Abstract
The trafficking of effectors, termed Yops, from Yersinia spp. into host cells is a multistep process that requires the type III secretion system (TTSS). The TTSS has three main structural parts: a base, a needle, and a translocon, which work together to ensure the polarized movement of Yops directly from the bacterial cytosol into the host cell cytosol. To understand the interactions that take place at the interface between the tip of the TTSS needle and the translocon, we developed a screen to identify mutations in the needle protein YscF that separated its function in secretion from its role in translocation. We identified 25 translocation-defective (TD) yscF mutants, which fall into five phenotypic classes. Some classes exhibit aberrant needle structure and/or reduced levels of Yop secretion, consistent with known functions for YscF. Strikingly, two yscF TD classes formed needles and secreted Yops normally but displayed distinct translocation defects. Class I yscF TD mutants showed diminished pore formation, suggesting incomplete pore insertion and/or assembly. Class II yscF TD mutants formed pores but showed nonpolar translocation, suggesting unstable needle-translocon interactions. These results indicate that YscF functions in Yop secretion and translocation can be genetically separated. Furthermore, the identification of YscF residues that are required for the assembly of the translocon and/or productive interactions with the translocon has allowed us to initiate the mapping of the needle-translocon interface.
Many gram-negative bacterial pathogens, including Yersinia spp., Shigella spp., Salmonella spp., enteropathogenic Escherichia coli, Burkholderia spp., and Pseudomonas spp., use a type III secretion system (TTSS) to inject effectors from bacteria into a host cell (19, 22, 46). Once inside the cell, the effectors, termed Yops in the case of Yersinia, disable the host immune response, allowing the bacteria to multiply and spread throughout the host. A functional TTSS is essential for a productive infection, as TTSS mutants cause minimal damage to the host (1a, 43).
Yop delivery from the bacterial cytosol into the host cytosol by the TTSS can be genetically separated into two distinct functional processes: (i) secretion of effectors through the bacterial membranes into the extracellular environment and (ii) translocation of effectors across the host cell plasma membrane (61, 66). The secretion of effectors out of the bacteria requires the function of the structural components of the secretion machinery, termed the needle complex. The needle complex is composed of ∼25 proteins and forms a syringe-like structure with a base imbedded within the bacterial membranes and a long hollow tube or “needle” protruding from the outside of the bacterium (3, 34, 63, 70). Effectors appear to be secreted through the inside of the needle (10, 29) and are believed to travel in a partially unfolded state. Also required are chaperones, which maintain effectors in a secretion-competent state in the bacterial cytosol (for recent reviews, see references 52 and 53).
The needle structures of Yersinia, Salmonella, Shigella, E. coli, Burkholderia, and Pseudomonas are each primarily composed of a single protein (YscF, PrgI, MxiH, EscF, BsaL, and PscF, respectively) that polymerizes to form a tube (23, 35, 63, 70, 74, 76). The needle proteins from different species share only between 20 and 30% sequence identity but are all small (7 to 10 kDa), are neutral in charge, and are mostly alpha helical in structure (11, 69, 76). Using a combination of X-ray fiber diffraction and electron microscopy, the first three-dimensional structure of a needle filament was solved to ∼16 Å by using purified needles from Shigella (9). The filament was composed of individual subunits, arranged with 5.6 subunits per turn in a helical fashion to form an extended cylinder with a central channel of ∼20 Å (9). Purified needles from Yersinia, Salmonella, Shigella, E. coli, and Pseudomonas show similar helical structures and dimensions by negative staining and transmission electron microscopy (23, 34, 54, 63, 70).
When observed by electron microscopy, needles on the outside of Yersinia, Salmonella, and Shigella show consistent lengths (30, 35, 71). In fact, needle length is critical for productive translocation of Yops. Elegant experiments have shown that increasing the distance between the needle tip and the host cell either by decreasing the length of the needle or by increasing the length of the bacterial adhesion molecule YadA results in a reduction in Yop translocation (45). Needle length is controlled by the YscP protein (30). It is hypothesized that YscP is a molecular ruler attached to both the TTSS base and the needle tip that allows needle polymerization until YscP is stretched out in an extended state (30).
Secretion of Yops in the presence of host cells is tightly regulated, occurring only upon cell contact. A number of regulatory proteins are required for maintaining cell contact-mediated secretion, including YopN, TyeA, SycN, YscB, and LcrG (12, 25, 26, 28, 75). Secretion regulation mutants constitutively secrete Yops in the absence of a cell contact-mediated signal (18, 75), resulting in uncoordinated secretion and translocation, which ultimately lead to a secondary translocation defect. Though YopN and LcrG are secreted (18, 64), all regulatory proteins are thought to control secretion from the inside the bacterium (15, 50). It has been proposed that the needle itself senses contact with the host cell and then transduces the signal to the bacterium to begin secretion, perhaps through slight changes in the packing of the needle subunits (9). Consistent with this model, Torruellas et al. recently identified mutations in the Yersinia needle protein YscF that result in unregulated secretion (73), suggesting some communication between the needle and the cytosolic regulatory complexes.
Effector translocation into host cells involves additional proteins which form a pore, termed the translocon, in the host plasma membrane. In Yersinia, the YopB, YopD, and LcrV proteins are required for translocation (5, 21, 38, 51, 56, 60). YopB and YopD are found in the host cell plasma membrane and thus are likely to form the translocation pore (21, 48, 72). LcrV is required for pore assembly (24) but is not found in the host membrane (20). LcrV was recently localized to the distal ends of needles (47), where it multimerizes to form a “tip complex.” In the absence of either YopB, YopD, or LcrV, Yops are secreted out of the bacteria but fail to enter the host cell (16, 37, 56). Secretion of all three proteins from the same bacterium is necessary for pore formation and subsequent Yop translocation (41, 48).
During infection of eukaryotic cells, the processes of secretion and translocation are coupled such that effectors are transported directly from the bacterium into the host cell (61, 66). How the needle complex, LcrV tip, and pore work together to ensure the polar movement of effectors is not known. We hypothesized that the needle is likely to interact with the translocon uniquely such that we could identify mutants that altered translocation but not secretion. To this end, we devised a screen to identify mutations in the Yersinia pseudotuberculosis needle protein YscF which retained functions required for needle assembly and for Yop secretion out of the bacterium but were specifically defective for Yop translocation into host cells. We identified five classes of yscF mutants with translocation defects, two of which were specifically altered in translocation but not Yop secretion or needle assembly.
MATERIALS AND METHODS
Bacterial strains and culture.
All Yersinia pseudotuberculosis strains are in the IP2666 background (27) except for ΔlcrV, which is in the YPIII background. IP2666 ΔyopB was a gift from M. Fisher, and ΔlcrV was a gift from J. M. Balada of the Mecsas laboratory. Yersinia bacteria were grown in 2× YT (yeast extract and Bacto Tryptone) medium. Escherichia coli strains SY327 λpir (44) and DH5α (7) were used for routine cloning. E. coli strain SM10 λpir (44) was used for introduction of plasmids into Yersinia by conjugation. E. coli BL21-DE (68) was used for overexpression of fusion proteins. E. coli bacteria were grown in LB. Carbenicillin or ampicillin (Amp) was used at 100 μg/ml for plasmid maintenance. Previously described Y. pseudotuberculosis and E. coli strains are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| SM10 λPir | Conjugation-competent strain | 44 |
| SY327 λPir | Routine cloning | 44 |
| DH5α | Routine cloning | 7 |
| BL21-DE | Protein expression in E. coli | 68 |
| Yersinia pseudotuberculosis | ||
| IP2666 pYV (pIBI) | Wild-type Y. pseudotuberculosis | 27 |
| ΔyopB | Deletion of yopB (codons 19-346) in IP2666 | M. Fisher, unpublished |
| ΔyscF | Deletion of yscF (codons 2-86) in IP2666 | This study |
| ΔyopN | Deletion of yopN (codons 2-287) in IP2666 | This study |
| ΔlcrV | Deletion of lcrV (codons 19-346) in YPIII | J. M. Balada, unpublished |
| Plasmids | ||
| pGEM-T easy | Cloning of PCR products, Apr | Promega |
| pCR2.1 | Cloning of PCR products, Apr, Kmr | Invitrogen |
| pCVD442 | Suicide vector, Apr | 14 |
| pTRC99A | IPTG-inducible expression vector, Apr | 1 |
| pGEX-5X-1 | GST fusion expression vector, Apr | Amersham |
| pGEM-yscF | Template for error-prone PCR | This study |
| pCVD442-ΔyscF | Suicide vector for deleting yscF | This study |
| pCVD442-ΔyopN | Suicide vector for deleting yopN | This study |
| pGEX-yscF | GST-YscF fusion expression vector | This study |
| pGEX-YopD | GST-YopD fusion expression vector | This study |
| pTRC99A-yscF | Expresses yscF from tac promoter | This study |
Yersinia deletion strains.
Deletion strains were constructed using the suicide vector pCVD442 by allelic exchange (14). DNA fragments (400 bp) flanking each side of the open reading frame (ORF) were amplified by PCR and sewn together using fusion PCR. The resulting PCR product was cloned into pCVD442, introduced into E. coli SM10 λpir, and then conjugated into Y. pseudotuberculosis IP2666 pYV (pIBI), as previously described (40). All oligonucleotides used for cloning are listed in Table S1 in the supplemental material.
ΔyscF.
The entire yscF ORF was replaced by the StuI restriction endonuclease sequence (underlined: ATGAGGCCTTAA). Flanking DNA fragments were amplified by PCR with primer pair AD1 and AD2 and primer pair AD3 and AD4 (see Table S1 in the supplemental material) and were sewn together by fusion PCR using primers AD1 and AD4. This final PCR product was cloned into pCVD442 by using SalI and SphI, creating pCVD442-ΔyscF.
ΔyopN.
The yopN ORF was deleted. Flanking DNA fragments were amplified by PCR with primer pair AD37 and AD56 and primer pair AD57 and AD40 (see Table S1 in the supplemental material) and were sewn together by fusion PCR using primers AD37 and AD40. The final PCR product was cloned into pCVD442 by using SalI and SphI, creating pCVD442-ΔyopN.
Plasmids.
Previously described plasmids are listed in Table 1.
(i) pTRC99A-yscF.
pTRC99A-yscF expresses YscF under the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible tac promoter. The yscF open reading frame was amplified by PCR using primers AD5 and AD8, digested with AflIII and XbaI, and cloned into the NcoI and XbaI sites, creating pTRC99A-yscF. YscF protein levels are comparable to those in the wild type (WT) when expression from pTRC99A-yscF is induced with 10 μM IPTG.
(ii) pGEX-yscF.
A glutathione S-transferase (GST)-YscF fusion was made in pGEX-5X-1 (Amersham). yscF was amplified using primers AD9 and AD10 by PCR, and the resulting fragment was cloned into pGEX-5X-1 by using EcoRI and BamHI, creating pGEX-yscF.
(iii) pGEX-yopD.
yopD was amplified by PCR using primers AD64 and AD65, and the resulting fragment was cloned into pGEX-5X-1 by using BamHI and XhoI, creating pGEX-yopD.
Antibodies. (i) YscF.
Recombinant GST-YscF was overexpressed from the pGEX-yscF plasmid in BL21-DE cells by a 5-h induction at 30°C with 1 mM IPTG. Bacteria were lysed by sonication and the cell lysates clarified by centrifugation. GST-YscF was purified from cell lysates by using glutathione Sepharose 4B, following the manufacturer's instructions (Amersham). YscF was released from GST while bound to the glutathione column by using factor Xa protease (Sigma).
(ii) YopD.
Recombinant GST-YopD was overexpressed from the pGEX-YopD plasmid in BL21-DE cells by a 2-h induction at 37°C with 1 mM IPTG. Bacteria were lysed by sonication and the cell lysates clarified by centrifugation. GST-YopD was purified from cell lysates by using glutathione Sepharose 4B (Amersham) and was released from the resin with 10 mM glutathione. Proteolytic cleavage with factor Xa was inefficient, so we used the entire GST-YopD fusion protein as an antigen. Purified YscF and GST-YopD proteins were sent to Covance Research Products (Denver, PA) for antiserum preparation in rabbits.
yscF library construction.
To create the yscF mutant library, the yscF open reading frame was first amplified using primers AD5 and AD8 and then ligated into pGEM-T easy (Promega). pGEM-yscF was used as a template for error-prone PCR using SP6 and T7 primers. Two libraries were constructed; library D was amplified in the presence of 15 mM MgCl, 0.2 mM MnCl, and 0.2 mM deoxynucleoside triphosphates. Library E was amplified in the presence of 15 mM MgCl, 0.2 mM MnCl, 0.1 mM dATP, and 0.2 mM dCTP, dGTP, and dTTP. The mutation rates were approximately 1 bp change per ORF for library D and ∼4 to 7 bp per ORF for library E. The mutagenized PCR products were ligated into plasmid pCR2.1 (Invitrogen), digested with AflIII and XbaI, and subcloned into pTRC99A as described above, creating the pTRC99A-yscF D and E libraries. Sequencing a random sampling of the pTRC99A-yscF libraries showed full mutagenesis coverage throughout the whole gene (data not shown).
yscF translocation-defective (TD) screen. (i) Congo red assay.
The pTRC99A-yscF* libraries were electroporated into the ΔyscF strain and plated onto LB-Amp agar. Single transformants were replica patched onto both LB-Amp plates (for maintenance) and LB agar plates containing 20 mM Na oxalate, 20 mM MgCl2, and 0.005% Congo red (for selection). Congo red plates were incubated at 37°C overnight and scored for red or white colonies.
(ii) Cell rounding assay.
Congo red-positive strains were grown overnight at 26°C in 96-well deep-well plates in 2× YT media containing carbenicillin, with inclusion of ΔyscF+pTRC99A-yscF as a positive control and ΔyscF+pTRC99A as a negative control. Bacteria were diluted to an optical density (OD) of 0.2 in 2× YT media containing 20 mM sodium oxalate and 20 mM MgCl2 (secretion media) with carbenicillin in a 96-well plate and grown at 26°C for 2 h. yscF expression was induced with 10 μM IPTG, and the cultures were shifted to 37°C for 2 h. After growth, the OD600 values for eight of the wells were determined and averaged. Yersinia bacteria were diluted into warmed phosphate-buffered saline (PBS) to ∼1 × 107 bacteria/ml based on the average OD. HEp-2 cells were seeded the night before at 1 × 104 cells/well in 96-well plates. Bacteria (1 × 105) were added to the HEp-2 cells for an approximate multiplicity of infection (MOI) of 10:1 in the presence of 10 μM IPTG. Bacteria were centrifuged onto the HEp-2 cells, and infections proceeded at 37°C. One hour postinfection, HEp-2 cells were inspected for cell rounding at 10× magnification under a light microscope and scored for a round, flat, or intermediate phenotype. Bacterial inputs were plated for eight of the wells to determine the actual MOI of the infection. Because only a few of the ODs for the starting cultures were measured and averaged for each 96-well plate, the MOI of each individual strain differed from experiment to experiment. Therefore, each of the ∼1,000 strains was screened a minimum of three times. One hundred seventy-six strains scored flat or flat/round (F/R) cell rounding phenotypes two out of three times in the initial screen. yscF-containing plasmids from these strains were isolated, purified, and checked for the YscF fragment by restriction enzyme digestion. A number of the plasmids were corrupt and were discarded. The remaining 134 pTRC99A-yscF* plasmids were retransformed into a fresh ΔyscF strain and retested for Yops translocation by the cell rounding assay with HEp-2 cells. Thirty-six strains were flat or flat/round three out of three times.
In vitro secretion.
Yersinia bacteria were diluted from overnight cultures to an OD of 0.2 into 2-ml secretion media, grown for 2 h at 26°C, and then shifted to 37°C for 2 h. Yersinia bacteria containing pTRC99A plasmids were grown in the presence of 100 μg/ml carbenicillin, and protein production was induced with 10 μM IPTG at the 37°C shift. Some samples shown in Fig. 4 (also Fig. S2 in the supplemental material) were induced with 25 or 100 μM IPTG as indicated. One-milliliter aliquots of the cultures were centrifuged. Secreted proteins in the culture supernatant were precipitated with 10% trichloroacetic acid (TCA), washed with ice-cold acetone, and air dried. Bacterial pellets and precipitated protein pellets were resuspended in Laemmli sample buffer.
FIG. 4.
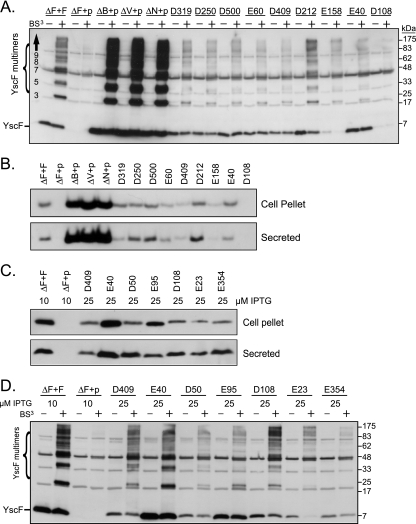
yscF TD mutants form external YscF polymers. Yersinia strains were grown in secretion media as in described in the legend to Fig. 3. (A) Chemical cross-linking. The chemical cross-linker BS3 or water was added to the bacteria and subsequently quenched with Tris-HCl. Bacteria were collected and pellets solubilized in sample buffer. Proteins were separated by SDS-PAGE, transferred to PVDF, and probed with antisera to YscF. Molecular mass standards are shown on the right in kDa. The positions of the YscF monomer (∼7 kDa) and YscF cross-linked multimers are shown on the left. A number of YscF antiserum cross-reactive background bands are visible in every lane (see ΔF+p, with [+] or without [−] BS3; +p is the ΔyscF strain containing vector pTRC99A alone) and are not cross-linker dependent. (B) YscF protein levels. Culture supernatants and bacterial pellets were collected from equal numbers of cells, and proteins were separated by SDS-PAGE. Cell-associated (top panel) and secreted (bottom panel) proteins were transferred to PVDF and probed with antisera to the YscF protein. (C) YscF protein levels after overexpression with increased amounts of IPTG. yscF mutants were induced with 25 μM IPTG and compared to the control strains induced with 10 μM IPTG. Culture supernatants and bacterial pellets were collected and processed as described for panel B. (D) Chemical cross-linking of overproduced yscF mutants. Strains were induced as described for panel C, cross-linked, and processed as described for panel B.
Tissue culture and infections.
HEp-2 cells were maintained in RPMI-glutamine (Cellgro) containing 5% heat-inactivated fetal bovine serum (Gibco) at 37°C under 5% CO2. HEp-2 cells were seeded into 6-, 24-, or 96- well plates ∼20 h before infection. For infections, Yersinia bacteria were washed in PBS, added to the tissue culture wells at the desired MOI, and centrifuged onto the cells for 5 min at 200 × g to initiate cell contact. Infections proceeded at 37°C under 5% CO2 for 1 h. IPTG (10 μM) was added to the tissue culture media for infections with strains carrying pTRC99A plasmids.
Secretion and translocation quantification.
HEp-2 cells were seeded at 6 × 105 cells/well in six-well tissue culture plates. Yersinia bacteria were diluted into secretion media, grown at 26°C for 2 h, and then grown for 2 h at 37°C after addition of 10 μM IPTG. Yersinia bacteria were centrifuged gently, resuspended in warmed PBS, and diluted for infection in PBS. Cells were infected with Yersinia at an MOI of 50:1 in the presence of 10 μM IPTG for 1 h at 37°C. After infection, the media were dumped off and the cells were washed twice gently in ice-cold PBS. All remaining steps were performed at 4°C. To disrupt the plasma membrane and release the HEp-2 cytosol, 400 μl of eukaryotic lysis buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and 5 μg/ml each of aprotinin, leupeptin, and pepstatin) was added to each well and the plates were rocked for 20 min. Lysates were transferred to microcentrifuge tubes and centrifuged for 15 min to remove bacteria and insoluble material. Lysate supernatant (300 μl) was removed and added to a fresh tube containing 60 μl 6× sample buffer. One-half of the sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with antisera to YopE (translocated protein) and actin (loading control). Chemiluminescence signals were captured on a Kodak digital science image station, and quantification was performed using Kodak 1D software. The amount of YopE translocated from WT (ΔyscF+pTRC99A-yscF) Yersinia was set to 100%. The amounts of YopE translocated from other strains were calculated with the following equation: (YopE translocated from mutant − uninfected)/(YopE translocated from WT − uninfected) × 100.
Chemical cross-linking.
Yersinia bacteria were grown under secretion-inducing conditions, centrifuged gently to pellet the bacteria (6,000 × g for 5 min), resuspended in PBS, and split into two equal aliquots. The chemical cross-linker BS3 (1 mM) or an equal volume of water was added to the bacteria and incubated at 37°C for 30 min. The cross-linker was then quenched with 5 mM Tris-HCl (pH 8.0) for 15 min at room temperature. Cross-linked bacteria were sedimented and the pellets resuspended in Laemmli sample buffer (36).
Nonpolar translocation assay.
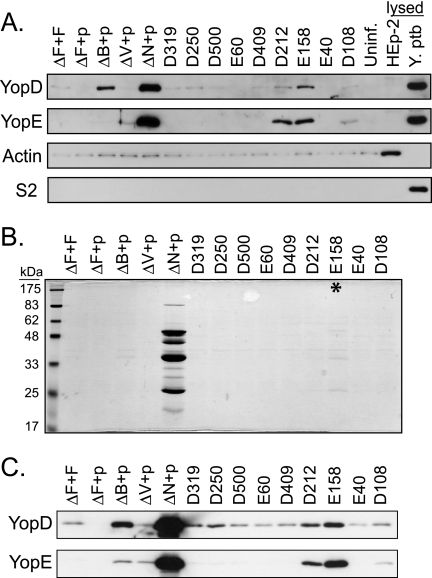
Yersinia bacteria were grown in 2× YT buffer containing 5 mM calcium and 100 mg/ml carbenicillin at 26°C for 1.5 h, induced with IPTG, and shifted to 37°C for 2 h. The bacteria were pelleted gently and resuspended in warmed PBS with calcium. HEp-2 cells were seeded the night before at 1.5 × 105 cells/well in a 24-well plate. Immediately before the infection, the HEp-2 cells were washed twice with warmed PBS, and 1 ml of RPMI-glutamine (no fetal bovine serum) was added to each well. Cells were infected with Yersinia at an MOI of 50:1 in the presence of 10 μM IPTG for 1 h at 37°C. For a HEp-2 lysis control, 0.1% Triton X-100 was added to one well of uninfected cells 10 min before the end of the infection. One milliliter of culture supernatant was removed from each well and transferred to a microcentrifuge tube. Culture supernatants were centrifuged for 15 min at 13,000 rpm in a microcentrifuge to remove bacteria. Proteins from 900 μl of the supernatant were precipitated with TCA and resuspended in sample buffer. As a control for lysed bacteria, the same number of WT bacteria used for infection (7.5 × 106) were solubilized in SDS-containing sample buffer. Proteins were separated by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and probed with antisera to YopD, YopE, actin, and S2.
Western blot analyses.
Antisera to the YscF, YopE, and YopD proteins were made in rabbits (see above) and were each used at a 1:10,000 dilution. Antiserum to the E. coli ribosomal subunit S2 (rabbit) was used at 1:10,000. Antibodies to mouse β-actin (Sigma) were used at 1:15,000. Goat anti-rabbit horseradish peroxidase (Bio-Rad) and goat anti-mouse horseradish peroxidase (Sigma) secondary antisera were used at 1:15,000. Chemiluminescence (Perkin Elmer Western Lightning kit) was used per the manufacturer's instructions.
Microscopy.
HEp-2 cells were seeded onto eight-well Lab-Tek chamber slides (Nalge Nunc, Naperville, IL) at 7 × 104 cells/well. Cells were infected with Yersinia at an MOI of 10:1, and infections proceeded for 1 h at 37°C. Micrographs were taken with a Nikon inverted TE2000-U microscope with a Photometrics charge-coupled-device camera at 10× magnification by using MetaVue software (Molecular Devices, Sunnyvale, CA).
RBC hemolysis assay.
Hemolysis assays were performed as described elsewhere (62). Briefly, Yersinia strains were grown in secretion media as described above and were sedimented for 5 min at 6,000 × g, and pellets were resuspended in warm RPMI at 2 × 109/ml. Sheep red blood cells (SRBCs; Innovative Research, Southfield, MI) were washed twice with ice-cold PBS and resuspended in RPMI at 2 × 109/ml. Equal volumes (50 μl) of red blood cells (RBCs) and bacteria suspensions were added to round-bottom 96-well plates in triplicate and centrifuged at room temperature for 15 min at 2,000 × g to initiate cell contact. Infections were incubated at 37°C under 5% CO2 for 3 h. Pellets were then resuspended with 150 μl ice-cold PBS, or dH2O for a lysis control, and reactions were centrifuged at 4°C for 15 min at 2,000 × g. Supernatants (150 μl) were transferred to a 96-well plate, and the absorbance value at 545 nm was read on a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA). RBC lysis caused by the ΔyscF+yscF strain was set to 100%, and RBC lyses for all other strains were calculated as percentages of those for the ΔyscF+yscF strain. All strains were tested at least twice.
RESULTS
Screen for translocation-deficient yscF mutants.
Upon host cell contact, Yops are transported directly from Yersinia into the target cell. The absence of detectable Yop effectors in the cell culture supernatant suggests coordinated and close interactions between the needle and the translocon. We hypothesized that the end of the needle is in direct contact with the translocon in the target cell plasma membrane, and thus, mutations in YscF that specifically disrupt the translocation process could be isolated. To separate YscF functions for Yop secretion from its potential functions in translocation, we designed a two-part screen to identify YscF mutants that secreted Yops across bacterial membranes normally but were defective in Yop translocation.
The first part of the screen exploited a quick and simple colorimetric plate assay to identify yscF mutants that could secrete Yops. Y. pseudotuberculosis secretes Yops when grown at 37°C in media containing low concentrations of calcium. Furthermore, in the presence of the dye Congo red, colonies of Y. pseudotuberculosis which are secreting Yops absorb the dye and turn red, whereas Y. pseudotuberculosis mutants that are not secreting Yops remain white (43, 58). Two randomly mutagenized pools of yscF genes, pool D and pool E, were created using error-prone PCR (39), inserted into the IPTG-inducible plasmid pTRC99A, and introduced into ΔyscF by transformation. The resulting library was termed yscF*. Single transformants of the yscF* libraries were patched onto Congo red plates, incubated at 37°C overnight, and scored for red or white color. Of the 2,900 strains screened, 1,058 bound to Congo red, indicating that they were competent to secrete Yops.
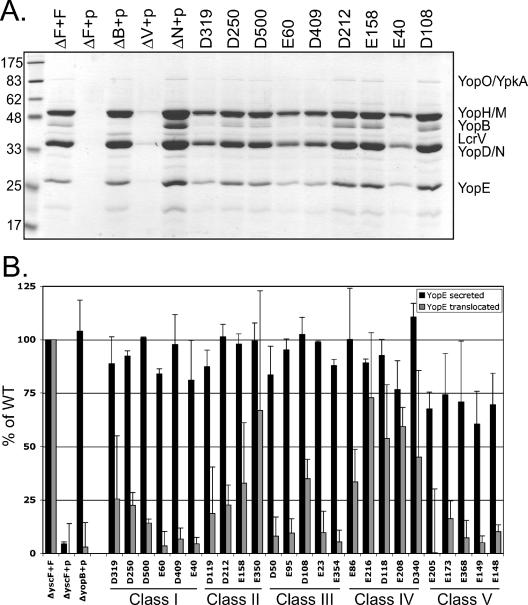
The pTRC99A promoter driving the expression of the yscF mutants was leaky enough such that addition of IPTG was not required for scoring red or white colonies on Congo red plates. However, the addition of 10 μM IPTG was required to obtain wild-type levels of YscF protein from the pTRC99A-yscF plasmid (Fig. 1A and Fig. S2A in the supplemental material), so this amount of inducer was used for YscF production in all strains for all subsequent experiments.
FIG. 1.
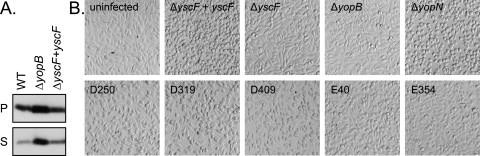
yscF secretion-positive mutants are defective for cell rounding during infection. HEp-2 cells were infected at an MOI of 10:1 with various Yersinia strains (ΔyscF+pTRC99A-yscF, ΔyscF+pTRC99A, ΔyopB+pTRC99A, ΔyopN+pTRC99A, ΔyscF+pD250, ΔyscF+pD319, ΔyscF+pD409, ΔyscF+pE40, and ΔyscF+pE354) grown in secretion media with IPTG to induce yscF production. Pictures were taken 1 hour postinfection.
The second part of the screen identified secretion-positive yscF strains that were translocation defective using a tissue culture infection assay. HEp-2 cells, which are normally flat and spread out (Fig. 1B, uninfected), round up when the Y. pseudotuberculosis effector protein YopE is translocated into the cytosol. Under our infection conditions, the vast majority of HEp-2 cells are round at 60 min postinfection (Fig. 1B, ΔyscF+yscF). HEp-2 cells infected with mutant Y. pseudotuberculosis strains that were not able to secrete effectors out of the bacteria (ΔyscF) or that were lacking a component of the translocon (ΔyopB) and cannot translocate effectors into the host cell remain flat and spread out (Fig. 1B). This “rounding up” assay was used to identify secretion-positive yscF* strains that inefficiently translocated Yops into a host cell. HEp-2 cells were infected with secretion-positive yscF* strains at a multiplicity of infection of ∼10:1. After 1 hour, the HEp-2 cells were inspected visually with a light microscope and were scored for flat, round, or intermediate F/R phenotypes and compared to ΔyscF+pTRC99A-YscF as a positive control for cell rounding and ΔyscF with the empty vector as a negative control. Of the 940 secretion-positive strains screened for cell rounding, 134 scored as F/R or flat two or three out of three times and were subsequently rescreened (see Materials and Methods). Thirty-six yscF* strains were consistently flat or F/R and were chosen for further studies. A few yscF* strains had completely flat cell rounding phenotypes (Fig. 1B, E354), suggesting a severe deficiency in Yop translocation. Most of the 36 yscF* strains showed F/R cell rounding phenotypes (Fig. 1B, D250, D319, D409, and E40), suggesting that low levels of Yops were translocated.
yscF translocation-defective mutations.
Plasmids from the 36 yscF* strains were sequenced to determine the nucleotide changes responsible for the translocation defect phenotype. Eight pairs of yscF* plasmids had identical mutations, and the duplicates were discarded. Three strains had more than one pTRC99A-yscF* plasmid; in each case, the translocation defect did not repeat when the individual pTRC99A-yscF* plasmids were tested. The remaining 25 yscF* strains were chosen for further phenotypic analysis and were named as a group yscF TD for translocation defective.
The TD mutations were spread throughout the entire yscF sequence (see Fig. S1 in the supplemental material, arrowheads). Seventeen of the yscF TD mutants had a single amino acid change, and 8 had two or three changes (Fig. 2). Some of the nucleotide changes resulted in very conservative amino acid substitutions, for example, D250 and E60 (A→V) and D319 and E208 (K→R), whereas some mutations resulted in changes in residue size, shape, and charge, for example, D119 (D→G) and E86 and D118 (K→E). There appeared to be one “hot spot” for translocation-defective mutations; changes in amino acids T22 through N35 were found in 12 of the 25 yscF TD strains (see Fig. S1 in the supplemental material).
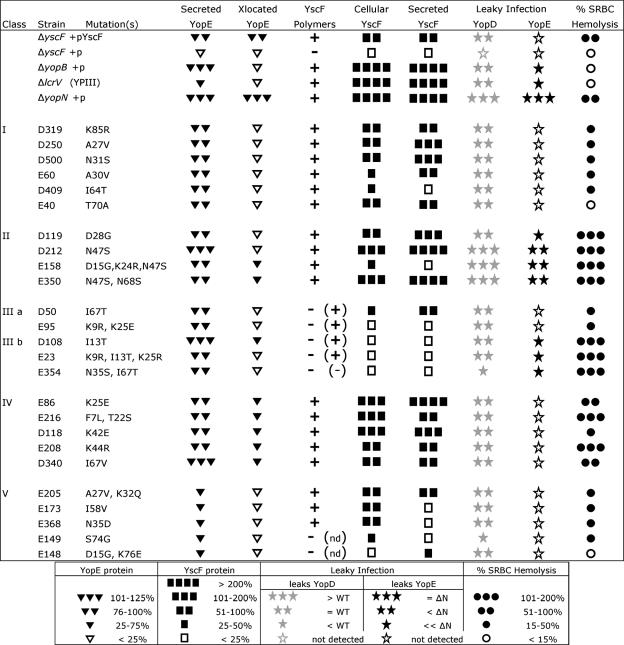
FIG. 2.
Compilation of yscF TD mutants and their phenotypes in various assays. All yscF TD mutants and control strains were tested for levels of secreted and translocated YopE (Fig. 3), YscF polymers (Fig. 4A), cellular and secreted YscF protein (Fig. 4B), YopD and YopE leaked during infection (Fig. 5A), and SRBC hemolysis (Fig. 6). (+) or (−) indicates detection of YscF polymers after overproduction of YscF protein (Fig. 4C). (nd), not done.
Most of the homology between YscF and needle proteins from other bacteria resided in the C-terminal half of the protein (see Fig. S1 in the supplemental material, shaded boxes) (4, 35, 63). Only two of the yscF TD mutants, N47S and K76E, contained a change in a highly conserved residue (see Fig. S1 in the supplemental material), suggesting that most of the conserved residues are important either for maintaining YscF structure or for some YscF function required prior to the effector translocation step, such as YscF secretion, YscF polymerization, or Yop secretion.
Translocation is the last stage of a multistep secretion process whose success depends on the proper function of each previous step. The known YscF-dependent steps include YscF secretion, assembly of the needle, and Yop secretion. To evaluate the molecular defects in the yscF TD mutants which led to poor translocation, our mutants were analyzed for YscF secretion, needle structure, and Yop secretion as well as secretion regulation, polarized Yop translocation, and pore formation (see below). In addition, the yscF TD strains were compared to several well-established mutants with translocation defects, ΔyopB, ΔlcrV, and ΔyopN, and grouped into different classes based on their most distinguishing phenotypes (Fig. 2, classes I through V). All yscF TD mutants were tested in these assays, and the results are summarized in Fig. 2. Representative mutants from classes I, II, and III are shown in most figures.
Most yscF TD mutants secrete Yops at or near WT levels.
Although a red-colony phenotype on Congo red plates correlates with the ability to secrete Yops, it is not a direct measure of the level of Yop secretion since strains with a functional secretion system that secrete lower levels or subsets of Yops still bind to Congo red (43). Therefore, it was plausible that yscF* strains had defects in Yop secretion and secreted either lower levels of all Yops or lower levels of specific Yops involved in translocation or cell rounding (59, 60) and thus had the TD phenotype in our screen.
To directly assess whether the yscF TD mutants secreted Yops, we used an in vitro liquid culture secretion assay. Bacteria were grown in media depleted of calcium at 37°C to induce secretion of Yops. Culture supernatants were collected, and secreted proteins were analyzed by SDS-PAGE and Coomassie blue staining. As expected, the complemented ΔyscF+pTRC99A-yscF strain secreted Yops, whereas the ΔyscF strain carrying the empty vector did not (Fig. 3A). All of the yscF TD strains secreted the full complement of Yops and translocon components (Fig. 2, secreted YopE, and 3A), ruling out the possibility that selective secretion of Yops was responsible for the translocation defect. However, the levels of Yop secretion in the yscF TD mutants were not all the same. Most yscF TD mutants secreted Yops at levels comparable to those for WT Y. pseudotuberculosis, but five strains (Fig. 3B, E205, E173, E368, E149, and E148) secreted 25 to 40% less YopE than the WT. These five yscF mutants showing more-pronounced defects in YopE secretion were placed in class V (Fig. 2).
FIG. 3.
Levels of secreted and translocated YopE from yscF TD mutants. Yersinia strains containing pTRC99A vector alone (+p) or pTRC99A expressing various yscF TD mutants were grown in secretion media for 2 h at 26°C, induced with IPTG, and shifted to 37°C for 2 h. (A) Bacteria were sedimented, and culture supernatants containing secreted proteins from equal numbers of cells were precipitated with TCA. Secreted proteins were separated by SDS-PAGE and stained with Coomassie blue. Molecular mass standards are shown on the left. Secreted Yops are indicated on the right. (B) Bacterial pellets were used to infect HEp-2 cells at an MOI of 50:1 for 1 hour. HEp-2 cells were gently washed in PBS, and plasma membranes were lysed with 0.1% NP-40. HEp-2 cytosol was collected, centrifuged to remove bacteria, and solubilized in sample buffer. Proteins in HEp-2 cytosol and bacterial culture supernatants were separated by SDS-PAGE, transferred to PVDF membranes, and probed with antisera to YopE. Chemiluminescent signals for YopE were quantified, and the amount secreted or translocated in the complemented ΔyscF+pTRC99A-yscF strain was set to 100% (ΔyscF+F). All strains were tested a minimum of three times, and the averages for secreted YopE (black bars) and translocated YopE (gray bars) are shown as percentages of the WT signal. Error bars indicate the standard deviations from the means.
yscF TD mutants are defective for Yop translocation.
Although the cell-rounding assay indicated that all yscF TD mutants were defective for some aspect of translocation, cell rounding is a qualitative assessment of the translocation process. To accurately evaluate the translocation abilities of the yscF TD strains, we quantified the amount of YopE in the host cell cytosol after infection and compared the amount of YopE translocated to the amount of YopE secreted in each mutant. Thus, we were able to determine the severity of the translocation defect even in yscF strains that secreted significantly lower levels of Yops than the WT.
To measure the amount of YopE secreted from each strain prior to infection, proteins in culture supernatants were precipitated and separated by SDS-PAGE as described for Fig. 3A. To measure the amounts of YopE translocated after infection with various Y. pseudotuberculosis strains, HEp-2 plasma membranes were gently washed and lysed and cytosolic proteins were collected and separated by SDS-PAGE. Secreted and translocated YopE was detected by Western blotting with YopE antisera, and chemiluminescent signals were quantified. All YopE signals were compared to that of the positive control, ΔyscF+pTRC99A-yscF, which was set to 100%. As expected, no YopE protein was detected in ΔyscF culture supernatants or ΔyscF-infected HEp-2 cytosol (Fig. 3B, ΔyscF+p). The ΔyopB strain, which is unable to make a pore, secreted WT levels of YopE in vitro, but very little YopE was detected in the HEp-2 cytosol. The residual 3% of translocated YopE detected in the ΔyopB mutant could reflect a small amount of nonspecific YopE adherence to the outside of the plasma membrane that remained after the infected HEp-2 cells were washed.
The results of this quantitative translocation assay validated the screening method, as most yscF TD mutants were significantly defective in Yop translocation compared to the WT (Fig. 3B). Of great interest, 14 yscF TD mutants in classes I, II, and III secreted Yops at levels comparable to the WT level but were greatly attenuated (<35% of the WT level) for translocation. Furthermore, the yscF TD class V mutants, which secreted low levels of Yops compared to the WT, appeared to have additional secretion-independent translocation defects because the ratio of YopE translocated to YopE secreted was very low (Fig. 3B, class V). For example, secretion of YopE in the E149 mutant was reduced to ∼60% compared to that in the WT, yet E149 translocated only 10% of the available secreted YopE. The class IV mutants, E86, E216, D118, D208, and D340, were grouped together because they showed near-WT levels of secretion but only a modest twofold or less reduction in Yop translocation (Fig. 3B, class IV), suggesting that translocation was only mildly compromised in these strains. The yscF TD mutant E350, which also has a weak translocation defect, was grouped into class II because of other predominant phenotypes (see below). In summary, our initial genetic screens and follow-up quantitative measurements of secretion and translocation indicated that a large number of yscF mutants secreted Yops at near-wild-type levels but had defects in translocation (classes I through III). Five yscF mutants had low levels of secretion (class V), and five had modest defects in translocation (class IV).
Most yscF TD mutants form extracellular YscF polymers.
All of the yscF TD mutants secreted Yops, so the needle formed was secretion competent. However, a needle that is competent for secretion will not necessarily succeed in translocation, since short or unstable needles secrete effectors but translocate effectors poorly (31, 45). To determine whether the inability to translocate Yops reflected less stable needles or less YscF protein, yscF TD mutants were examined for the presence of needles by chemical cross-linking (15). When exposed to the membrane-impermeable chemical cross-linker BS3, extracellular YscF protein cross-links to itself and forms a distinct, reproducible ladder pattern of multimers, as shown by SDS-PAGE and Western blotting with YscF antibodies (Fig. 4A, ΔF+F). Monomeric YscF ran at ∼7 kDa and consisted of both intracellular pools of YscF and extracellular, non-cross-linked YscF (Fig. 4A, YscF). Cross-linked YscF species began at molecular masses corresponding to that of a YscF trimer and increased in size in multiples of ∼7 and 14 kDa (Fig. 4A, YscF multimers). This cross-linking pattern was dependent on the presence of YscF and BS3 (Fig. 4A) and so represents external YscF protein polymers. Mutants were categorized by their abilities or inabilities to form detectable YscF cross-links (Fig. 2, YscF polymers). We did not compare levels of cross-links between mutants because this assay is inherently not quantitative.
Detectable YscF cross-links were observed in most of the yscF TD mutants, and thus, these mutants did not have severe defects in needle polymerization or stability (Fig. 2 and 4A). However, two groups of yscF TD mutants had lowered or undetectable levels of cross-linked YscF protein (see, for example, Fig. 4A, D108), indicating that these yscF alleles were unable to make stable polymers or that the polymers formed a significantly different structure which changed the availability of reactive groups for cross-linking. Alternatively, some of the yscF TD mutants that showed low or no YscF cross-links may have assembled the needle normally but lost a lysine residue critical for the amine-reactive cross-linking reaction. This may be the case for the lysines mutated in the E148 (K76E), E95 and E23 (K9R), and E205 (K32Q) strains. yscF TD mutants that showed normal levels of Yop secretion but low levels of YscF polymers were placed in class III. Some class V mutants, which had low levels of Yop secretion, also had low or undetectable levels of YscF polymers.
yscF TD mutants have various levels of YscF protein.
To assemble individual needle subunits into a needle, the subunits must be synthesized and secreted through the base of the TTSS (15, 32). To determine whether yscF TD mutants, particularly those with low levels of YscF polymers, were defective in YscF stability or secretion, the levels of cell-associated and secreted YscF TD proteins were measured. yscF TD strains were grown under secretion-inducing conditions, and YscF levels were ascertained by Western blotting with antibodies to the YscF protein in both bacterial pellets and culture supernatants (Fig. 2 and 4B).
In general, the amounts of total YscF protein observed in the cross-linking studies described above correlated with the amounts of cell-associated YscF but not necessarily with the levels of secreted YscF. However, a number of yscF TD strains contained low levels of both cell-associated and secreted YscF, including most of the class III and some of the class V mutants (Fig. 2 and 4B). To increase the amount of YscF protein produced, we added more IPTG to the growth medium (Fig. 4C and Fig. S2B in the supplemental material). Overexpression of the class III yscF alleles resulted in detectable amounts of both cell-associated and secreted YscF protein (Fig. 4C). Cross-linking the class III strains following overproduction of the mutant YscF proteins revealed that the D50, D108, and E23 mutants could polymerize (Fig. 4D). In contrast, the E95 mutant formed a trimer but was very inefficient at formation of higher-order cross-links, and E354 did not form any polymers detectable by cross-linking. Thus, we predict that the class III mutants translocate poorly because of aberrant needle numbers, lengths, or structures.
Curiously, some mutants, like E158, had very low levels of YscF protein yet appeared to make polymers as well as other yscF TD mutants (Fig. 4A, compare E158 to D409). The class V mutants, E173, E368, and E149, did not efficiently secrete YscF, suggesting that their general Yop secretion defect was reflective of a YscF secretion problem. Some of the class II and class IV strains made and/or secreted significantly more YscF than the WT and had abundant YscF cross-links, suggesting that they contained more and/or longer needles than the WT. Interestingly, this excessive YscF phenotype was shared with the translocation-negative control strains ΔyopB and ΔlcrV and with the secretion regulation-defective strain ΔyopN (Fig. 1A, 2, and 4B), even though these strains secreted various levels of Yops (Fig. 3), suggesting that YscF and Yop production are not coregulated. The class I and class II mutants appeared to have normal needle structures, and in general, the levels of YscF protein were equal to or higher than the WT level. The absence of an obvious structural needle problem in the class I and class II yscF mutants indicated that their translocation defects involved problems with secretion regulation, translocon assembly, or needle-translocon interactions.
Class II and class IIIb yscF TD mutants displayed nonpolar translocation.
Uncoordinated secretion and translocation in the presence of mammalian cells result in the release of Yops into the extracellular milieu, termed nonpolar translocation, and decreased translocation efficiency. Translocation-deficient mutants in translocon (ΔyopB and ΔlcrV) or regulatory (ΔyopN) components of the TTSS exhibit nonpolar translocation (17, 37, 49, 55, 61). Thus, a nonpolar translocation phenotype may indicate that yscF TD mutants had unstable or disrupted connections between the needle and the translocon, that the translocon was not assembled correctly, or that contact-mediated secretion regulation was malfunctioning. To assess “leaky” translocation, we looked for the presence of YopE in the tissue culture supernatant in the course of the infection. In addition, we also assessed the levels of the secreted translocon component YopD, as translocon components are normally found in low levels in the tissue culture supernatants during infection (37, 49). Y. pseudotuberculosis strains were grown at 37°C in high-calcium medium and then centrifuged onto HEp-2 cells to initiate cell contact and subsequent Yop secretion and translocation. After infection, tissue culture supernatants were collected and analyzed for the presence of YopD and YopE by SDS-PAGE and Western blotting. To monitor for HEp-2 and Y. pseudotuberculosis lysis, supernatants were also probed with antisera to the eukaryotic cytosolic marker actin and the bacterial cytosolic marker S2, respectively.
As expected, low levels of the translocon component YopD were found in culture supernatants for infections with WT Y. pseudotuberculosis, whereas YopE was absent (Fig. 5A). The YopD found in the tissue culture supernatant did not originate from disrupted bacteria or lysed HEp-2 cells, as the S2 and actin proteins were not readily detected (Fig. 5A, S2 and actin) unless HEp-2 cells or bacteria were solubilized with detergents (Fig. 5A, lysed HEp-2 and lysed Y. pseudotuberculosis). Y. pseudotuberculosis lacking the translocon component YopB or LcrV leaked low levels of YopE into the extracellular space during infection (Fig. 5A, ΔB+p and ΔV+p). The ΔyopB and ΔlcrV strains had higher and lower levels of YopD than the WT, respectively, in the tissue culture supernatant. This difference reflects the low levels of Yops secreted in the ΔlcrV strain (Fig. 3) (2, 65). Also as expected, the ΔyopN strain showed very large amounts of YopD and YopE in the culture supernatant, as Yops are secreted constitutively due to defective regulation from within the bacterial cytosol (15).
FIG. 5.
Most yscF TD mutants maintain tight needle-translocon connections during infection. Yersinia strains containing pTRC99A or pTRC99A expressing various yscF TD mutants were grown in 2× YT media with 5 mM calcium for 2 h at 26°C, induced with IPTG, and shifted to 37°C for 2 h. (A) Bacteria were added to monolayers of HEp-2 cells at an MOI of 50:1 and centrifuged to initiate cell contact and subsequent Yops translocation. After 1 hour of infection, tissue culture supernatants were collected and centrifuged to remove bacteria. Proteins were precipitated with TCA, separated by SDS-PAGE, and analyzed by Western blotting with antisera to the YopE and YopD proteins. Controls for disrupted HEp-2 cells and bacteria included uninfected cells lysed with Triton X-100 (lysed HEp-2) and bacteria solubilized in SDS-containing sample buffer (lysed Y. pseudotuberculosis [Y. ptb]). Antisera to actin were used to detect leakage of HEp-2 cytosol during infection, and antisera to the ribosomal subunit S2 were used to detect lysis of bacteria. (B) Secreted proteins in the presence of calcium. Y. pseudotuberculosis bacteria were centrifuged, and culture supernatants were collected. Secreted proteins were precipitated with TCA, separated by SDS-PAGE, and stained with Coomassie blue. Molecular mass standards are shown on the left. The asterisk points to the E158 mutant with barely detectable secreted Yops. (C) Western blot of secreted proteins from panel B, probed with antiserum to YopE and YopD.
Eighteen of the yscF TD mutants showed normal polarized translocation, with low levels of YopD and no YopE detected in the tissue culture supernatant (Fig. 2 and 5), suggesting that both the needle-translocon interactions and the secretion regulation functions remained intact.
Seven yscF TD mutants released YopE during infection, indicating a nonpolar translocation phenotype. Three yscF TD strains, D212, E158, and E350, showed an excess of both YopD and YopE proteins in the culture supernatant (Fig. 2 and 5A). Four yscF TD mutants, D108, D119, E354, and E23, leaked YopE, while YopD levels were comparable to those in the WT (Fig. 2 and 5A). Four yscF mutants that were leaky during infection but that appeared to have normal needle structures were grouped into class II. Three of the five class III mutants were also leaky and were subdivided into class IIIb. None of the yscF TD mutants displayed an exact ΔyopB or ΔlcrV phenotype, suggesting that needle-translocon protein interactions were not completely abolished.
Most yscF TD mutants regulate effector secretion normally.
The translocation defects in class II and class IIIb mutants was likely a result of inefficient transfer of Yops because of a poor connection between the needle and the translocon or because they had secretion regulation defects and were constitutively secreting Yops in the absence of cell contact or both. To test the yscF TD mutants for constitutive Yop secretion, strains were grown at 37°C in media containing calcium in the absence of eukaryotic cells and secreted Yops were detected by both Coomassie blue staining and Western blotting. As expected, WT Yersinia did not secrete effector Yops in the presence of calcium (Fig. 5B and C, ΔF+F). The 18 yscF TD strains that did not leak Yops during infection of tissue culture cells also showed normal calcium-regulated secretion (Fig. 2 and 5C). Initial observations by Coomassie blue staining suggested that only the E158 and D212 yscF TD mutants secreted Yops in high-calcium media, although at extremely low levels and considerably less than the ΔyopN strain (Fig. 5B). However, when examined by Western blotting with YopE antisera, all class II and class IIIb strains that leaked YopE during infection of HEp-2 cells were also shown to secrete YopE in the presence of calcium (Fig. 5C). Thus, leakage of YopE during contact-mediated translocation correlated with a malfunction in calcium-regulated secretion. Because the ΔlcrV, ΔyopB, and ΔyopN mutants also secreted YopE in high-calcium medium, class II and class IIIb yscF TD needle interactions with LcrV, translocon components, or regulatory components may be altered, but we could not distinguish between the possibilities by using this assay.
Class I mutants have reduced pore-forming abilities, while class II mutants have normal or enhanced pore-forming activities.
Translocation of Yops into the host cell cytosol requires LcrV-dependent assembly of YopB and YopD into a functional pore in the host plasma membrane. All of the yscF TD strains secreted YopB, YopD, and LcrV, so these components are present to form pores. However, weakened or disrupted interactions of mutant yscF needles with LcrV may have altered the ability of LcrV to assemble the pore.
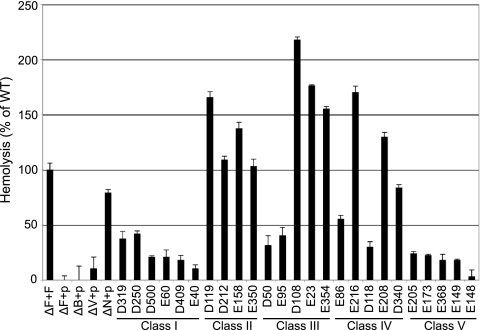
We tested the yscF TD mutants for their abilities to form pores in SRBCs by detection of hemoglobin released into the culture supernatant. The amount of RBC lysis caused by each of the yscF TD mutant strains was normalized to the WT level (Fig. 6, ΔF+F). Generally, the pore-forming abilities of the yscF TD mutants were consistent within each class, with the exception of the class IV strains. The class I yscF TD mutants showed reduced RBC lysis, ∼10 to 40% of the WT level (Fig. 6, class I), similar to the levels of Yop translocation observed in these strains (Fig. 3B), indicating that the class I yscF TD mutants had problems with pore insertion or assembly. Because the class I mutants secreted WT levels of Yops, showed normal needles, and did not leak Yops during translocation, decreased pore formation was the defining phenotype for this class and thus was likely to be the direct cause of the translocation defect in these strains.
FIG. 6.
Hemolysis of sheep red blood cells by yscF TD strains. Yersinia bacteria were grown in secretion media for 2 h at 26°C, induced with IPTG, and shifted to 37°C for 2 h. Bacteria were mixed 1:1 with sheep red blood cells and gently centrifuged to initiate cell contact. Infections were allowed to occur for 3 h at 37°C, and hemoglobin released into the culture supernatant was detected by reading absorbance values at 545 nm. The amount of RBC hemolysis conferred by the ΔyscF+pTRC99A-yscF strain was set to 100% (ΔF+F), and hemolysis levels in all other strains were normalized to WT levels. Experiments were performed in triplicate at least twice, and results for one representative experiment are shown. Error bars show the standard deviations from the means for the triplicate samples.
In contrast, the class II and class IIIb yscF TD strains lysed RBCs as well as or better than the WT (Fig. 6, class II and class III, D108, E23, and E354), indicating that these strains inserted YopB and YopD into the RBC membrane and that LcrV was able to construct a pore of sufficient diameter to lyse the RBC. The combined observations that the class II and class IIIb yscF TD strains all leak Yops during infection, yet form pores, strongly suggest that the needle and the pore cannot maintain a tight connection following pore insertion.
The yscF TD class IIIa and class V strains were almost entirely defective for pore formation (Fig. 6, class III, D50 and E95, and class V), consistent with their very low levels of Yop translocation and small amounts of YscF polymers, indicating that these yscF mutant needle structures were not competent to interact with a host cell membrane to initiate pore insertion. The class IV yscF TD strains showed various abilities for lysis of RBCs (Fig. 6, class IV) which generally mirrored their levels of Yop translocation. The pore-forming defects in these strains should contribute to the mild decrease in translocation but may be slightly overcome by an increase in needle numbers.
DISCUSSION
To determine the regions of the needle required for translocation of Yops, we isolated YscF mutants that were defective for translocation but not secretion. Previous mutagenesis studies on YscF and MxiH were designed to examine the role of the needle in secretion regulation and targeted acidic residues or conserved residues among needle proteins of different pathogens (31, 73). These studies successfully identified residues important for secretion regulation and also found mutations that resulted in defects in needle structure and effector secretion. By generating randomly mutagenized yscF strains and screening for Yop translocation, we uncovered mutations in YscF residues that were not broadly conserved but were specifically important for translocation. The identification of needle mutants that are defective for translocation but regulate secretion normally reveals a novel phenotype.
Translocation of effectors into a host cell requires coordination between the needle and the translocon. Although the needle itself may dictate this coordination by directly interacting with the translocon (YopB and YopD), the recent localization of LcrV to the distal tips of needles (47) and the fact that LcrV cross-links to YscF on purified needles (47) suggest that LcrV is poised to form a bridge connecting the needle with the translocon (Fig. 7, WT). Furthermore, LcrV is required for the formation of functional pores and regulates pore size (6, 24) but is not itself found in pores (24); all of these observations are consistent with the idea that LcrV functions at the tip of the needle. Mutations in the needle protein may disrupt the assembly, stability, or function of the LcrV tip complex, leading to a decrease in translocation. Alternatively, the needle may connect directly to YopB and/or YopD during translocation, if LcrV is released from the end of the needle after translocon assembly. Thus, mutations in the needle protein could then disrupt the needle-translocon interface.
FIG. 7.
Models of proposed mechanisms for translocation-defective yscF mutations. Needle, LcrV, and translocon components are shown before and after cell contact. (WT) The needle, LcrV tip, and translocon coordinate for Yop translocation. (A) Model A. Disrupted LcrV tip assembly leads to poor translocon formation. (B) Model B. Disrupted interactions between the LcrV tip and/or the translocon leads to leakage of Yops and inefficient translocation. (C) Model C. Unstable YscF-YscF interactions lead to needle breakage before or after cell contact. (D) Model D. LcrV binding to the needle is abolished and no pores are formed. See Discussion for details.
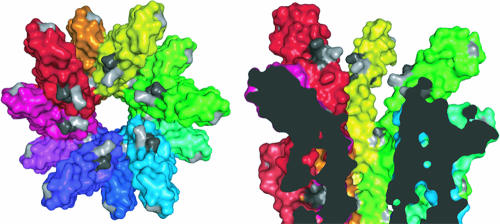
A model of the LcrV tip complex on the assembled needle has been proposed (11) based on the crystal structures of LcrV (13) and the Shigella needle protein MxiH (11). The MxiH monomer has a hairpin-like shape composed of two alpha helices connected by a conserved four-residue ordered loop, termed the P-(S/D)-(D/N)-P turn, and has a slight bend in one helix that divides the monomer into head and tail domains. Fitting the MxiH monomer into the structure of the assembled needle revealed that the tail domain formed the core of the tube, while the head domain angled outwards, with the PSNP turn exposed on the surface (11) (Fig. 8). The tip complex-needle model showed five LcrV molecules interdigitated between the heads of the five needle monomers at the top ring of the needle (11). In this model, the head domain of each needle monomer at the distal end is proposed to make multiple direct contacts with the tip complex (11).
FIG. 8.
Mapping of YscF residues A27, D28, A30, N31, and N47 onto the surface of a T3SS needle. (Left) Surface representation of the end-on view of the Shigella T3SS needle, with each subunit colored differently. The equivalent residues (T23, Q24, L26, Q27, and N43) of the Shigella subunit protein MxiH have been highlighted in gray. (Right) Cutaway surface representation of the side-on view of the Shigella T3SS, with the equivalent Yersinia YscF residues that were mutated highlighted in gray. Class I mutation equivalents A27, A30, and N31 are colored light gray, and class II mutation equivalents D28 and N47 are colored dark gray. This figure was prepared using the PyMOL program (11).
Twelve of the 25 yscF TD alleles have mutations that reside within a short region of the N terminus from residue 22 to residue 35 (Fig. 2 and Fig. S1 in the supplemental material), including three class I and two class II mutants. Mapping these class I and class II yscF mutations onto the Shigella needle reveals a continuous patch on the head domain that faces the central channel, where we might expect the LcrV tip to bind (Fig. 8, gray residues). Interestingly, the sequence of residues 27 to 31 of YscF in Yersinia (ADDAN) is similar to those in needle proteins from Pseudomonas, Aeromonas spp., Photorhabdus lumenescens, and some Vibrio spp. and shows the consensus sequence (A[D/N/K]XAN)(Table 2). Each of these species contains a protein with sequence homology to LcrV, and at least two homologs complement some LcrV functions in Yersinia (6, 20, 24, 47). Furthermore, this region is more divergent in needle proteins from other TTSS-containing bacteria, including Shigella, Salmonella, E. coli, and Burkholderia bacteria, none of which contain a protein with sequence homology to LcrV (Table 2), though functional homologs are present (8, 33, 57, 67). Thus, our genetic screen revealed a region of the needle protein critical for translocation of Yops, located at the end of the needle where LcrV binds, that correlates with the potential of the bacterium to form a tip complex.
TABLE 2.
Proteins with sequence homologies to LcrVa
| Organism | Protein | Sequenceb | Homology to LcrV |
|---|---|---|---|
| Yersinia spp. | YscF | ADDAN | Yes |
| Pseudomonas aeruginosa | PscF | ANAAN | Yes |
| Photorhabdus luminescens | LscF | ADTAN | Yes |
| Aeromonas spp. | AscF | ANDAN | Yes |
| Vibrio spp. | hypoth. | AKDAN | Yes |
| Shigella flexneri | MxiH | TQTLQ | No |
| Salmonella spp. | PrgI | VDNLQ | No |
| Burkholderia spp. | BsaL | VKDLN | No |
| E. coli O157 | EscF | GKTLS | No |
| Citrobacter rodentium | EscF | ADTLA | No |
| Sodalis glossinidius | hypoth. | LETAR | No |
| Chromobacterium spp. | hypoth. | VEQAG | No |
| Bordetella spp. | BscF | LNAHE | No |
Needle protein NCBI accession numbers: Yersinia pseudotuberculosis, YP_068491; Pseudomonas aeruginosa, PA01 AAG05108; Photorhabdus luminescens, AA018031; Aeromonas hydrophila, AAV30245; Vibrio parahaemolyticus, NP_798073; Shigella flexneri, NP_858270; Salmonella enterica serovar Typhimurium, NP_461794; Burkholderia mallei, YP_106136; E. coli O157, NP_290252; Citrobacter rodentium, AAL06385; Sodalis glossinidius, YP_454238; Chromobacterium violaceum, NP_902255; and Bordetella pertussis, CAC79556. LcrV homolog NCBI accession numbers: Yersinia pseudotuberculosis, YP_068466; Pseudomonas aeruginosa, AAO91771; Photorhabdus luminescens, AAO18053; Aeromonas hydrophila, AAV67437; Vibrio parahaemolyticus, NP_798038.
Boldface indicates a match with the consensus sequence (A[D/N/K]XAN).
We envision at least four models in which needle protein mutations could disable translocation by decreased tip complex functioning, poor needle-translocon connections, or needle instability (Fig. 7). In model A, the yscF mutation does not disturb LcrV binding but alters the conformation of the tip complex such that YopB and YopD are not inserted, pores are not assembled correctly, or the needle and translocon are misaligned and the pores are blocked. In model B, the mutation weakens interactions between the needle and the LcrV tip and/or the pore. Pores are formed, but the needle does not remain connected to the pore. In model C, the mutation destabilizes needle monomer-monomer interactions such that the needles fall apart before or after cell contact. In model D, the mutation disrupts the binding site for LcrV entirely, so the tip complex is not formed and LcrV cannot assemble the pore. All five classes of yscF TD mutants have phenotypes that are consistent with one or more of models A, B, and C. None of the yscF mutant phenotypes are consistent with model D.
By all assays thus far, the class I mutants form normally regulated secretion-competent needles, and yet they show a deficiency in pore formation, strongly indicating decreased LcrV functioning. We expect that LcrV still localizes to the tips of class I needles and that a connection between the needle and the translocon remains intact for three reasons: first, Yops are not leaked during translocation; second, some pores are formed; and third, some Yop translocation does occur, consistent with model A (Fig. 7). Class I needle ends may be altered such that the tip complex cannot function properly, resulting in inadequate pore insertion or assembled pores that are not competent for translocation. As discussed above, the class I mutants D250 (A27V), D500 (N31S), and E60 (A30V) all map to the predicted LcrV binding pocket and thus are likely to directly interfere with tip complex binding or assembly (Fig. 8, dark gray residues). The other three class I mutants, D319 (K85R), D409 (I64T), and E40 (T70A), all map to regions of YscF expected to be important for YscF monomer-monomer interactions (11). These mutants may have slight changes in overall needle structure, leading to defects at the tip that interfere with LcrV functioning or translocon binding. Alternatively, these mutants could have defects in polymerization such that normal amounts of YopE and YscF are secreted and secretion is regulated normally, but the needle is not long enough to reach and insert the pore into SRBCs or HEp-2 cells. Testing the class I mutants for the presence of LcrV at the needle end, examining the amounts of YopB and YopD inserted into the membrane and the sizes of the pores formed, and measuring needle length will help distinguish between these possibilities.
The class II yscF TD mutants show normal Yop secretion and needle formation but generally have higher YscF levels than the WT, have faulty secretion regulation, and leak Yops during infection. These phenotypes are similar to that seen with ΔlcrV and ΔyopB mutants, suggesting disrupted contact between the needle and LcrV or the needle and the pore. However, the class II yscF mutants formed pores as well as or better than WT Yersinia, suggesting that the pore assembly function of LcrV is intact. We propose that the yscF class II mutants have poor needle-translocon complex stabilities following cell contact because of altered tip complex conformation, consistent with model B (Fig. 7). Both of the class II yscF TD single mutants, N47S and D28G, are predicted to be surface exposed at the end of the assembled needle and important for needle-tip complex interactions: N47 is part of the PSNP turn, and D28 is within the predicted LcrV binding pocket (Fig. 8, light gray residues). Mutations in the PSNP turn and the D28 residue have also been identified by other groups and lead to nearly identical phenotypes (31, 73). In Yersinia pestis, D46A, D46C, D28A, and D28C mutations lead to constitutive secretion and to decreases in Yop translocation (73). Multiple mutations in the PSNP turn of MxiH lead to unregulated secretion and low levels of invasion, yet these mutants form pores normally (31). Intriguingly, a double mutant, D28A D46A, was null for translocation (73), suggesting that the combination of the two defects may lead to a complete loss of the tip complex.
The class IIIa and class V yscF TD mutants do not make normal needles as assessed by cross-linking or Yop secretion. Abnormalities could result from low YscF levels, poor YscF polymerization, or unstable needles breaking off, all causing short needles and/or blocked needles. Needle polymerization is observed in class IIIa mutants when YscF levels are raised artificially, suggesting that low YscF protein levels or polymer instability is their likely defect. Needle length is important for translocation (45), and short needles do not form pores in SRBCs (31). The lack of pore formation and the less defined or absent needles support a short needle phenotype in the class IIIa and class V strains. Short needles in the yscF TD mutants are not likely to be caused by defects in YscP functioning, as ΔyscP strains make long needles (30) which are competent for translocation (45). Interestingly, the class IIIa and class V needles do not leak Yops when in contact with host cells, indicating that interactions with LcrV and/or members of the YopN regulation complex are intact despite the structural malfunctions. Both of the class IIIa and three of the class V alleles have mutations within the tail domain of the needle monomer, which may lead to unstable YscF-YscF interactions (11). The Yop secretion defect of the yscF class V mutants suggests that in addition to having the structural defects, these needles may be partially obstructed.
The yscF TD class IIIb mutants also have low YscF protein levels and make aberrant needles, but in addition, they are not capable of regulating secretion. Two of the class IIIb strains, D108 and E23, carry an I13T mutation. A cysteine substitution at YscF residue 13 also led to constitutive secretion in Y. pestis; however, an alanine substitution resulted in WT levels for secretion regulation, needle formation, and translocation (73), suggesting that hydrophobicity but not necessarily size is important at this location. Though the amino termini of MxiH and the Burkholderia needle protein BsaL were not ordered in either atomic structure (11, 76), I13 lies on the modeled helix that lines the inner wall of the needle channel (11). Because of their secretion regulation defects, class IIIb mutants may have disrupted interactions with the YopN complex and/or LcrV (see below). Intriguingly, the class IIIb mutants are still able to form pores in RBC membranes despite these proposed defects in needle structure, suggesting that their translocation defect occurs after pore formation, analogous to model C (Fig. 7).
Both class II and class IIIb mutants are defective for secretion regulation, which could be due to defects in either the cytosol, the tip, or the translocon, since unregulated secretion is observed in the absence of YopN, LcrV, and YopB. Recent work has shown that YopN functions in a complex with TyeA, YscB, and SycN within the bacterial cytosol (15) and likely acts close to the mouth of the TTSS base, as mutations in YopN that result in opposing phenotypes of either constitutive secretion or blocked secretion have been identified (15). Cryo-electron microscopy structures of the Salmonella needle complex suggest that the needle begins at the outer membrane ring of the TTSS base (42). The needle and the YopN complex are then separated by as much as 200 Å and are not likely to have direct access to each other. Based on the location of some class II mutations at the predicted interface between the needle and LcrV (Fig. 8), it is more likely that these mutations are disrupting LcrV or translocon functioning than disrupting YopN functioning. Alternatively, the needle itself could be sensing contact with the host cell and transmitting a signal to begin secretion through other base components to the YopN complex, as suggested previously (9, 73). Indeed, the class IIIb yscF TD mutants may fall into this category, as each strain contains a mutation in a residue predicted to reside in the tail domain of YscF, which may change the needle structure and disrupt needle signaling.
In conclusion, using an unbiased genetic approach, we identified specific YscF residues that are required for the assembly of the translocon and/or productive interactions with the translocon, allowing us to initiate the mapping of the needle-translocon interface. Many of the class I and class II mutations reside in an area of YscF that, once assembled into a needle, is predicted to form a pocket for interactions with LcrV. These mutations could not have been predicted based on the primary sequence of the needle protein. Future studies investigating the defects of these yscF TD classes should provide insights into the molecular events required for translocation.
Supplementary Material
Acknowledgments
We thank Janet Deane and Susan Lea (University of Oxford) for sharing unpublished data, for helpful discussions, and for the creation of Fig. 8; Hyuk-Kyu Seoh and Cathy Squires (Tufts University) for the kind gift of the S2 antiserum; J. M. Balada for the construction of the ΔlcrV strain; M. Fisher for the ΔyopB strain; and the members of the Mecsas laboratory and Andrew Bohm (Tufts University) for helpful discussions.
This work was supported by NIH NRSA GM67517 (to A.J.D.), NIH R01 AI056068 (to J.M.), and the Center for Gastroenterology Research on Absorptive and Secretory Processes (GRASP), NIDDK P3034928.
Footnotes
Published ahead of print on 27 October 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 68:301-315. [DOI] [PubMed] [Google Scholar]
- 1a.Balada-Llasat, J. M., and J. Mecsas. 2006. Yersinia has a tropism for B and T cell Zones of lymph nodes that is independent of the type III secretion system. PLoS Pathogens 2:816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman, T., S. Hakansson, A. Forsberg, L. Norlander, A. Macellaro, A. Backman, I. Bolin, and H. Wolf-Watz. 1991. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J. Bacteriol. 173:1607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blocker, A., P. Gounon, E. Larquet, K. Niebuhr, V. Cabiaux, C. Parsot, and P. Sansonetti. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 5.Boland, A., M. P. Sory, M. Iriarte, C. Kerbourch, P. Wattiau, and G. R. Cornelis. 1996. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15:5191-5201. [PMC free article] [PubMed] [Google Scholar]
- 6.Broms, J. E., C. Sundin, M. S. Francis, and A. Forsberg. 2003. Comparative analysis of type III effector translocation by Yersinia pseudotuberculosis expressing native LcrV or PcrV from Pseudomonas aeruginosa. J Infect. Dis. 188:239-249. [DOI] [PubMed] [Google Scholar]
- 7.Cambau, E., F. Bordon, E. Collatz, and L. Gutmann. 1993. Novel gyrA point mutation in a strain of Escherichia coli resistant to fluoroquinolones but not to nalidixic acid. Antimicrob. Agents Chemother. 37:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24:747-756. [DOI] [PubMed] [Google Scholar]
- 9.Cordes, F. S., K. Komoriya, E. Larquet, S. Yang, E. H. Egelman, A. Blocker, and S. M. Lea. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278:17103-17107. [DOI] [PubMed] [Google Scholar]
- 10.Crepin, V. F., R. Shaw, C. M. Abe, S. Knutton, and G. Frankel. 2005. Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J. Bacteriol. 187:2881-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deane, J. E., P. Roversi, F. S. Cordes, S. Johnson, R. Kenjale, S. Daniall, F. Booy, W. D. Picking, W. L. Picking, A. Blocker, and S. M. Lea. 2006. Molecular model of a type three secretion system needle: implications for host cell sensing. Proc. Natl. Acad. Sci. USA 103:12529-12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBord, K. L., V. T. Lee, and O. Schneewind. 2001. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 183:4588-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derewenda, U., A. Mateja, Y. Devedjiev, K. M. Routzahn, A. G. Evdokimov, Z. S. Derewenda, and D. S. Waugh. 2004. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure 12:301-306. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferracci, F., F. D. Schubot, D. S. Waugh, and G. V. Plano. 2005. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol. Microbiol. 57:970-987. [DOI] [PubMed] [Google Scholar]
- 16.Fields, K. A., M. L. Nilles, C. Cowan, and S. C. Straley. 1999. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 67:5395-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields, K. A., and S. C. Straley. 1999. LcrV of Yersinia pestis enters infected eukaryotic cells by a virulence plasmid-independent mechanism. Infect. Immun. 67:4801-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg, A., A. M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goure, J., P. Broz, O. Attree, G. R. Cornelis, and I. Attree. 2005. Protective anti-V antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J Infect. Dis. 192:218-225. [DOI] [PubMed] [Google Scholar]
- 21.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 22.He, S. Y., K. Nomura, and T. S. Whittam. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206. [DOI] [PubMed] [Google Scholar]
- 23.Hoiczyk, E., and G. Blobel. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. USA 98:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmstrom, A., J. Olsson, P. Cherepanov, E. Maier, R. Nordfelth, J. Pettersson, R. Benz, H. Wolf-Watz, and A. Forsberg. 2001. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol. Microbiol. 39:620-632. [DOI] [PubMed] [Google Scholar]
- 25.Iriarte, M., and G. R. Cornelis. 1999. Identification of SycN, YscX, and YscY, three new elements of the Yersinia Yop virulon. J. Bacteriol. 181:675-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iriarte, M., M. P. Sory, A. Boland, A. P. Boyd, S. D. Mills, I. Lambermont, and G. R. Cornelis. 1998. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 17:1907-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov, M. I., J. A. Stuckey, H. L. Schubert, M. A. Saper, and J. B. Bliska. 2005. Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence. Mol. Microbiol. 55:1346-1356. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, M. W., J. B. Day, and G. V. Plano. 1998. YscB of Yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180:4912-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, Q., and S. Y. He. 2001. Role of the Hrp pilus in type III protein secretion in Pseudomonas syringae. Science 294:2556-2558. [DOI] [PubMed] [Google Scholar]
- 30.Journet, L., C. Agrain, P. Broz, and G. R. Cornelis. 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302:1757-1760. [DOI] [PubMed] [Google Scholar]
- 31.Kenjale, R., J. Wilson, S. F. Zenk, S. Saurya, W. L. Picking, W. D. Picking, and A. Blocker. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280:42929-42937. [DOI] [PubMed] [Google Scholar]
- 32.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 35.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lee, V. T., and O. Schneewind. 1999. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol. Microbiol. 31:1619-1629. [DOI] [PubMed] [Google Scholar]
- 38.Lee, V. T., C. Tam, and O. Schneewind. 2000. LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275:36869-36875. [DOI] [PubMed] [Google Scholar]
- 39.Leung, D. W., E. Y. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 40.Logsdon, L. K., and J. Mecsas. 2003. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect. Immun. 71:4595-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marenne, M. N., L. Journet, L. J. Mota, and G. R. Cornelis. 2003. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb. Pathog. 35:243-258. [DOI] [PubMed] [Google Scholar]
- 42.Marlovits, T. C., T. Kubori, M. Lara-Tejero, D. Thomas, V. M. Unger, and J. E. Galan. 2006. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441:637-640. [DOI] [PubMed] [Google Scholar]
- 43.Mecsas, J., I. Bilis, and S. Falkow. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mota, L. J., L. Journet, I. Sorg, C. Agrain, and G. R. Cornelis. 2005. Bacterial injectisomes: needle length does matter. Science 307:1278. [DOI] [PubMed] [Google Scholar]
- 46.Mota, L. J., I. Sorg, and G. R. Cornelis. 2005. Type III secretion: the bacteria-eukaryotic cell express. FEMS Microbiol. Lett. 252:1-10. [DOI] [PubMed] [Google Scholar]
- 47.Mueller, C. A., P. Broz, S. A. Muller, P. Ringler, F. Erne-Brand, I. Sorg, M. Kuhn, A. Engel, and G. R. Cornelis. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674-676. [DOI] [PubMed] [Google Scholar]
- 48.Neyt, C., and G. R. Cornelis. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971-981. [DOI] [PubMed] [Google Scholar]
- 49.Nilles, M. L., K. A. Fields, and S. C. Straley. 1998. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J. Bacteriol. 180:3410-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nilles, M. L., A. W. Williams, E. Skrzypek, and S. C. Straley. 1997. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J. Bacteriol. 179:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nordfelth, R., and H. Wolf-Watz. 2001. YopB of Yersinia enterocolitica is essential for YopE translocation. Infect. Immun. 69:3516-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 53.Parsot, C., C. Hamiaux, and A. L. Page. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7-14. [DOI] [PubMed] [Google Scholar]
- 54.Pastor, A., J. Chabert, M. Louwagie, J. Garin, and I. Attree. 2005. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 253:95-101. [DOI] [PubMed] [Google Scholar]
- 55.Persson, C., R. Nordfelth, A. Holmstrom, S. Hakansson, R. Rosqvist, and H. Wolf-Watz. 1995. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 56.Pettersson, J., A. Holmstrom, J. Hill, S. Leary, E. Frithz-Lindsten, A. von Euler-Matell, E. Carlsson, R. Titball, A. Forsberg, and H. Wolf-Watz. 1999. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol. Microbiol. 32:961-976. [DOI] [PubMed] [Google Scholar]
- 57.Picking, W. L., H. Nishioka, P. D. Hearn, M. A. Baxter, A. T. Harrington, A. Blocker, and W. D. Picking. 2005. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 73:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riley, G., and S. Toma. 1989. Detection of pathogenic Yersinia enterocolitica by using congo red-magnesium oxalate agar medium. J. Clin. Microbiol. 27:213-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 60.Rosqvist, R., A. Forsberg, and H. Wolf-Watz. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosqvist, R., K. E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skryzpek, E., and S. C. Straley. 1993. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J. Bacteriol. 175:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skrzypek, E., and S. C. Straley. 1995. Differential effects of deletions in lcrV on secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J. Bacteriol. 177:2530-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 67.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 68.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 69.Swietnicki, W., B. S. Powell, and J. Goodin. 2005. Yersinia pestis Yop secretion protein F: purification, characterization, and protective efficacy against bubonic plague. Protein Expr. Purif. 42:166-172. [DOI] [PubMed] [Google Scholar]
- 70.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamano, K., E. Katayama, T. Toyotome, and C. Sasakawa. 2002. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J. Bacteriol. 184:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tardy, F., F. Homble, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Torruellas, J., M. W. Jackson, J. W. Pennock, and G. V. Plano. 2005. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol. 57:1719-1733. [DOI] [PubMed] [Google Scholar]
- 74.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell. Microbiol. 3:753-762. [DOI] [PubMed] [Google Scholar]
- 75.Yother, J., and J. D. Goguen. 1985. Isolation and characterization of Ca2+-blind mutants of Yersinia pestis. J. Bacteriol. 164:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, L., Y. Wang, W. L. Picking, W. D. Picking, and R. N. De Guzman. 2006. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 359:322-330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.