Abstract
Several characteristics of the 16S rRNA gene, such as its essential function, ubiquity, and evolutionary properties, have allowed it to become the most commonly used molecular marker in microbial ecology. However, one fact that has been overlooked is that multiple copies of this gene are often present in a given bacterium. These intragenomic copies can differ in sequence, leading to identification of multiple ribotypes for a single organism. To evaluate the impact of such intragenomic heterogeneity on the performance of the 16S rRNA gene as a molecular marker, we compared its phylogenetic and evolutionary characteristics to those of the single-copy gene rpoB. Full-length gene sequences and gene fragments commonly used for denaturing gradient gel electrophoresis were compared at various taxonomic levels. Heterogeneity found between intragenomic 16S rRNA gene copies was concentrated in specific regions of rRNA secondary structure. Such “heterogeneity hot spots” occurred within all gene fragments commonly used in molecular microbial ecology. This intragenomic heterogeneity influenced 16S rRNA gene tree topology, phylogenetic resolution, and operational taxonomic unit estimates at the species level or below. rpoB provided comparable phylogenetic resolution to that of the 16S rRNA gene at all taxonomic levels, except between closely related organisms (species and subspecies levels), for which it provided better resolution. This is particularly relevant in the context of a growing number of studies focusing on subspecies diversity, in which single-copy protein-encoding genes such as rpoB could complement the information provided by the 16S rRNA gene.
Molecular microbial ecology commenced in 1990 with the direct amplification and sequencing of 16S rRNA genes from the environment (17). This procedure revolutionized microbial ecology and permanently changed the way we study prokaryotes in the environment. The adoption of molecular tools by microbial ecologists has rapidly enhanced our knowledge of prokaryote abundance, diversity, and function. Certain molecular tools are now commonly used, including clone libraries, fluorescent in situ hybridization (FISH), and fingerprinting methods, such as denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP) analysis. Each of these methods measures a different aspect of the community (diversity, in situ detection, and community dynamics, respectively), but all of them generally utilize a single gene to differentiate operational taxonomic units (OTUs), namely, the 16S rRNA gene.
Pauling and Zuckerkandl (45) first proposed the use of gene sequences as a molecular clock to decipher phylogenetic relationships. Woese and Fox (43, 44) introduced the use of rRNA genes for this purpose, which served as the basis for their definition of the three domains of life. The notion that rRNA genes could identify an organism by reconstructing its phylogeny, along with the possibility of storing sequences in databases, resulted in the rapid adoption of the 16S rRNA gene by microbiologists. This gene has now established itself as the “gold standard,” not only in bacterial phylogeny but also in microbial ecology studies.
However, none of the 16S rRNA-based molecular methods allows for an accurate representation of microbial communities. While bias is introduced into molecular community analysis by many mechanical factors, such as sample handling, fixation, DNA extraction, and PCR (9, 12, 13, 15, 28, 42), it is also created by the existence of multiple heterogeneous copies of the 16S rRNA gene within a genome (10, 11).
The implications of using a gene displaying intragenomic heterogeneity for fingerprinting methods used in molecular community analysis were first described by Dahllöf et al. (11). They demonstrated that single species could produce complex banding patterns with DGGE, similar to those reported for whole communities. Subsequently, Crosby and Criddle (10) used an artificial community of bacteria whose whole genomes have been sequenced completely to determine the factors causing both over- and underestimates of diversity with various fingerprinting methods.
Most researchers currently adopt an OTU definition based on 16S rRNA gene sequence identity, usually considering organisms displaying 97 to 98% identity in this gene to be part of the same OTU (16). However, recent evidence suggests that even when taking into account intragenomic heterogeneity, clusters of sequences with 99% sequence identity reveal extensive ribotype microdiversity, which potentially underlies important ecological differentiation (2). For example, in a natural Vibrio splendidus population, even single ribotypes (unique 16S rRNA gene sequences) present important genotypic variations (38). The case for greater diversity than can be detected using current 16S rRNA gene-based molecular community analysis techniques is also supported by multilocus sequence analysis (MLSA) studies, which involve the sequencing of a number of genes coding for proteins with housekeeping functions to assess diversity in collections of isolates (23, 36). Such studies have identified organisms with identical 16S rRNA gene sequences that have significant sequence divergence in protein-encoding genes. Although the 16S rRNA gene is by far the most frequently used gene, methods in molecular microbial ecology are not limited to this gene. Alternative core housekeeping genes, such as the RNA polymerase β subunit gene (rpoB), have been used with DGGE (7, 11, 25, 31, 32).
The use of a single-copy gene for community analysis is an important milestone in microbial ecology, as it could allow for the accurate measurement of diversity and phylogenetic relationships, avoiding a loss in phylogenetic resolution and biases in diversity measurements due to the presence of intragenomic heterogeneity. However, the criteria for which the 16S rRNA gene was originally selected must be established for alternative markers. In this study, a comparison between the 16S rRNA and rpoB genes is performed in order to evaluate the use of an alternative gene as a marker for molecular microbial ecology.
This study addresses four issues. Firstly, the localization of intragenomic heterogeneity within specific regions of the 16S rRNA molecule is explored. Secondly, we compare how well the 16S rRNA and rpoB genes and their fragments used for DGGE reconstruct bacterial phylogenies. Thirdly, the influence of 16S rRNA gene intragenomic heterogeneity on bacterial phylogeny at the subspecies level is determined. Finally, we address whether the rpoB gene fulfills the criteria required for a molecular marker in microbial ecology.
MATERIALS AND METHODS
Sequence retrieval and alignment.
rpoB and 16S rRNA gene DNA sequences were retrieved for 111 completely sequenced bacterial genomes from the NCBI Microbial Genome Database (data not shown). DNA sequences of the rpoB gene were translated to create an amino acid sequence data set. For each taxonomic group analyzed, amino acid alignments (RpoB protein) and DNA alignments (rpoB and 16S rRNA genes) were constructed using CLUSTALW (37) and edited manually to remove ambiguous positions. The 16S rRNA gene alignment was edited to begin and end at the universal primer sites 27F and 1492R, which are most commonly used for clone library construction. Taxonomic groups for which alignments were compiled cover multiple taxonomic levels, including the domain (Bacteria), phylum (Actinobacteria, Firmicutes, and Proteobacteria), order (Bacillales, Enterobacteriales, and Lactobacillales), class (Alphaproteobacteria and Gammaproteobacteria), family (Chlamydiaceae and Mycoplasmataceae), genus (Streptococcus), and species/subspecies (Escherichia coli/Shigella flexneri) levels.
Recoding of alignment positions displaying intragenomic heterogeneity.
Multiple copies of the 16S rRNA gene present in a single organism were condensed to a single consensus sequence. When heterogeneity was present between multiple copies found in a given genome, the specific positions at which this heterogeneity occurred were recoded to denote the multiple character states observed for that position. The following code, which is recognized by the PAUP* program (35) used to compile phylogenetic trees, was used for recoding: R = AG, Y = CT, M = AC, K = GT, W = AT, S = CG, B = CGT, D = AGT, H = ACT, V = ACG, and N = ACGT.
Mutational saturation analysis.
The first and third positions within codons, which are synonymous for many amino acids, display higher mutational rates than the second position. If the sequences compared are sufficiently distant, then multiple consecutive nucleotide substitutions at the same position can occur after differentiation, making the position uninformative and/or misleading with regards to phylogenetic analyses. We therefore tested whether the rpoB data sets used for phylogenetic analysis were affected by mutational saturation. Saturation analysis was done for each codon position of the rpoB gene, using comp_mat in MUST, version 3.0 (27). The proportion of observed pairwise differences between rpoB sequences was calculated with an uncorrected neighbor-joining method using MUST 3.0 and was plotted against the proportion of inferred substitutions estimated by maximum parsimony using PAUP* 4.0b10 (35). If no linear fit could be found between observed and inferred nucleotide substitutions, the codon position tested was considered to present some degree of mutational saturation.
Phylogenetic analyses.
Phylogenetic analyses were performed at the DNA level for the 16S rRNA gene data sets and some of the rpoB data sets. For the rpoB gene, third codon positions that did not display mutational saturation were included in the analyses. The trees were constructed with PAUP* 4.04b, applying the heuristic search option and using the TBR branch-swapping algorithm. Maximum likelihood was used as the tree reconstruction method, with the nucleotide substitution model (GTR), among-sites rate variation parameter α (G), proportion of invariable sites (I), and nucleotide frequencies determined using MODELTEST (29). The confidence of each node was determined by building a consensus tree of 100 maximum likelihood trees from bootstrap pseudoreplicates of the original data set.
Maximum likelihood phylogenetic analyses of the rpoB gene amino acid translation were performed using PROML with the JTT amino acid substitution matrix, a rate heterogeneity model with gamma-distributed rates over four categories, with the α among-sites rate variation parameter estimated using TREE-PUZZLE, global rearrangements, and randomized input order of sequences (10 jumbles). Bootstrap support values represent a consensus (obtained using CONSENSE) of 100 Fitch-Margoliash distance trees (obtained using PUZZLEBOOT and FITCH) from pseudoreplicates (obtained using SEQBOOT) of the original alignment. The settings of PUZZLEBOOT were the same as those used for PROML, except that no global rearrangements and randomized input order of sequences are available in this program. PROML, CONSENSE, FITCH, and SEQBOOT are from the PHYLIP package, version 3.6a (http://evolution.genetics.washington.edu/phylip.html). TREE-PUZZLE and PUZZLEBOOT can be obtained from the program's website (http://www.tree-puzzle.de).
Random taxon sampling analysis.
To evaluate the influence of intragenomic heterogeneity in a 16S rRNA gene phylogeny at the subspecies level, a random taxon sampling analysis of 16S rRNA paralogous gene copies from multiple Escherichia coli and Shigella flexneri strains was performed. Shigella strains, although bearing a separate genus name, have been shown to be part of the E. coli species (30). Sequences from the more distant organism Salmonella enterica serovar Typhi LT2 were also included as an outgroup for the analysis. All strains of E. coli, S. flexneri, and S. enterica serovar Typhi LT2 analyzed here possess seven copies of the 16S rRNA gene. In this analysis, one of these seven paralogous copies was randomly sampled from each strain to create a data set, for which a maximum likelihood tree was computed. The procedure was repeated 1,000 times, giving a set of 16S rRNA gene trees, with each including one randomly chosen 16S rRNA gene copy from each E. coli and S. flexneri strain. Trees were systematically rooted with randomly sampled 16S rRNA gene copies from S. enterica serovar Typhi LT2.
Determination of evolutionary rates across sites (sliding-window analysis).
To better define regions of the 16S rRNA and rpoB genes with higher evolutionary rates, a sliding-window analysis was performed on a data set composed of the evolutionary rates at each position in the sequence alignments. These evolutionary rates were calculated for each position of the RpoB amino acid alignment and the 16S rRNA gene alignment of sequences from 50 representative species of the domain Bacteria. TREE-PUZZLE was used to assign an evolutionary rate category from 1 to 8 for each position (1 = slowest, 8 = fastest). These position and rate values were copied into an Excel spreadsheet, in which a macro was used to calculate the average evolutionary rate category for each 10 neighboring positions. The 10-position window was moved along the data set by increments of a single position, recalculating the average evolutionary rate category each time.
RESULTS
rpoB and 16S rRNA gene copy numbers and intragenomic heterogeneity.
An alternative marker to the 16S rRNA gene for molecular microbial ecology studies is the single-copy gene encoding the RNA polymerase β subunit, rpoB. This study compares the rpoB and 16S rRNA genes by sampling all gene copies from 111 bacterial genomes (Table 1). Four hundred sixty copies of the 16S rRNA gene (giving an average of 4.2 copies per genome) and 111 copies of the rpoB gene were recovered, with the latter being found as a single-copy gene in every genome. The extent of intragenomic 16S rRNA gene heterogeneity among bacteria ranges from 0 to 11.6% sequence divergence for 1 to 15 16S rRNA gene copies. Sixty-two percent of bacteria with more than one 16S rRNA gene copy were found to display some degree of intragenomic heterogeneity for this gene (Table 1).
TABLE 1.
16S rRNA gene intragenomic heterogeneity among a variety of bacterial taxonomic groups
| Taxonomic group | No. of sequenced genomes | No. of genomes with multiple 16S rRNA genes | 16S rRNA gene copy no. | Mean 16S rRNA gene copy no. ± SDa | No. of heterogeneous positions | Mean no. of heterogeneous positions ± SDa | 16S rRNA gene length (nt) |
|---|---|---|---|---|---|---|---|
| Aquificae | 1 | 1 | 2 | ND | 0 | ND | 1545 |
| Thermotogae | 1 | 0 | 1 | ND | 0 | ND | 1519 |
| Deinococci | 1 | 1 | 3 | ND | 2 | ND | 1463 |
| Cyanobacteria | 4 | 2 | 1-4 | 2 ± 0.41 | 0 | 0 ± 0 | 1446-1451 |
| Chlorobi | 1 | 1 | 2 | ND | 0 | ND | 1463 |
| Proteobacteria | 1446-1523 | ||||||
| Alphaproteobacteria | 9 | 6 | 1-4 | 2.22 ± 0.09 | 0 | 0 ± 0 | 1446-1461 |
| Betaproteobacteria | 4 | 3 | 1-4 | 3.25 ± 0.5 | 0 | 0 ± 0 | 1498-1503 |
| Gammaproteobacteria | 29 | 26 | 1-11 | 5.31 ± 0.85 | 0-30 | 5.69 ± 0.89 | 1498-1523 |
| Epsilonproteobacteria | 4 | 3 | 1-3 | 2 ± 0.82 | 0-1 | 0.5 ± 0.58 | 1457-1474 |
| Firmicutes | |||||||
| Bacillales | 11 | 11 | 5-13 | 7.45 ± 0.73 | 4-34 | 10.27 ± 0.76 | 1502-1525 |
| Lactobacillales | 12 | 12 | 4-6 | 5.42 ± 0.08 | 0-3 | 0.92 ± 0.00 | 1507-1528 |
| Clostridiales | 3 | 3 | 5-11 | 8.67 ± 0.21 | 6-19 | 13 ± 0.56 | 1467-1474 |
| Thermoanaerobacteriales | 1 | 1 | 4 | ND | 188 | ND | 1496-1589 |
| Mollicutes | 6 | 2 | 1-2 | 1.33 ± 0.52 | 0-3 | 0 ± 0 | 1473-1486 |
| Actinobacteria | 11 | 5 | 1-6 | 2.91 ± 0.26 | 0-8 | 1.73 ± 0.83 | 1481-1506 |
| Chlamydiae | 7 | 2 | 1-2 | 1.29 ± 0.49 | 0 | 0 ± 0 | 1513-1515 |
| Spirochaetes | 3 | 2 | 1-2 | 1.67 ± 0.58 | 0-1 | 0.33 ± 0.58 | 1470-1509 |
| Bacteroidetes | 1 | 1 | 5 | ND | 34 | ND | 1487 |
| Fusobacteria | 1 | 1 | 5 | ND | 3 | ND | 1481 |
| Planctomycetes | 1 | 0 | 1 | ND | 0 | ND | 1496 |
ND, not done.
Localization of intragenomic heterogeneity in the 16S rRNA molecule.
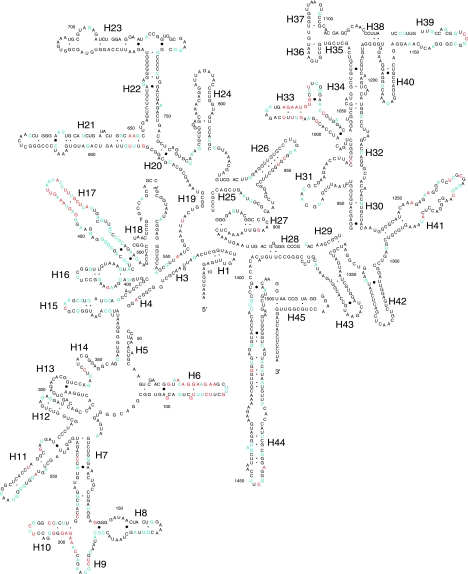
Heterogeneous positions between intragenomic copies of the 16S rRNA gene were mapped on the Escherichia coli 16S rRNA secondary structure for all 111 genomes (Fig. 1). Positions that display heterogeneity in a single genome are shown in blue, while positions displaying heterogeneity in more than one genome are shown in red. Nucleotide positions displaying intragenomic heterogeneity between paralogous copies were found throughout the length of the 16S rRNA gene (Fig. 1). However, such heterogeneous positions are concentrated in specific regions. These regions are called “heterogeneity hot spots” here and occur within helices 6, 9 to 11, 17, 33, 34, 39, and 41 (Fig. 1).
FIG. 1.
Mapping of the 16S rRNA positions where intragenomic heterogeneity was observed among the 111 bacterial species analyzed. Intragenomic heterogeneity is represented by nucleotide substitutions between multiple copies of the 16S rRNA gene present in a single organism. Heterogeneous positions were mapped on the secondary structure of one of the E. coli K-12 16S rRNAs. Helices (H) are numbered as described by Cannone et al. (8). Positions indicated in blue are heterogeneous in one species, and those in red are heterogeneous in two or more species.
Evolutionary characteristics of the rpoB and 16S rRNA genes.
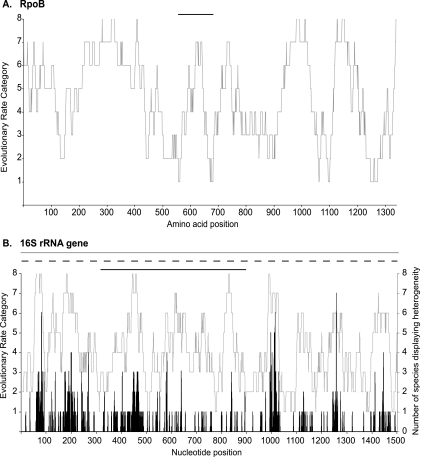
A good phylogenetic marker displays some conserved regions, which are used as targets for universal probes and primers and are useful for deciphering phylogenetic relationships between distant organisms, as well as more variable regions, which are used as sites for genus- and family-level probes and primers and to differentiate closely related organisms. Figure 2A and B plot the evolutionary rate categories of positions along the amino acid translation of the rpoB gene and the DNA sequence of the 16S rRNA gene. This allows the visualization of conserved and variable regions, both of which can be observed for either the RpoB protein or the 16S rRNA gene. An amino acid translation of the rpoB gene was used because there is mutational saturation of the first, second, and third codon positions (Table 2). The evolutionary characteristics of these markers can also be compared directly when it is possible to use the rpoB nucleotide sequence (rather than its amino acid translation) for phylogenetic analysis. The nucleotide sequence of a protein-encoding gene can be used for such analysis when mutational saturation is absent at all codon positions or when it is present at the third (synonymous) position and the latter is removed from the data set. We tested rpoB data sets representing various taxonomic levels for mutational saturation at all three codon positions, and only the domain-level (Bacteria) data set and one phylum-level data set (Proteobacteria) displayed saturation at codon positions other than the third position. Tables 2 and 3 present the evolutionary parameters of rpoB and 16S rRNA gene data sets that have been compiled for phylogenetic analyses at various taxonomic levels. When only considering nucleotide data sets (i.e., excluding the Bacteria and Proteobacteria data sets, for which analyses were performed at the amino acid level), the rpoB gene displays greater variation in the evolutionary rates of nucleotide positions across its length than does the 16S rRNA gene. This is illustrated by higher gamma rate parameters (G) and lower proportions of invariable sites (I) for most rpoB data sets (Tables 2 and 3). Such variation in evolutionary rates across sites indicates that phylogenetically informative positions (i.e., not saturated or invariable) can be found for large-scale (domain/phylum) or fine-scale (species/subspecies) analyses.
FIG. 2.
Evolutionary rates and intragenomic heterogeneity at specific sites across the lengths of the bacterial RpoB protein and the 16S rRNA gene. (A) Across-site evolutionary rate variation for the amino acid translation of the rpoB gene (gray line). Sites that are slowly evolving or invariable have low rate categories, and quickly evolving sites have high rate categories. The black bar above the graph illustrates the fragment targeted in DGGE analysis (11). (B) Across-site evolutionary rate variation for the 16S rRNA gene (gray line) and mapping of the number of species displaying intragenomic heterogeneity at all sites across the length of this gene (black line). Bars above the graph illustrate fragments of the 16S rRNA gene targeted for clone libraries (gray bar), T-RFLP analysis (dashed bar indicating variable length), and DGGE (black bar) (34).
TABLE 2.
Characteristics of rpoB data sets and phylogenies for different taxonomic levelsa
| Taxonomic group | Taxonomic level | No. of taxa | Saturated codon position(s) |
rpoB (complete gene)
|
Resolutionb |
rpoB (DGGE fragment)
|
Resolutionb | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of nucleotide positions | G | I | No. of nucleotide positions | G | I | ||||||
| Bacteria | Domain | 50 | All | 774 (aa) | 0.67 | 0.05 | 26/47 | 126 (aa) | 0.65 | 0.08 | 8/47 |
| Actinobacteria | Phylum | 11 | None | 3,336 | 1.73 | 0.30 | 6/8 | 390 | 2.83 | 0.34 | 5/8 |
| Chlamydiaceae | Family | 7 | None | 3,756 | 0.59 | 0.30 | 2/4 | 387 | 1.13 | 0.56 | 2/4 |
| Firmicutes | Phylum | 33 | 3rd | 1,978 | 0.82 | 0.16 | 19/30 | 262 | 1.61 | 0.30 | 10/30 |
| Bacillales | Order | 11 | 3rd | 2,308 | 1.64 | 0.62 | 4/8 | 262 | 0.39 | 0.24 | 5/8 |
| Lactobacillales | Order | 12 | 3rd | 2,336 | 0.74 | 0.37 | 5/9 | 262 | 0.87 | 0.36 | 4/9 |
| Streptococcus | Genus | 9 | None | 3,546 | 0.71 | 0.37 | 4/6 | 393 | 0.49 | ND | 6/6 |
| Mycoplasmataceae | Family | 6 | None | 2,679 | 1.7 | 0.18 | 2/3 | 354 | 1.34 | 0.15 | 1/3 |
| Proteobacteria | Phylum | 46 | 1st, 3rd | 1,147 (aa) | 0.57 | 0.08 | 35/43 | 127 (aa) | 0.52 | 0.10 | 23/43 |
| Alphaproteobacteria | Class | 9 | None | 3,993 | 0.4 | ND | 4/6 | 387 | 1.88 | 0.31 | 2/6 |
| Gammaproteobacteria | Class | 29 | 3rd | 2,660 | 0.89 | 0.36 | 19/26 | 258 | 0.89 | 0.36 | 11/26 |
| Enterobacteriales | Order | 14 | None | 4,020 | 1.27 | 0.34 | 11/11 | 387 | 0.56 | 0.22 | 5/11 |
| E. coli/S. flexneri | Species | 5 | None | 4,026 | ND | ND | 2/2 | 387 | ND | ND | 1/2 |
ND, value cannot be defined (data set is too small and/or lacks informative positions).
Number of nodes resolved (>80% bootstrap support value) out of the total nodes, with bold type indicating the gene (rpoB or 16S rRNA) displaying the most resolved node.
TABLE 3.
Characteristics of 16S rRNA gene data sets and phylogenies for different taxonomic levelsa
| Taxonomic group | Taxonomic level | No. of taxa | 16S rRNA (complete gene)
|
Resolutionb | 16S rRNA (DGGE fragment)
|
Resolutionb | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of nucleotide positions | G | I | No. of nucleotide positions | G | I | |||||
| Bacteria | Domain | 50 | 1,241 | 0.69 | 0.33 | 20/47 | 511 | 0.69 | 0.33 | 11/47 |
| Actinobacteria | Phylum | 11 | 1,428 | 0.67 | 0.49 | 6/8 | 563 | 0.67 | 0.49 | 5/8 |
| Chlamydiaceae | Family | 7 | 1,512 | ND | 0.76 | 2/4 | 588 | ND | 0.76 | 2/4 |
| Firmicutes | Phylum | 33 | 1,362 | 0.68 | 0.43 | 17/30 | 537 | 0.68 | 0.43 | 13/30 |
| Bacillales | Order | 11 | 1,498 | 0.54 | 0.56 | 4/8 | 585 | 0.54 | 0.56 | 5/8 |
| Lactobacillales | Order | 12 | 1,477 | 0.53 | 0.53 | 4/9 | 582 | 0.53 | 0.53 | 5/9 |
| Streptococcus | Genus | 9 | 1,510 | ND | 0.84 | 4/6 | 588 | ND | 0.84 | 4/6 |
| Mycoplasmataceae | Family | 6 | 1,453 | 0.64 | 0.32 | 3/3 | 575 | 0.64 | 0.32 | 2/3 |
| Proteobacteria | Phylum | 46 | 1,329 | 0.72 | 0.48 | 28/43 | 533 | 0.72 | 0.48 | 20/43 |
| Alphaproteobacteria | Class | 9 | 1,420 | 0.87 | 0.59 | 2/6 | 559 | 0.87 | 0.59 | 1/6 |
| Gammaproteobacteria | Class | 29 | 1,462 | 0.82 | 0.55 | 20/26 | 577 | 0.82 | 0.55 | 10/26 |
| Enterobacteriales | Order | 14 | 1,494 | 0.60 | 0.52 | 7/11 | 588 | 0.60 | 0.52 | 6/11 |
| E. coli/S. flexneri | Species | 5 | 1,505 | ND | ND | 0/2 | 588 | ND | ND | 0/2 |
ND, value cannot be defined (data set is too small and/or lacks informative positions).
Number of nodes resolved (>80% bootstrap support value) out of total nodes, with bold type indicating the gene (rpoB or 16S rRNA) displaying the most resolved node.
The rpoB DGGE fragment (Fig. 2A) contains a variable region allowing it to provide phylogenetic resolution between relatively closely related bacteria (Table 2). The rpoB primers used to amplify this fragment are found at conserved positions (10). The 16S rRNA gene universal primer sites used for clone libraries, T-RFLP analysis, and DGGE were also shown to occur at conserved positions (Fig. 2B). However, the internal regions of all these 16S rRNA gene fragments contain one or more heterogeneity hot spots, making all methods susceptible to the influence of using a gene displaying intragenomic heterogeneity. These heterogeneity hot spots are located in quickly evolving regions of the 16S rRNA gene, but not all quickly evolving regions of the 16S rRNA gene contain heterogeneity hot spots (Fig. 2B).
Comparing rpoB and 16S rRNA gene phylogenies.
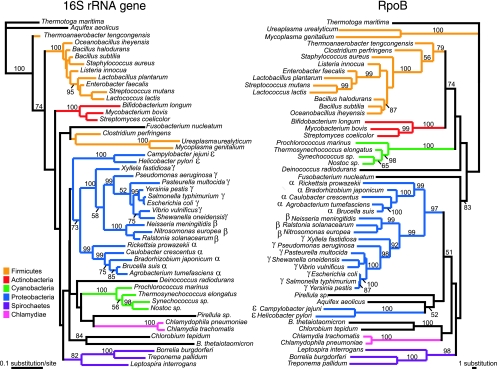
To compare the phylogenetic resolution of the rpoB and 16S rRNA genes, both full-length and DGGE fragments were used to reconstruct bacterial phylogenies at various taxonomic levels (Tables 2 and 3). The phylogeny of T-RFLP analysis-generated DNA fragments was not included, as this method generates DNA fragments of various sizes. In order to reduce overrepresentation of some phyla and make computing-intensive methods such as maximum likelihood phylogeny and maximum likelihood distance bootstrapping feasible, 50 bacterial species were selected to reconstruct the phylogeny of the bacterial domain (Fig. 3). To compare the capacities of rpoB and 16S rRNA genes to reconstruct phylogenies at the subspecies level, data sets of these genes from E. coli and Shigella flexneri strains were compiled and analyzed using various phylogenetic tools (Fig. 4).
FIG. 3.
Comparison of the best maximum likelihood trees for the 16S rRNA gene and the RpoB protein for the domain Bacteria.
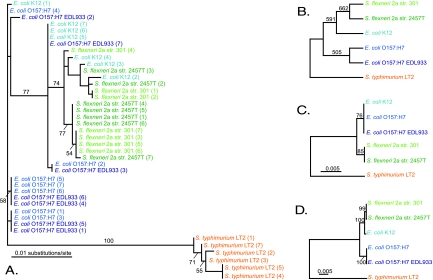
FIG. 4.
Evaluation of 16S rRNA and rpoB genes as phylogenetic markers at the subspecies level. (A) Best maximum likelihood tree for the 16S rRNA genes of Escherichia coli and Shigella flexneri strains (all seven copies from each strain are included). (B) Random sampling analysis of 16S rRNA paralogous gene copies from multiple E. coli and S. flexneri strains. One thousand trees were constructed by randomly choosing one 16S rRNA gene copy from each strain for every tree. Values above the nodes represent the numbers of trees in which the given nodes were recovered. (C) Best maximum likelihood tree for the 16S rRNA genes of E. coli and S. flexneri strains for which positions displaying intragenomic heterogeneity were recoded as ambiguous. (D) Best maximum likelihood tree for the rpoB genes of E. coli and S. flexneri strains.
Both the RpoB protein and 16S rRNA gene trees reconstruct the major bacterial phyla (Fig. 3), with a few exceptions. The Firmicutes do not form a monophyletic group in the 16S rRNA gene tree, while they do so in the RpoB tree. Mycoplasma species are not found within the Firmicutes for the RpoB protein or 16S rRNA gene tree, as the parasitic intracellular lifestyle of these bacteria imposes strong evolutionary biases on their genes, which in turn cause phylogenetic artifacts. The RpoB tree also reconstructs a tentative relationship between the gram-positive Actinobacteria and Firmicutes. The Proteobacteria are monophyletic in both the RpoB and 16S rRNA gene trees, with the exception of the Epsilonproteobacteria, which make the Proteobacteria polyphyletic in the RpoB tree. The origins of the Betaproteobacteria also differ between the trees. In the 16S rRNA gene tree, the Betaproteobacteria arise from within the Gammaproteobacteria, and in the RpoB tree, the Betaproteobacteria are a sister group to the Gammaproteobacteria.
Tables 2 and 3 summarize the comparisons of phylogenetic resolution using the full-length rpoB and 16S rRNA genes and their DGGE fragments at various taxonomic levels (domain, phylum, family, order, class, and species). The full-length rpoB and 16S rRNA genes showed equal resolution (number of tree nodes showing significant statistical support) in four of the comparisons. The rpoB gene offered better resolution in 7 of the 13 comparisons, while the 16S rRNA gene only provided enhanced resolution in 2 of the comparisons. The rpoB and 16S rRNA DGGE fragments showed similar phylogenetic resolution powers, with both fragments having improved resolution in 5 of the 13 comparisons and 3 of the comparisons having equal phylogenetic resolution. Interestingly, the rpoB and 16S rRNA-DGGE fragments showed comparable phylogenetic resolution to that of the full-length rpoB and 16S rRNA genes for several data sets, including those for the Actinobacteria, Chlamydiaceae, Bacillales, Lactobacillales, Streptococcus, Mycoplasmataceae, Alphaproteobacteria, and Enterobacteriales.
Intragenomic heterogeneity displayed by the 16S rRNA gene is most likely to affect the fine-scale phylogeny of closely related organisms, as their 16S rRNA gene intergenomic heterogeneity could be comparable to intragenomic heterogeneity. For this reason, we used the more exhaustively sampled species E. coli/S. flexneri (Shigella strains are part of the E. coli species [29]) as a data set for various phylogenetic methods to determine if their fine-scale phylogeny could be resolved. We recovered 17 unique 16S rRNA gene sequences from the five E. coli/S. flexneri strains in the data set (Fig. 4A), which would cause diversity to be grossly overestimated when 100% 16S rRNA gene sequence identity is used to define an OTU. Additionally, intragenomic and intergenomic heterogeneity levels appear to be of comparable importance, given that the 16S rRNA gene copies from each strain do not form monophyletic clades. There is also little bootstrap support for the nodes, with the outgroup alone being supported with statistical significance as a monophyletic clade. If a single 16S rRNA gene is chosen to represent a strain, the phylogeny obtained can change significantly depending on which copy was chosen. This is illustrated by the results of our random taxon sampling analysis (Fig. 4B). In this analysis, one of the seven 16S rRNA paralogous gene copies found in each E. coli/S. flexneri genome was randomly chosen for phylogenetic analysis, and this procedure was replicated 1,000 times. The trees obtained in these analyses differed significantly from each other, as a consensus of these trees was poorly supported (Fig. 4B). Recoding nucleotide positions that displayed intragenomic heterogeneity as ambiguous also yielded a poorly resolved tree (Fig. 4C). All of the methods employed here clearly show that the 16S rRNA gene cannot resolve the relationships among E. coli/S. flexneri strains. The rpoB gene, for its part, can resolve the relationships between some of these strains with strong statistical support (Fig. 4D).
DISCUSSION
Frequency and distribution of intragenomic heterogeneity between 16S rRNA gene copies.
In this study, the 16S rRNA gene copy number ranged from 1 to 15, with an average of 4.2 copies per genome. This is consistent with the rRNA Operon Copy Number Database, which calculates an average of 4.1 ribosomal operons per bacterial genome (21). Recently, it was suggested that the average rRNA operon copy number should be revised down due to the overrepresentation of generalist bacteria among bacteria whose genomes have been sequenced (3). However, while it is clear that generalist bacteria have higher ribosomal operon numbers than do specialist bacteria (20), this does not translate to their overrepresentation among bacteria whose genomes have been sequenced. We suggest the opposite, that genomics, since its early days, has been biased toward sequencing bacteria with specialist lifestyles (such as pathogens and symbionts), which tend to display fewer copies of the rRNA operon.
There are two main hypotheses used to explain the existence of multiple rRNA operons within a genome, as follows: (i) multiple rRNA operons provide a multiplier effect on translation, allowing a bacterium to grow rapidly in response to environmental change (20); and (ii) functional differentiation between rRNA operons allows for differential expression of rRNA operons in response to environmental change (18). For the first hypothesis to be true, genetic drift would have to be counteracted by gene conversion to give rise to rRNA operons with little sequence heterogeneity distributed randomly across their length. The second hypothesis predicts that selection of functionally differentiated rRNA operons would lead to the concentration of heterogeneous positions in specific regions and to differentiated stem and loop structures between rRNA molecules.
We have found evidence for both scenarios. The 38% of bacterial genomes which contain more than one 16S rRNA gene copy but display no sequence heterogeneity between copies suggest that multiple rRNA operons can exist because of their multiplier effect on translation. However, we have not examined promoters, ribosomal proteins, or the 5S and 23S rRNA genes, which could also be important for functional differentiation between ribosomal operons. For genomes with more than one rRNA operon, 62% display some degree of sequence divergence (0 to 11.6%) between the intragenomic 16S rRNA gene copies. While heterogeneous positions are found throughout the length of 16S rRNA gene copies, localized regions display above-average numbers of heterogeneous positions (helices 6, 9 to 11, 17, 33, 34, 39, and 41) (Fig. 1). Such hot spots could result from the accumulation of neutral nucleotide substitutions or recombination. In haloarchaea, hot spots have been found to result from recombination (6). For the firmicute Thermoanaerobacter tengcongensis, about half of the 11.6% pairwise sequence difference between its two most divergent 16S rRNA gene copies is a result of large insertions in one of these copies. These large inserts form secondary structures (3) suggestive of a functional role in the ribosome.
It has yet to be shown whether paralogous copies of rRNA operons are co- or differentially expressed under various environmental conditions in prokaryotes. However, such a link between functionality and intragenomic rRNA divergence has been observed in the apicomplexan parasite Plasmodium berghei (18). Its two types of 18S rRNA genes (which differ at 5.0% of their nucleotide positions) are preferentially expressed in different stages of the life cycle of this eukaryotic parasite (18).
Comparing the rpoB and 16S rRNA genes as phylogenetic markers.
The rpoB and 16S rRNA genes were compared at various taxonomic levels for the ability to resolve bacterial phylogeny. For a total of 13 data sets, the rpoB gene provided more phylogenetic resolution than the 16S rRNA gene in 7 cases, equal resolution in 4 cases, and lower resolution in 2 cases (Table 2). Importantly, it resolved more of the relationships in the comparison at the domain level (Bacteria), which is the level at which most molecular microbial ecology studies occur.
A different picture emerges from a comparison of the 16S rRNA- and rpoB-DGGE fragments, with each yielding better resolution in five of the comparisons and being equal in three of them. The 16S rRNA-DGGE fragment produced enhanced resolution in the comparison at the domain level (Bacteria), although both fragments had poor resolution at this level. This is not surprising, as the rpoB- and 16S rRNA-DGGE fragments are very small (380 and 550 bp, respectively).
It should be noted that neither the rpoB nor 16S rRNA gene can resolve all, and that generally each resolves only about half, of the phylogenetic relationships in a given data set with either their full length or DGGE fragment. Phylogenetic studies should therefore be cautious in arriving at conclusions on the origins of environmentally derived sequences and should employ rigorous phylogenetic methods involving maximum likelihood-based algorithms to determine relationships between sequences. Also, a measure of statistical confidence, such as bootstrapping, should always be included in phylogenetic analyses.
The influence of intragenomic heterogeneity displayed by the 16S rRNA gene on bacterial phylogeny was also assessed, as most heterogeneous positions are localized within fragments commonly used for microbial ecology studies (namely, clone libraries, DGGE, and T-RFLP analyses) (Fig. 2). Phylogenetic analysis of all 16S rRNA gene copies present in the five strains of E. coli/S. flexneri of our data set (17 unique sequences) presented multiple clusters containing sequences from different strains. This clustering of sequences from various strains demonstrates a lack of correlation between the origin of 16S rRNA genes and their nucleotide sequences at the subspecies level for the E. coli/S. flexneri group. Intragenomic heterogeneity is therefore likely to affect the fine-scale phylogeny of closely related bacteria. For such closely related organisms, intragenomic heterogeneity can be as significant as intergenomic heterogeneity, as we have shown here for the E. coli/S. flexneri strains. This suggests that one should use caution in interpreting environmental 16S rRNA gene sequence-derived phylogenies at the species level or lower. For some prokaryotes, such as the archaeal order Halobacteriales, levels of 16S rRNA intragenomic heterogeneity are so high (>5.0%) that they can even affect phylogeny at the genus level (6, 40).
The rpoB gene did resolve several of the relationships among the E.coli/S. flexneri strains with strong statistical support (Fig. 4D). We therefore suggest rpoB as an additional or alternative marker in ecological studies, as it is capable of deciphering fine-scale phylogenetic relationships that go undetected using the 16S rRNA gene. Indeed, the use of a marker such as rpoB would be especially important for studies focusing on the genus level or lower because of the increased resolution it would provide as well as the absence of intragenomic heterogeneity as a source of bias.
Establishing rpoB as an alternative gene marker for microbial ecology.
Protein-encoding genes, such as rpoB, have several advantages over RNA-encoding genes as molecular markers. They can be used at both the amino acid and nucleotide levels for phylogenetic analysis. Protein alignments allow for resolution of relationships at higher taxonomic levels (domain or phylum) when one or more codon positions are saturated. Nucleotide-level alignments allow for fine-scale resolution, with synonymous first and third codon positions allowing for nearly neutral mutations which can be detected between very closely related organisms (species level or lower). This is likely the reason why rpoB performed better than the 16S rRNA gene in resolving relationships at the subspecies level. The resolution provided by rpoB can also be increased further by sequencing other protein-encoding genes and examining allelic profiles of different isolates rather than using single-gene sequence comparisons (36, 38). Although such MLSA cannot be done directly on an environmental sample, a marker used to describe diversity in an environmental sample (such as rpoB) can also be included in the set of genes used for MLSA of a collection of isolates. The improved phylogenetic resolution at the subspecies level provided by rpoB seems to be of increasing importance, with recent studies suggesting important ecological or genotypic differences in organisms with identical 16S rRNA gene sequences (ribotypes) (e.g. see reference 23).
Our analyses also demonstrate that rpoB displays other important characteristics as an ecological marker, including (i) its universal presence in all prokaryotes; (ii) the presence of slowly and quickly evolving regions for the design of probes and primers of differing specificities; (iii) having a housekeeping function, making it less susceptible to some forms of lateral gene transfer; and (iv) a large enough size to contain phylogenetic information, even after removal of regions that are difficult to align.
There are a few disadvantages in using rpoB (or another protein-encoding gene with similar characteristics) over the 16S rRNA gene as a molecular marker for microbial ecology studies. The single major obstacle resides in a fundamental property of protein-encoding genes, i.e., the saturation of all third codon positions over a long evolutionary timescale, which makes it more difficult to design universal primers for rpoB. Nonetheless, rpoB primers that can amplify a range of bacterial groups have been successfully used for DGGE studies (7, 11, 25, 31, 32). Alternative rpoB primers targeting the most conserved amino acid motifs of the RpoB protein have been developed and tested on most culturable bacterial phyla (R. J. Case, unpublished data). Additionally, Santos and Ochman (33) have developed a set of 10 primer pairs targeting protein-encoding housekeeping genes (including rpoB) to overcome limitations of the 16S rRNA genes to identify and classify bacteria.
A distinct advantage of using 16S rRNA as an ecological marker is the high concentration of targets in cells for FISH analyses. However, the development of more sensitive methods, such as catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH), should also allow for detection of organisms using protein-encoding genes (14, 26). Although mRNA for the rpoB gene would not be as abundant as rRNA in prokaryotic cells, it has been shown that this mRNA is relatively stable and should be present at significant concentrations, regardless of the growth phase of the host (24). Interestingly, we noticed that heterogeneous regions between the seven 16S rRNA gene copies in E. coli coincided with regions of poor FISH probe accessibility (data not shown) described by Behrens et al. (5), where probe accessibility was measured as a function of probe signal. However, a poor probe signal could result from a heterogeneous pool of 16S rRNAs, as Behrens et al. later showed that poor accessibility of probes did not result from the ribosome's tertiary structure (4).
There is little doubt of the scientific contributions made by studies using the 16S rRNA gene, as molecular studies have circumvented the need to culture bacteria. Central to this contribution is our appreciation of the extent of and ability to catalogue microbial diversity by storing 16S rRNA gene sequences in free and accessible databases. However, the growing number of sequenced genomes and environmental metagenomic libraries provides us with the necessary reference sequences to use other genes for molecular microbial ecology. Also contributing to the accumulation of sequences for alternative molecular markers are MLSA studies using protein-encoding genes.
Alternative markers in molecular microbial ecology—the way forward.
Lateral gene transfer has uncoupled function from phylogeny for prokaryotes, so the identity of a bacterium responsible for a function is unlikely to be uncovered through diversity studies. This has already led researchers to use functional genes, such as the [NiFe] hydrogenase gene, amoA, pmoA, nirS, nirK, nosZ, and pufM, in conjunction with DGGE (1, 19, 22, 39, 41) and to detect pmoA and dysB mRNAs with CARD-FISH (14, 26), leading the way for in situ identification of protein-encoding genes. The potential for alternative housekeeping genes to study diversity or the use of operational genes in molecular microbial ecology is an exciting prospect. This study demonstrates that some of them, with characteristics similar to those of rpoB, would have the potential to yield the same phylogenetic information as the 16S rRNA gene, with enhanced resolution for fine-scale analyses.
Footnotes
Published ahead of print on 27 October 2006.
REFERENCES
- 1.Achenbach, L. A., J. Carey, and M. T. Madigan. 2001. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microbiol. 67:2922-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 3.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens, S., B. M. Fuchs, F. Mueller, and R. Amann. 2003. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 69:4935-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boucher, Y., C. J. Douady, A. K. Sharma, M. Kamekura, and W. F. Doolittle. 2004. Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J. Bacteriol. 186:3980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne, D. G., and C. B. Munn. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7:1162-1174. [DOI] [PubMed] [Google Scholar]
- 8.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. S. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. D. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, D. P. 1998. Redefining relativity: quantitative PCR at low template concentrations for industrial and environmental microbiology. J. Ind. Microbiol. Biotechnol. 21:128-140. [Google Scholar]
- 10.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-794. [DOI] [PubMed] [Google Scholar]
- 11.Dahllöf, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lipthay, J. R., C. Enzinger, K. Johnsen, J. Aamand, and S. J. Sorensen. 2004. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol. Biochem. 36:1607-1614. [Google Scholar]
- 13.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flatt, P., J. Gautschi, R. Thacker, M. Musafija-Girt, P. Crews, and W. Gerwick. 2005. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar. Biol. 147:761-774. [Google Scholar]
- 15.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 16.Gevers, D., F. M. Cohan, J. G. Lawrence, B. G. Spratt, T. Coenye, E. J. Feil, E. Stackebrandt, Y. Van de Peer, P. Vandamme, F. L. Thompson, and J. Swings. 2005. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733-739. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 18.Gunderson, J. H., M. L. Sogin, G. Wollett, M. Hollingdale, V. F. Delacruz, A. P. Waters, and T. F. Mccutchan. 1987. Structurally distinct, stage specific ribosomes occur in Plasmodium. Science 238:933-937. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickx, B., W. Dejonghe, F. Faber, W. Boenne, L. Bastiaens, W. Verstraete, E. M. Top, and D. Springael. 2006. PCR-DGGE method to assess the diversity of BTEX mono-oxygenase genes at contaminated sites. FEMS Microbiol. Ecol. 55:262-273. [DOI] [PubMed] [Google Scholar]
- 20.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 23.Papke, R. T., J. E. Koenig, F. Rodriguez-Valera, and W. F. Doolittle. 2004. Frequent recombination in a saltern population of Halorubrum. Science 306:1928-1929. [DOI] [PubMed] [Google Scholar]
- 24.Passador, L., and T. Linn. 1989. Autogenous regulation of the RNA polymerase beta subunit of Escherichia coli occurs at the translational level in vivo. J. Bacteriol. 171:6234-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peixoto, R. S., H. L. D. Coutinho, N. G. Rumjanek, A. Macrae, and A. S. Rosado. 2002. Use of rpoB and 16S rRNA genes to analyse bacterial diversity of a tropical soil using PCR and DGGE. Lett. Appl. Microbiol. 35:316-320. [DOI] [PubMed] [Google Scholar]
- 26.Pernthaler, A., and J. Pernthaler. 2005. Simultaneous fluorescence in situ hybridization of mRNA and rRNA for the detection of gene expression in environmental microbes. Methods Enzymol. 397:352-371. [DOI] [PubMed] [Google Scholar]
- 27.Philippe, H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21:5264-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 30.Pupo, G. M., R. T. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rantsiou, K., G. Comi, and L. Cocolin. 2004. The rpoB gene as a target for PCR-DGGE analysis to follow lactic acid bacterial population dynamics during food fermentations. Food Microbiol. 21:481-487. [Google Scholar]
- 32.Renouf, V., O. Claisse, C. Miot-Sertier, and A. Lonvaud-Funel. 2006. Lactic acid bacteria evolution during winemaking: use of rpoB gene as a target for PCR-DGGE analysis. Food Microbiol. 23:136-145. [DOI] [PubMed] [Google Scholar]
- 33.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 34.Schafer, H., L. Bernard, C. Courties, P. Lebaron, P. Servais, R. Pukall, E. Stackebrandt, M. Troussellier, T. Guindulain, J. Vives-Rego, and G. Muyzer. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34:243-253. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1993. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- 36.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 39.Throback, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK, and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 40.Walsh, D. A., E. Bapteste, M. Kamekura, and W. F. Doolittle. 2004. Evolution of the RNA polymerase B subunit gene (rpoB) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol. Biol. Evol. 21:2340-2351. [DOI] [PubMed] [Google Scholar]
- 41.Wawer, C., and G. Muyzer. 1995. Genetic diversity of Desulfovibrio spp. in environmental samples analyzed by denaturing gradient gel electrophoresis of [Nife] hydrogenase gene fragments. Appl. Environ. Microbiol. 61:2203-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster, G., C. J. Newberry, J. C. Fry, and A. J. Weightman. 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55:155-164. [DOI] [PubMed] [Google Scholar]
- 43.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuckerkandl, E., and L. Pauling. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357-366. [DOI] [PubMed] [Google Scholar]