Abstract
During Pseudomonas aeruginosa flow cell biofilm development, the cell population differentiates into a nonmotile subpopulation which forms microcolonies and a migrating subpopulation which eventually colonizes the top of the microcolonies, resulting in the development of mushroom-shaped multicellular structures. The cap-forming subpopulation was found to develop tolerance to membrane-targeting antimicrobial agents, such as the cyclic cationic peptide colistin and the detergent sodium dodecyl sulfate. The stalk-forming subpopulation, on the other hand, was sensitive to the membrane-targeting antibacterial agents. All biofilm-associated cells were sensitive to the antibacterial agents when tested in standard plate assays. A mutation eliminating the production of type IV pili, and hence surface-associated motility, prevented the formation of regular mushroom-shaped structures in the flow cell biofilms, and the development of tolerance to the antimicrobial agents was found to be affected as well. Mutations in genes interfering with lipopolysaccharide modification (pmr) eliminated the biofilm-associated colistin tolerance phenotype. Experiments with a PAO1 strain harboring a pmr-gfp fusion showed that only the cap-forming subpopulation in biofilms treated with colistin expresses the pmr operon. These results suggest that increased antibiotic tolerance in biofilms may be a consequence of differentiation into distinct subpopulations with different phenotypic properties.
Bacteria living associated with surfaces display a number of phenotypes which apparently differ from those of cells growing planktonically, despite identical genotypes (6, 33, 35). It has therefore been suggested that cells growing as a biofilm express genes that are distinct from those in cells growing planktonically (4, 38). This concept has been further developed through detailed studies of a number of model bacteria, which have shown that there seem to be defined steps of progression in surface colonization to a fully mature complex biofilm structure (6, 34, 42). In particular, Pseudomonas aeruginosa has attracted considerable interest as a model organism due to its significance in medical contexts as a pathogen and also due to its genomic complexity, which reflects excellent adaptability. This high degree of versatile colonization is based on the genomic content of a large number of catabolic genes and a corresponding very high number of regulatory systems (24).
P. aeruginosa biofilm development on solid surfaces is best described as a multistep developmental process which involves cellular attachment (49), firm association of adhered cells to the substratum (36), clonal microcolony growth of nonmotile cells (21), and, under specific laboratory conditions, colonization of the microcolonies by a motile subpopulation that forms the cap of mature mushroom-shaped structures (21). All these steps involve distinct gene expression and individual regulatory components (8, 34). Stabilization of the microcolony structures and the subsequent mushroom-shaped structures requires extracellular polymeric substances, such as polysaccharides (10, 11, 14, 17, 29, 51), cup fimbriae (23, 47, 48), and excreted high-molecular-weight DNA (50).
In connection with the emerging scheme of biofilm development involving differential gene expression, it is becoming increasingly interesting to investigate possible phenotypic traits associated with the biofilm state. One of these biofilm traits is increased tolerance to antibiotics and biocides (41), which is a severe practical problem in many sectors of our society (medicine, industry, water supply systems, foods, etc.). Many explanations for the frequently observed increase in tolerance to antimicrobial agents have been offered (41): poor penetration of the compound due to the encasement of the cells in a polymer matrix, biofilm-induced high-level resistance, slow or no growth of a large part of the biofilm population, lack of oxygen, electrostatic charge of the biofilm surface, and others. Depending on the specific antimicrobial compound, each of these factors could be relevant. It is therefore not possible to develop a consensus explanation for biofilm-associated antibiotic tolerance. Apart from the complexity of this problem, taking into account that so many factors may play a role, it is also not trivial to determine if cells in a biofilm are sensitive or tolerant.
In light of the well-established biofilm model for P. aeruginosa, it is an intriguing task to unravel the mechanism of antibiotic resistance/tolerance for this organism, and the focus could obviously be on the role of any step in biofilm development and its underlying regulatory activities. From previous observations documented in the literature, it seems clear that glucose-grown P. aeruginosa biofilms are composed of two easily distinguishable subpopulations: one which forms the top of mushroom-shaped multicellular structures and one which constitutes the stalk of these structures (21). The phenotypic difference between the two subpopulations has also been documented to encompass the synthesis of rhamnolipids (25). The genes for this group of quorum-sensing-controlled surfactants were found to be mainly induced in the stalk cells beginning at a time before the cap was formed, and considering the impact of rhamnolipids in maintaining the channels between the mushroom structures (7), the differentiation into two subpopulations with distinct phenotypic properties appears to be related directly to the various steps of this complex process. We thought it would be interesting to determine the level of antibiotic tolerance of these two distinct populations of cells, assuming that there could be a correlation between structural development and properties and tolerance to antimicrobial agents. We chose to test two compounds—colistin and sodium dodecyl sulfate (SDS)—both of which target the cell envelope, which would make both of them compatible with the use of propidium iodide as an in situ indicator of cell death.
Colistin is a cyclic cationic decapeptide linked to a fatty acid side chain (19). It belongs to a group of similarly structured bacterial antimicrobial peptides, the polymyxins (43). Its use as antibiotic in human therapy has been hampered by its nephrotoxicity, but in recent years the increasing development of multiresistant pathogens has stimulated renewed interest in this antimicrobial peptide (9, 18, 26). Although the polymyxins were identified more than 50 years ago, their modes of action as antimicrobials have not been completely elucidated. In general, the mechanism of action involves binding of the cationic compounds to the anionic membranes of bacteria (32), and it is established that lipid A located at the end of the lipopolysaccharide (LPS) is a primary target for the polymyxins (52, 53). This binding causes an apparent induced disruption of the cell membrane, leading to leakage of cell contents and cell death (13). It has been proposed that, as a first step, the peptides mediate their own uptake through binding to lipid A, after which it enters the periplasm of the cell and inserts into the cytoplasmic membrane (53). The actual killing may involve several targets in the cell, and it occurs within a very short time in the case of most of the polymyxin derivatives (seconds to a few minutes) (52, 53).
Induced tolerance to colistin has been documented for several bacterial species, including P. aeruginosa (27, 28, 30, 54). A major route of adaptive polymyxin resistance involves the two-component signal transduction systems phoPQ and pmrAB. The phoPQ system is a global regulatory system which among other environmental conditions responds to limiting levels of divalent cations such as Mg2+ (12, 40). Interestingly, Ramsey and Whiteley (36) recently reported that PhoQ is necessary for mushroom formation in P. aeruginosa biofilms (36). In Salmonella and Escherichia coli, the pmrAB system is linked to the phoPQ transduction system, which means that low Mg2+ indirectly activates the pmrAB-controlled genes (39, 45). The operon pmrH-M is directly controlled by pmrAB, and the gene products are responsible for the synthesis of N4-aminoarabinose, which binds to lipid A and reduces binding of the polymyxins (making the cells resistant to the peptides) (46).
Here, we present evidence that biofilm formation of P. aeruginosa grown in flow chambers with glucose as the carbon source results in the development of a subpopulation which shows greatly increased phenotypic tolerance to both colistin and SDS and that this subpopulation seems to be identical to the one constituting the cap of the mushroom-shaped structures.
MATERIALS AND METHODS
Bacterial strains and media.
Pseudomonas aeruginosa PAO1 (16) tagged with green fluorescent protein (GFP) or yellow fluorescent protein (YFP) (20) and a ΔpilA derivative constructed by allelic displacement tagged with GFP or cyan fluorescent protein (CFP) (20) as well as two pmr mutants, pmrB and pmrF, and the MPAO1 background strain as a control (Pseudomonas Genome Project [44]), all tagged with GFP, were used in this study. Modified FAB medium (15) was supplemented with 10 mM glucose for batch overnight cultures and with 0.3 mM glucose for biofilm cultivation. Biofilms and batch cultures were grown at 30°C.
For monitoring of pmr operon expression in biofilms, a transcriptional fusion to GFP inserted at the Tn7 insertion site on the P. aeruginosa chromosome was constructed. The intergenic region between PA3551 and PA3552 was PCR amplified from P. aeruginosa PAO1 using the primers P1(PA3551) (5′-AAT TAA GAA TTC GAG CGG CAA AAA GCT AAC AC-3′), containing an EcoRI restriction site, and P2(PA3552) (5′-AAT TAA TCT AGA GGG AAG GCG TAA TCA CTT CA-3′), containing an XbaI restriction site. The amplification was performed in a T3 Thermocycler PCR machine from Biometra using the Expand high-fidelity system (Roche). The PCR fragment was inserted into the corresponding sites in the multiple cloning site of plasmid pJBA27 (1) and electroporated into Escherichia coli MT102, resulting in the transcriptional GFP reporter plasmid pSM2472. This plasmid was further digested with NotI, and the fragment containing the P2(PA3552)-gfp fusion was cloned into the mini-Tn7 delivery vector pSM1959 (22), resulting in the plasmid pSM2476. Mini-Tn7 mobilization into the P. aeruginosa chromosome was performed according to the method of Koch et al. (22). Electroporation was performed by using a Gene Pulser (Bio-Rad) as recommended by the manufacturer with 50 μl bacterial cells and 10 to 100 ng DNA. The cells were then added to 1 ml of SOC medium (0.5% yeast extract, 2.0% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM MgSO4, 20 mM glucose), incubated for 1 h at 37°C, and then spread onto selective agar plates. DNA extraction, treatment with modifying enzymes and restriction endonucleases, ligation of DNA fragments with T4 ligase, and transformation of E. coli were performed using standard methods (2).
Cultivation of biofilms.
Biofilms were grown at 30°C in flow chambers with individual channel dimensions of 1 by 4 by 40 mm. The flow system was assembled and prepared as described previously (31). The flow chambers were inoculated by injecting 250 μl of overnight culture diluted to an optical density at 600 nm of 0.001 into each flow channel with a small syringe. After inoculation, flow channels were left without flow for 1 h, after which medium flow was started using a Watson Marlow 205S peristaltic pump. Each channel was supplied with a flow of 3 ml/h of FAB medium containing the appropriate carbon source. The mean flow velocity in the flow cells was 0.2 mm/s.
Exposure of biofilms to colistin and sodium dodecyl sulfate.
The bacterial red fluorescent viability stain propidium iodide (PI) was added to biofilm medium 15 min before and during exposure to 10 μg/ml, 25 μg/ml, or 100 μg/ml colistin (H. Lundbeck, Copenhagen, Denmark) or 0.01% SDS (Sigma).
Red fluorescing and green fluorescing cells from a colistin-treated biofilm were sorted by fluorescence-activated cell sorting (Becton Dickinson FACS Vantage SE) and plated on Luria-Bertani plates to verify that red fluorescing cells were not viable and green fluorescing cells were viable.
Microscopy, fluorescent recovery after photobleaching (FRAP), and image processing.
All microscopic observations were completed using a Zeiss LSM510 confocal laser scanning microscope (CLSM; Carl Zeiss, Jena, Germany) equipped with an argon and a NeHe laser and detectors and filter sets for simultaneous monitoring of CFP (excitation, 458 nm; emission, 475 nm), GFP (excitation, 488 nm; emission, 517 nm) YFP (excitation, 514 nm; emission, 527 nm), and red fluorescence emitted from the PI (excitation, 543 nm; emission, 565 nm). Images were obtained using a 63×/1.4 Plan-APOChromat differential interference contrast objective or a 40×/1.3 Plan-Neofluar oil objective.
Photobleaching of GFP was done by scanning an area of interest with a 15% attenuated 488-nm line from an argon laser. PI was used as a control for laser-induced cell death.
Multichannel simulated fluorescence projection (shadow projection) images and vertical cross sections were generated using the IMARIS software package (Bitplane AG, Zürich, Switzerland).
RESULTS
Visualization of two distinctly different subpopulations in monoclonal Pseudomonas aeruginosa biofilms.
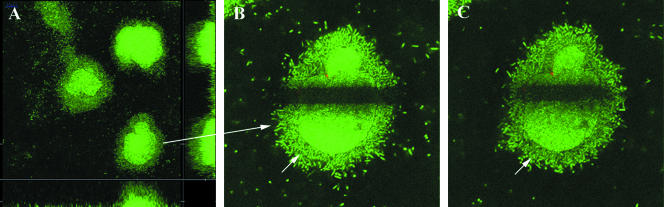
P. aeruginosa often forms biofilms with mushroom-shaped multicellular structures in flow chambers (for example, see reference 21). We have previously shown that in flow chambers irrigated with glucose minimal medium, these structures develop based on interactions between motile and nonmotile subpopulations (21). In addition, investigations using several other P. aeruginosa background strains have shown the same pattern of development (data not shown). Microscopic examination of wild-type glucose-grown P. aeruginosa biofilms showed that 2-day-old microcolonies comprise two visually different populations (Fig. 1A). The two populations could be distinguished by both light and fluorescent microscopy. In light of the documented presence of both motile and nonmotile cells in these biofilms, and taking into account their specific roles in the development of mushroom-shaped microcolonies, it was thought that the difference between the two observed subpopulations was associated with motility. In order to test this hypothesis, we used FRAP to identify motile and nonmotile subpopulations of the microcolonies. FRAP is typically used to study protein dynamics and activity within single living cells (37), but in the present study it was used to investigate the dynamics of bacteria within multicellular biofilm structures.
FIG. 1.
Two-day-old P. aeruginosa biofilm showing motile and nonmotile subpopulations in a microcolony. (A) Three-dimensional CLSM image projection and an orthogonal section image of the biofilm structure (xyz dimensions, 146.2 μm by 146.2 μm by 24.7 μm); (B) bleached microcolony indicated by the arrow in panel A; (C) the same microcolony 15 min after bleaching. (B and C) xy dimensions, 73.1 μm by 73.1 μm. Arrows in panels B and C indicate the motile population.
A rectangular area across the middle of a GFP-fluorescent microcolony was bleached by the use of an argon laser (Fig. 1B). To ensure that the laser did not kill the cells but only left them nonfluorescent, PI was added to the medium before, during, and after bleaching. No difference in cell death was observed between the bleached and nonbleached areas (data not shown). These results also showed that the nonmotile cells were alive. Movement of nonbleached cells into the bleached area was assessed by acquiring pictures every 15 min after photobleaching for 6 h and 15 min. As early as the first picture after bleaching, fluorescent cells had moved into the bleached area of the looser outer layer of the microcolony (Fig. 1C). In contrast, no cell movement was observed in the inner denser core of the colony during the entire recording time (more than 6 h) (See Timelapse01 at www.cbm.biocentrum.dtu.dk/english/services/supplement/Haagensen1). The horizontal optical section depicted in Fig. 1 was positioned at a short distance from the substratum. We also used FRAP to study cell motility on the substratum/microcolony interface of a 2-day-old microcolony. Again, two subpopulations were present: Single cells moving along the substratum were readily observed in the outer part of the microcolony, whereas cells in the center of the microcolony were nonmotile (see Timelapse02 at www.cbm.biocentrum.dtu.dk/english/services/supplement/Haagensen2). Use of FRAP was possible only on cell layers relatively close to the substratum.
The FRAP results support the suggestion that the mushroom-shaped P. aeruginosa structures in flow chamber biofilms indeed are composed of two distinct subpopulations which are easily distinguished by their microscopic structural features and by their motility phenotypes. This leaves open the possibility that other phenotypes may be associated with these two subpopulations in the biofilm.
Biofilm induced tolerance to colistin and SDS.
A major objective of this investigation was to identify specific P. aeruginosa cells or subpopulations in flow chamber-grown biofilms which displayed tolerance to addition of antimicrobial agents. Since this type of analysis depends to a large degree on direct imaging of the biofilm populations and their phenotypes, it was decided to use antimicrobial agents affecting the cell envelope directly—an activity assumed to be more compatible with the use of the in situ indicator of dead cells propidium iodide. In control experiments, it was shown that when an agent such as 25 μg/ml colistin or 0.01% SDS is added to cultures of P. aeruginosa (whether growing as suspended cultures or in biofilms), cellular uptake of propidium iodide correlated to loss of viability upon plating for viable colony formation (not shown). Cellular susceptibility to these two antimicrobial agents was therefore assessed in flow cell biofilms.
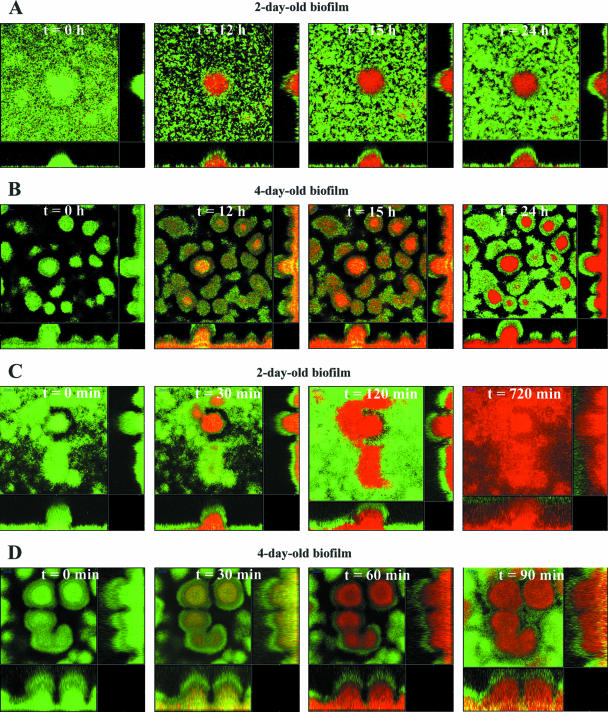
For the experiments investigating tolerance development, GFP expression (green fluorescence) and uptake of PI (red fluorescence) were used as live-cell and dead-cell indicators, respectively. PI was added to the medium of 2- and 4-day-old biofilm 15 min before and during exposure to colistin or SDS, and the effect was followed with time-lapse CLSM. Upon addition of 25 μg/ml colistin to the medium of a 2-day-old biofilm, the cells in the middle of the microcolonies were killed after 12 to 15 h exposure (Fig. 2A), whereas the cells in the outer layer were still alive even after several days of exposure (data not shown). Interestingly, the relative positions of the susceptible and tolerant subpopulations in the biofilms correlated with the positions of the motile and nonmotile subpopulations identified by FRAP in this study and by use of various mixtures of CFP- and YFP-tagged cells by Klausen et al. (21). A similar pattern was observed when 4-day-old biofilms were treated with 25 μg/ml colistin (Fig. 2B). Again, the top layer of cells in the mushroom-shaped structures was tolerant, whereas the subpopulation positioned in the mushroom stalk was susceptible to the peptide. In control experiments, it was shown that tolerant cells isolated from biofilms never displayed any change in sensitivity to the antibiotics used in the present investigation when subsequently tested in suspended cells and on plates (not shown); consequentially, no genetic resistance can account for any of the presented observations.
FIG. 2.
Effect of colistin and SDS on P. aeruginosa PAO1 biofilms. Green fluorescent P. aeruginosa cells were grown in flow cells supplemented with 0.3 mM glucose for 2 or 4 days. The cell death indicator PI was added to the medium 15 min before exposure to 25 μg/ml colistin (A and B) or 0.01% SDS (C and D), and the effect was monitored with time-lapse CLSM. Both colistin and SDS were unable to kill a tolerant subpopulation in the outer layer of the biofilm.
The effect of addition of SDS to P. aeruginosa biofilms showed similarities to the effects of colistin treatment. When medium with 0.01% SDS was added to 2-day-old biofilms, cells in the middle of the microcolonies were killed after 15 to 30 min, whereas an outer layer of cells remained alive (Fig. 2C). SDS killed cells faster than colistin and quickly disrupted the biofilm structure, so that cells were eventually washed out of the flow chamber. The same pattern was observed when 4-day-old biofilms were treated with 0.01% SDS (Fig. 2D).
Correlation between type IV pilus-driven motility and differentiation after colistin or SDS treatment.
Because the nonmigrating and migrating subpopulations, described by Klausen et al. (21) and further characterized in this paper, seemed to correlate with the two subpopulations identified by colistin and SDS treatment of live/dead-stained biofilms, the role of type IV pilus-driven motility in the development of the tolerance phenotype was further examined. When a 2-day-old biofilm formed by P. aeruginosa pilA mutants (unable to produce type IV pili) was treated with 25 μg/ml colistin, mainly single tolerant cells were found occurring randomly within the microcolonies (Fig. 3). An analogous pattern of randomly distributed tolerant cells in biofilm microcolonies of this mutant strain was observed when a 2-day-old biofilm was treated with 0.01% SDS (not shown). Type IV pilus-mediated motility thus seems to be important for the structural organization of colistin- and SDS-tolerant subpopulations in P. aeruginosa PAO1 biofilms.
FIG. 3.
Effect of colistin on a pilA mutant. Green fluorescent pilA mutant cells were grown in flow cells supplemented with 0.3 mM glucose for 2 days and then exposed to 25 μg/ml colistin. The cell death indicator PI was added to the medium 15 min before exposure to 25 μg/ml colistin, and the effect was monitored with CLSM. Tolerant cells were distributed randomly in the biofilm. (A) Two-day-old biofilm before exposure to 25 μg/ml colistin; (B) biofilm exposed to 25 μg/ml colistin for 24 h.
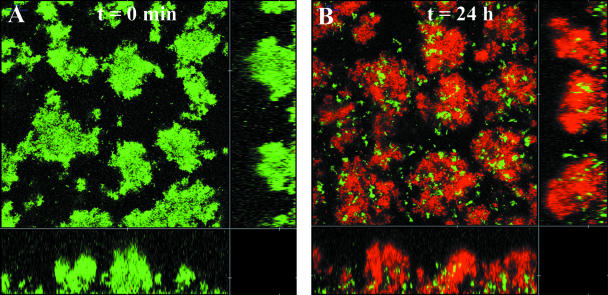
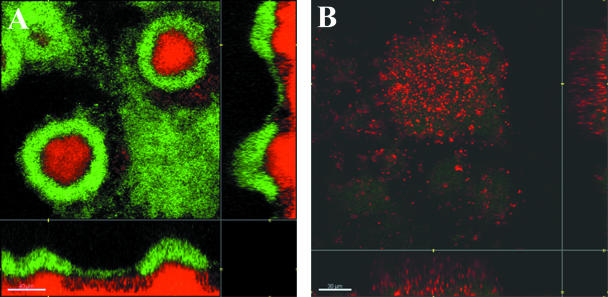
To further determine the requirement for type IV pilus-driven motility in connection with development of colistin-tolerant mushroom caps in P. aeruginosa biofilms, a mixed biofilm containing wild-type PAO1 cells tagged with YFP and PAO1 pilA cells tagged with CFP was grown in glucose minimal medium. We have previously shown that wild-type P. aeruginosa PAO1 cells can colonize and form mushroom caps on P. aeruginosa pilA mutant microcolonies via a pathway that depends on type IV pili (21). When mixed 3-day-old biofilms were subjected to 25 μg/ml colistin, the wild-type cells on top of the pilA microcolonies were tolerant to the antibiotic treatment, whereas the stalk-forming pilA cells were sensitive (Fig. 4).
FIG. 4.
(A) Two-day-old biofilm established using a 1:1 mixture of YFP-tagged wild-type P. aeruginosa PAO1 and CFP-tagged nonmotile pilA mutant PAO1. (B) Treatment with 25 μg/ml colistin at day 3 for 24 h. Red color represents dead cells detected by PI staining.
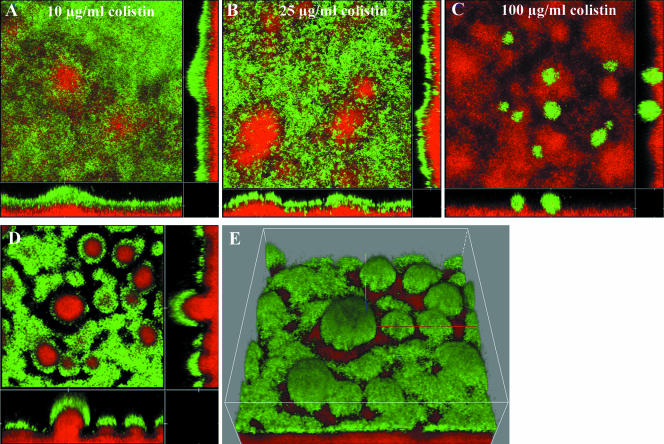
The colistin MIC for planktonic P. aeruginosa cells was 1 μg/ml, and in biofilms, tolerant subpopulations were observed when 10, 25, or 100 μg/ml colistin was used (Fig. 5A, B, and C). However, when the biofilm was challenged with the highest concentration, only a few cell clusters seemed to be resistant to the antibiotic (Fig. 5C). In contrast, increasing the colistin concentration to 100 μg/ml in a PAO1 biofilm that had already been treated with 25 μg/ml did not seem to have any significant effect on the live subpopulation (Fig. 5D and E).
FIG. 5.
Green fluorescent wild-type P. aeruginosa biofilms were established and after 2 days exposed to 10 μg/ml (A), 25 μg/ml (B), or 100 μg/ml (C) colistin. The cell death indicator PI was added to the medium 15 min before exposure. Colistin at 10 mg/ml resulted in the same distribution of a tolerant subpopulation as 25 μg/ml, whereas 100 mg/ml gave only a few cells the ability to survive and develop in the biofilm. (D and E) A PAO1 biofilm was treated at day 4 with 25 μg/ml colistin for 3 days, and then the concentration was increased to 100 μg/ml. Images were taken at day 8. Glucose minimal medium was used for growth of the biofilms.
Tolerance to colistin is associated with the pmr operon.
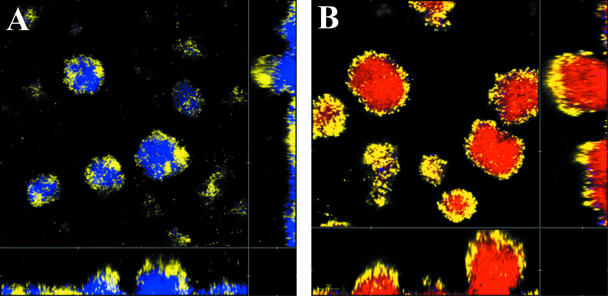
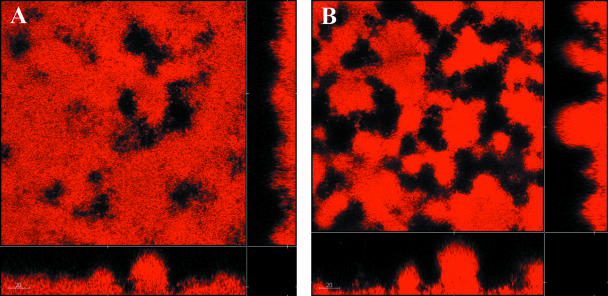
The pmr operon encodes a pathway for the synthesis of 4-amino-4-deoxyarabinose and its binding to lipid A in the LPS of P. aeruginosa. Expression of the operon is controlled by a two-component regulatory system encoded by the pmrAB genes. When induced, the lipid A modification prevents colistin binding, and the cells become resistant to the peptide. It was therefore of interest to investigate whether mutations in the pmr genes had any effect on the development of colistin tolerance in a flow chamber biofilm. Two mutants, a pmrB (encoding the sensor kinase of the two-component signal transduction regulatory system) mutant and a pmrF (one of the structural genes in the pathway) mutant, both tagged with GFP, were grown in biofilms for 4 days before exposure to 25 μg/ml colistin. After 24 h of exposure to colistin, PI was added to the medium, and the distribution of live and dead cells was monitored using CLSM. As seen in Fig. 6, all cells were killed by the colistin treatment, suggesting that the pmr operon, and hence modification of LPS, is directly involved in the development of colistin tolerance in PAO1 biofilms. The pmr mutants display type IV pilus-driven motility comparable to that of PAO1, as determined by plate assay (data not shown).
FIG. 6.
Two pmr mutants, a pmrB mutant (A) and a pmrF mutant (B), tagged with GFP were grown in glucose minimal medium in flow chambers. At day 4 the biofilms were treated with 25 μg/ml colistin for 24 h and stained with PI (red) for monitoring of dead cells.
Furthermore, we investigated whether the tolerant subpopulation seen in colistin-treated PAO1 biofilms expresses the pmr operon. A fusion of this operon to gfp was constructed in PAO1, and the reporter strain was introduced in the flow chamber system. After 4 days of growth, the biofilm was treated with colistin for 24 h, and then GFP expression from the reporter fusion was monitored. As shown in Fig. 7A, there was clear induction of the top part of the biofilm structure and the mushroom cap, which correlates with the tolerant subpopulation obtained in PAO1 biofilms after colistin treatment. Figure 7B shows an image of a biofilm without colistin addition and without GFP expression, demonstrating that colistin is required for the pmr operon to be induced.
FIG. 7.
PAO1 (pmr-gfp fusion) biofilm grown in glucose minimal medium in flow chambers. (A) Biofilm at day 4, after treatment with 25 μg/ml colistin for 24 h and staining with PI (red) for monitoring of dead cells. Green cells show induction of the pmr-gfp fusion. (B) Biofilm at day 4, without colistin treatment and with PI staining. No cells are induced.
The role of the pmr operon in the induction of colistin tolerance was further verified by a DNA array analysis, in which global gene expression in biofilms grown with and without colistin was investigated. After 24 h of colistin treatment, during which all cells in the non-cap-forming part of the biofilm were killed, the pmr genes were found to be expressed at a level 6- to 10-fold above that of the untreated control (data not shown), representing expression in the live cap-forming population.
DISCUSSION
It is clear from many investigations that biofilm-associated cells display high-level tolerance to many antibiotics and other antimicrobial agents, creating considerable problems in removing biofilms from both abiotic and biotic surfaces in various settings, including in patients with infections (6). However, it is less clear if antibiotic tolerance is a shared feature of all biofilm-associated cells or if this property is associated with only parts of the biofilm populations. It is also not clear whether the biofilm-associated antibiotic tolerance is a direct consequence of the biofilm lifestyle per se or whether indirect induction of tolerance occurs in ways similar to what may even be the case for planktonic cells grown under special conditions. In order to obtain a more direct identification of the survivors after antibiotic treatment of biofilms, it is necessary to visualize them in situ. In the present context, we found it particularly interesting to investigate whether the stalk- and cap-forming subpopulations observed in P. aeruginosa biofilms displayed any difference in antibiotic susceptibility.
In the present studies we observed that both colistin and SDS preferentially killed cells forming the core/stalk of the wild-type mushroom structures. The two distinct subpopulations in the PAO1 mushroom-shaped structures were killed with significantly different efficiencies under the conditions used. When the cells were harvested from the flow cells and plated on various concentrations of colistin, they all showed wild-type sensitivity, demonstrating that the biofilm-associated tolerance to quite high colistin concentrations was not caused by genetic changes but rather seemed to be induced by the specific biofilm conditions. Tolerance induction was only rarely observed in planktonic cells (data not shown). The observation that killing was most efficient in the inner parts of the biofilm structures was unexpected and suggests further that, at least for these antimicrobials, penetration of the drugs into the biofilm plays no role in the antibiotic tolerance. Recent studies by Boles et al. (5) showed that exogenous addition of rhamnolipid or SDS to flow chamber-grown P. aeruginosa biofilms caused central hollowing of the microcolonies and ultimate detachment of the biofilms (5), and Banin et al. (3) showed that also the metal chelator EDTA preferentially kills cells from the inner parts of the microcolonies in flow chamber-grown P. aeruginosa biofilms (3). We have found that colistin-tolerant cells in fact appear very early after adhesion of the cells to the glass surface, before any microcolonies have formed, showing that the difference in exposure to the antibiotic does not seem to be relevant (data not shown).
The role of the pmr modification pathway in the development of colistin tolerance in P. aeruginosa biofilms was investigated by growing biofilms of two different pmr mutants and monitoring the development of colistin tolerance. For both mutants, no colistin-tolerant subpopulations were formed, strongly suggesting that there is a direct link between this specific genetic determinant of resistance and the biofilm-associated colistin tolerance. Furthermore, it was shown that the cap-forming subpopulation of the biofilm structures indeed expresses the pmr operon after colistin treatment, verifying the link between tolerance development and LPS modification. DNA microarray data comparing colistin-treated and untreated biofilms of PAO1 and obtained in the same way as the image data presented in this work showed significant up-regulation of all the genes in the pmr operon in response to colistin treatment, which suggests a role for LPS modification in the development of colistin tolerance (J. A. J. Haagensen, unpublished data). The up-regulation of pmr was observed after 24 h of colistin treatment, at which time only the mushroom cap cells were alive (Fig. 2), which means that the increased pmr expression must have taken place in these cells (the stalk cell population was dead at this time). This implies that colistin-induced expression of the pmr operon is limited to the P. aeruginosa biofilm subpopulation constituting the cap and related domains.
When a high concentration of colistin (100 μg/ml) was added to the biofilms of PAO1, killing was very efficient and the few live cell clusters seemed to be derived from single cells rather than being associated with a subpopulation (not investigated further). However, biofilms initially treated with a lower concentration of colistin (25 μg/ml) and subsequently receiving a very high concentration were not affected further, indicating that the previously induced tolerance to the lower concentration was sufficient to allow the cells to withstand even higher colistin concentrations (100 times the MIC). This is compatible with the pmr induction observed in Fig. 7.
That colistin and other peptides may induce increased peptide resistance in P. aeruginosa was previously documented by McPhee et al. (30). It has been shown that in P. aeruginosa, the pmrAB genes in many ways are functionally similar to those described for Salmonella enterica serovar Typhimurium, but there are also interesting differences (30). One of these is that many antimicrobial peptides (including colistin), when present in low concentrations, activate the pmrAB genes, resulting in induced resistance to the very same peptides. This induction of resistance is apparently independent of the normal signal transduction pathways, indicating that an alternative mode of regulation is involved. The finding that pregrowth of a P. aeruginosa biofilm in the presence of a low colistin concentration results in full tolerance to a much higher concentration, which would otherwise nearly eradicate the entire population, is fully consistent with this type of induced tolerance.
We have previously shown that development of mushroom-shaped structures in P. aeruginosa biofilms depends on type IV pili as well as on the particular carbon source used to support growth (21). On one hand, a P. aeruginosa pilA mutant, which is deficient in the biosynthesis of type IV pili and hence type IV pilus-driven motility, produced microcolony structures without caps; on the other hand, changing the growth conditions, such as replacing glucose with citrate as the carbon source, caused the P. aeruginosa wild type to form flat unstructured biofilms, probably due to increased type IV pilus-driven motility. In the present context it was of interest to assess the impacts of these changes on colistin tolerance in the resulting biofilms. The scattered appearance of surviving cells found in the microcolonies of 2-day-old pilA mutant biofilms after treatment with colistin was probably a result of the lack of cap formation due to the lack of type IV pili in these cells. In citrate-grown biofilms of wild-type cells, however, there was no apparent correlation between type IV pilus-driven motility and colistin tolerance (data not shown).
In this study, we present data which show that development of P. aeruginosa biofilms in flow cells involves an early differentiation of the attached population into at least two distinct subpopulations with clear phenotypic differences. These two subpopulations also differ in their tolerance to antimicrobial agents. Addition of normally lethal concentrations of either colistin or SDS efficiently killed all cells of the microcolony-forming subpopulations, whereas the migrating dynamic subpopulations which eventually colonized the microcolonies and formed mushroom caps displayed a high level of tolerance to such treatments. We found no permanently resistant cells in such biofilms, suggesting that the tolerance was transient and induced in the biofilm. This finding has several interesting implications. It adds to the current concept of biofilm development, showing that surface growth may result in a separation of the original population into several isogenic but clearly distinguishable subpopulations. It also points to a possible mechanism behind the finding that biofilm-associated cells are often much more resistant to antibiotics than otherwise identical planktonic cells. At least in the case of colistin (and SDS) tolerance, there seems to be a link between surface attachment via induced LPS modification (genetically determined by a two-component signal transduction pathway, pmr) and subpopulation-associated tolerance to greatly increased concentrations of the peptide. Due to the functional similarity in modes of action of many antimicrobial peptides, our results may also explain why infecting bacteria organized in biofilm-like aggregates may show little susceptibility to even high concentrations of peptides produced by the host innate defense system.
In conclusion, this work substantiates the observation that the stalks of the mushroom-shaped microcolonies in P. aeruginosa biofilms are formed by a nonmotile subpopulation and that the caps of these microcolonies are formed by a motile subpopulation. We provide evidence that the stalks of the mushroom are sensitive to colistin and SDS, while the caps are tolerant to both agents. Whether there is a link between tolerance development and motility or whether the apparent correlation is indirect is subject to further study in our laboratory. However, we show clear results pointing at LPS modification as an explanation for the observed tolerance development.
Acknowledgments
This work was supported by grants to Søren Molin from the Danish Research Councils and to Robert K. Ernst and Samuel I. Miller (AI47938).
We thank Tove Johansen and Paula Ragas for expert technical assistance.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., M. R. Kingston, R. E. Moore, D. D. Seidman, J. G. Smith, and K. Struhl. 1996. Current protocols in molecular biology. John Wiley, New York, N.Y.
- 3.Banin, E., K. M. Brady, and E. P. Greenberg. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 72:2064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beloin, C., and J. M. Ghigo. 2005. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 13:16-19. [DOI] [PubMed] [Google Scholar]
- 5.Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210-1223. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333-1341. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 11.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heydorn, A., B. K. Ersboll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow-chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 16.Holloway, B. W., and A. F. Morgan. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40:79-105. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasiakou, S. K., A. Michalopoulos, E. S. Soteriades, G. Samonis, G. J. Sermaides, and M. E. Falagas. 2005. Combination therapy with intravenous colistin for management of infections due to multidrug-resistant gram-negative bacteria in patients without cystic fibrosis. Antimicrob. Agents Chemother. 49:3136-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz, E., and A. L. Demain. 1977. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol. Rev. 41:449-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 21.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 22.Koch, B., L. E. Jensen, and O. Nybroe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods. 45:187-195. [DOI] [PubMed] [Google Scholar]
- 23.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 24.Lazdunski, A. M., I. Ventre, and J. N. Sturgis. 2004. Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2:581-592. [DOI] [PubMed] [Google Scholar]
- 25.Lequette, Y., and E. P. Greenberg. 2005. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., R. L. Nation, R. W. Milne, J. D. Turnidge, and K. Coulthard. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents. 25:11-25. [DOI] [PubMed] [Google Scholar]
- 27.Macfarlane, E. L., A. Kwasnicka, and R. E. Hancock. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146:2543-2554. [DOI] [PubMed] [Google Scholar]
- 28.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 29.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 31.Møller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton, B. A. 1956. The properties and mode of action of the polymyxins. Bacteriol. Rev. 20:14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 35.Purevdorj-Gage, B., W. J. Costerton, and P. Stoodley. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151:1569-1576. [DOI] [PubMed] [Google Scholar]
- 36.Ramsey, M. M., and M. Whiteley. 2004. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 53:1075-1087. [DOI] [PubMed] [Google Scholar]
- 37.Reits, E. A., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145-E147. [DOI] [PubMed] [Google Scholar]
- 38.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soncini, F. C., V. E. Garcia, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 42.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 43.Storm, D. R., K. S. Rosenthal, and P. E. Swanson. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:723-763. [DOI] [PubMed] [Google Scholar]
- 44.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 45.Trent, M. S. 2004. Biosynthesis, transport, and modification of lipid A. Biochem. Cell Biol. 82:71-86. [DOI] [PubMed] [Google Scholar]
- 46.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 47.Vallet, I., S. P. Diggle, R. E. Stacey, M. Camara, I. Ventre, S. Lory, A. Lazdunski, P. Williams, and A. Filloux. 2004. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J. Bacteriol. 186:2880-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasseur, P., I. Vallet-Gely, C. Soscia, S. Genin, and A. Filloux. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985-997. [DOI] [PubMed] [Google Scholar]
- 50.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 51.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, M., E. Maier, R. Benz, and R. E. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235-7242. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., P. Dhillon, H. Yan, S. Farmer, and R. E. Hancock. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3317-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwir, I., D. Shin, A. Kato, K. Nishino, T. Latifi, F. Solomon, J. M. Hare, H. Huang, and E. A. Groisman. 2005. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 102:2862-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]