR tended to be higher than the CFU counts (Fig. 3), there was no significant difference between the values; the differences were at most fourfold. Therefore, we suggest that the rRNA-targeted RT-qPCR method is suitable for quantification of a bacterial population irrespective of the growth phase. However, it is important to consider the changeable metabolic activity of bacteria in order to obtain correct data by this procedure.
Recently, RNA molecules have been used as an indicator of bacterial cell viability as an alternative to colony-forming ability or DNA molecules (6, 11, 14, 32). rRNA has been recognized as more labile and is more susceptible to degradation caused by adverse treatment than DNA, and its level is positively correlated with viability under some bacterial killing regimens (34). In E. coli, the decrease in the level of rRNA during 60 h of incubation showed a good correlation with the decrease in the number of CFU (Fig. 3A). In addition, the RT-qPCR counts for Enterobacteriaceae and P. aeruginosa in human feces were highly correlated with the CFU counts (Fig. 5). However, as it has been reported that severe stress, such as heat shock (33), UV irradiation (34), or ethanol treatment (39), increases the dissociation between the CFU count and the rRNA content, further analysis of whether rRNA can be used as an accurate indicator of bacterial viability under any possible biological conditions is still needed. In any case, the results suggest that the amount of rRNA can be used as an indicator of viable bacterial population size at least in physiologic circumstances such as in gastrointestinal tracts.
The CFU counts of C. perfringens were significantly lower than the RT-qPCR counts after 42 h of incubation, and the viable cell counts determined with a LIVE/DEAD BacLight bacterial viability kit were nearly equal to the RT-qPCR counts rather than the CFU counts (Fig. 4). The viable but not culturable state of bacteria has been reported to be a survival mechanism for bacteria that allows them to face environmentally stressful conditions, such as starvation, incubation outside the temperature range for growth, elevated osmotic concentrations, or exposure to white light (37). When in such a state, bacteria often do not grow on conventional culture media but still have metabolic activity, maintain pathogenicity (24), and, in some cases, may return to active growth when optimal conditions are restored (7, 25). The viability of nonculturable cells is typically determined by the substrate responsive assay (direct viable count assay) (22), by detection of respiratory activity (CTC assay) (19), by monitoring the membrane potential (26), or by determining the presence of an intact cytoplasmic membrane (3, 4, 15, 27). Although approximately 60 species have now been reported to demonstrate this physiological response, there have been no descriptions of clostridia (37). Therefore, for this phenomenon in C. perfringens, further analysis of the bacterial cell status from several perspectives, such as metabolic activity and membrane potential, in addition to the presence of nucleic acids, membrane integrity, and cultivability, is still needed.
The new rRNA-targeted RT-qPCR technique developed in the present study enables detection of minor bacterial species, such as members of the Enterobacteriaceae, Enterococcus, Staphylococcus, and C. perfringens, with sensitivity equal to that of the culture method (102 to 103 CFU per g feces), as well as detection of the predominant populations in the intestines. Because of its high sensitivity and convenience, the RT-qPCR assay targeting rRNA may be useful for a wide variety of bacteriological examinations. It can be used for detection of opportunistic infections in clinical settings. Quick evaluation of contamination is essential in clinical examinations, but even the molecular methods previously reported require a cultivation step to increase the number of cells several days prior to the assay (23). The entire RT-qPCR assay, including the RNA extraction step developed in this study, can be completed in 5 h, and its sensitivity may allow omission of the cultivation step and eliminate the risk of false-positive results. In addition to assessment of specific bacterial counts in feces and peripheral blood, it can be used for rapid detection of potential bacterial contamination in tissue specimens and smaller bacterial populations, such as oral and vaginal microfloras. Moreover, the RT-qPCR assay might also be an effective tool for examining environmental microbial populations, such as those in water and soil, and for quick evaluation of food contamination. The method may be especially valuable for detecting noncultivable, subdominant members of bacterial communities or for examining samples that have been frozen and therefore are not suitable for culture-based examination. On the other hand, identification of certain functions of bacteria is the next objective after determination of the exact population levels by RT-qPCR, leading to information about what commensal bacteria do in the corresponding environments. By using the same RNA specimens used for rRNA quantification, various information concerning bacterial functions should be available from the viewpoint of mRNA. For example, specific pathogens have unique virulence factors, such as the production of toxins, an apparatus for invasion, and drug resistance, which can also be targets for quantitative analysis of the corresponding mRNA expression.
In conclusion, we developed an RT-qPCR detection method targeting rRNA to enumerate bacteria in human feces and peripheral blood. Specific primers for rRNA sequences of Enterobacteriaceae, Enterococcus, Staphylococcus, Pseudomonas, and C. perfringens were used in conjunction with RT-qPCR, which allowed sensitive and accurate quantification of the target bacteria. The sensitivity was approximately 100-fold higher than that of the existing PCR methods and nearly equivalent to that of conventional culture methods. This RT-qPCR method should be an effective tool for sensitive quantification of viable bacterial populations.
Acknowledgments
We thank Toshihiko Takada and Takahiro Matsuki for their technical advice. We also thank Rie Fujioka for her assistance with this research.
Footnotes
Published ahead of print on 27 October 2006.
REFERENCES
- 1.Arfvidsson, C., and K. G. Wahlund. 2003. Time-minimized determination of ribosome and tRNA levels in bacterial cells using flow field-flow fractionation. Anal. Biochem. 313:76-85. [DOI] [PubMed] [Google Scholar]
- 2.Asato, Y. 2005. Control of ribosome synthesis during the cell division cycles of E. coli and Synechococcus. Curr. Issues Mol. Biol. 7:109-117. [PubMed] [Google Scholar]
- 3.Auty, M. A., G. E. Gardiner, S. J. McBrearty, E. O. O'Sullivan, D. M. Mulvihill, J. K. Collins, G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2001. Direct in situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl. Environ. Microbiol. 67:420-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banning, N., S. Toze, and B. J. Mee. 2002. Escherichia coli survival in groundwater and effluent measured using a combination of propidium iodide and the green fluorescent protein. J Appl. Microbiol. 93:69-76. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ Microbiol. 70:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleve, G., L. Rizzotti, F. Dellaglio, and S. Torriani. 2003. Development of reverse transcription (RT)-PCR and real-time RT-PCR assays for rapid detection and quantification of viable yeasts and molds contaminating yogurts and pasteurized food products. Appl. Environ Microbiol. 69:4116-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaveerach, P., A. A. ter Huurne, L. J. Lipman, and F. van Knapen. 2003. Survival and resuscitation of ten strains of Campylobacter jejuni and Campylobacter coli under acid conditions. Appl. Environ. Microbiol. 69:711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis, P. P., M. Ehrenberg, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68:639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreier, J., M. Stormer, and K. Kleesiek. 2004. Two novel real-time reverse transcriptase PCR assays for rapid detection of bacterial contamination in platelet concentrates. J. Clin. Microbiol. 42:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Favier, C. F., E. E. Vaughan, W. M. De Vos, and A. D. Akkermans. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl. Environ. Microbiol. 68:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felek, S., A. Unver, R. W. Stich, and Y. Rikihisa. 2001. Sensitive detection of Ehrlichia chaffeensis in cell culture, blood, and tick specimens by reverse transcription-PCR. J. Clin. Microbiol. 39:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardiner, G. E., E. O'Sullivan, J. Kelly, M. A. Auty, G. F. Fitzgerald, J. K. Collins, R. P. Ross, and C. Stanton. 2000. Comparative survival rates of human-derived probiotic Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl. Environ. Microbiol. 66:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueimonde, M., S. Tolkko, T. Korpimaki, and S. Salminen. 2004. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, M. C., A. K. Nielsen, S. Molin, K. Hammer, and M. Kilstrup. 2001. Changes in rRNA levels during stress invalidates results from mRNA blotting: fluorescence in situ rRNA hybridization permits renormalization for estimation of cellular mRNA levels. J. Bacteriol. 183:4747-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatzinger, P. B., P. Palmer, R. L. Smith, C. T. Penarrieta, and T. Yoshinari. 2003. Applicability of tetrazolium salts for the measurement of respiratory activity and viability of groundwater bacteria. J Microbiol. Methods 52:47-58. [DOI] [PubMed] [Google Scholar]
- 20.Jansen, G. J., A. C. Wildeboer-Veloo, R. H. Tonk, A. H. Franks, and G. W. Welling. 1999. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215-221. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, E., Y. Miyamoto, S. Narushima, and K. Itoh. 2002. Design of species-specific primers to identify 13 species of Clostridium harbored in human intestinal tracts. Microbiol. Immunol. 46:353-358. [DOI] [PubMed] [Google Scholar]
- 22.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:415-420. [DOI] [PubMed] [Google Scholar]
- 23.Kurupati, P., C. Chow, G. Kumarasinghe, and C. L. Poh. 2004. Rapid detection of Klebsiella pneumoniae from blood culture bottles by real-time PCR. J. Clin. Microbiol. 42:1337-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lleo, M. M., B. Bonato, C. Signoretto, and P. Canepari. 2003. Vancomycin resistance is maintained in enterococci in the viable but nonculturable state and after division is resumed. Antimicrob. Agents Chemother. 47:1154-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lleo, M. M., B. Bonato, M. C. Tafi, C. Signoretto, M. Boaretti, and P. Canepari. 2001. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J. Appl. Microbiol. 91:1095-1102. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Amoros, R., S. Castel, J. Comas-Riu, and J. Vives-Rego. 1997. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide, and CTC. Cytometry 29:298-305. [DOI] [PubMed] [Google Scholar]
- 27.Lowder, M., A. Unge, N. Maraha, J. K. Jansson, J. Swiggett, and J. D. Oliver. 2000. Effect of starvation and the viable-but-nonculturable state on green fluorescent protein (GFP) fluorescence in GFP-tagged Pseudomonas fluorescens A506. Appl. Environ. Microbiol. 66:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto, M., M. Sakamoto, H. Hayashi, and Y. Benno. 2005. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colonic microbiota. J Microbiol. Methods 61:305-319. [DOI] [PubMed] [Google Scholar]
- 32.McKillip, J. L., and M. Drake. 2004. Real-time nucleic acid-based detection methods for pathogenic bacteria in food. J. Food Prot. 67:823-832. [DOI] [PubMed] [Google Scholar]
- 33.McKillip, J. L., L. A. Jaykus, and M. Drake. 1999. Nucleic acid persistence in heat-killed Escherichia coli O157:H7 from contaminated skim milk. J. Food Prot. 62:839-844. [DOI] [PubMed] [Google Scholar]
- 34.McKillip, J. L., L. A. Jaykus, and M. Drake. 1998. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagashima, K., T. Hisada, M. Sato, and J. Mochizuki. 2003. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl. Environ. Microbiol. 69:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43(Spec. No.):93-100. [PubMed] [Google Scholar]
- 38.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J. Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan, G. E., C. I. Masters, J. A. Shallcross, and B. M. MacKey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stocks, S. M. 2004. Mechanism and use of the commercially available viability stain, BacLight. Cytometry 61A:189-195. [DOI] [PubMed] [Google Scholar]
- 41.Suau, A. 2003. Molecular tools to investigate intestinal bacterial communities. J. Pediatr. Gastroenterol. Nutr. 37:222-224. [DOI] [PubMed] [Google Scholar]
- 42.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takada, T., K. Matsumoto, and K. Nomoto. 2004. Development of multi-color FISH method for analysis of seven Bifidobacterium species in human feces. J Microbiol. Methods 58:413-421. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small-colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoetendal, E. G., C. T. Collier, S. Koike, R. I. Mackie, and H. R. Gaskins. 2004. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 134:465-472. [DOI] [PubMed] [Google Scholar]