Abstract
Flagellar motility is an important determinant of Campylobacter jejuni that is required for promoting interactions with various hosts to promote gastroenteritis in humans or commensal colonization of many animals. In a previous study, we identified a nonmotile mutant of C. jejuni 81-176 with a transposon insertion in Cj1026c, but verification of the role of the encoded protein in motility was not determined. In this study, we have determined that Cj1026c and the gene immediately downstream, Cj1025c (here annotated as flgP and flgQ, respectively), are both required for motility of C. jejuni but not for flagellar biosynthesis. FlgP and FlgQ are not components of the transcriptional regulatory cascades to activate σ28- or σ54-dependent expression of flagellar genes. In addition, expression of flgP and flgQ is not largely dependent on σ28 or σ54. Immunblot analyses revealed that the majority of FlgP in C. jejuni is associated with the outer membrane. However, in the absence of FlgQ, the amounts of FlgP in the outer membrane of C. jejuni are greatly reduced, suggesting that FlgQ may be required for localization or stability of FlgP at this location. This study provides insight into features of FlgP and FlgQ, two proteins with previously undefined functions that are required for the larger, multicomponent flagellar system of C. jejuni that is necessary for motility.
Campylobacter jejuni is the second leading cause of bacterial gastroenteritis in humans in the United States and a leading cause of diarrheal disease throughout the world (7). In contrast, C. jejuni is able to promote a natural, commensal colonization of the gastrointestinal tracts of many domesticated and agriculturally important animals, including poultry (18, 23). These commensal relationships, while harmless to the animal hosts, create large reservoirs of the bacterium that can result in contamination of the human food supply, leading to a high incidence of C. jejuni gastroenteritis (9).
The production of flagella and flagellar motility are important determinants of C. jejuni for promoting interactions with various hosts. Nonmotile flagellar mutants of C. jejuni colonize chicks at reduced levels in a 1-day-old chick model of commensalism (14, 20, 26, 28). In addition, flagellar motility is required for interactions with the human intestinal tract to promote infection which can culminate in a productive gastroenteritis (3).
C. jejuni produces a single flagellum at one or both poles of the bacterium. To produce functional flagella, bacteria must coordinate both the temporal expression of over 40 flagellar genes and the ordered production and secretion of the encoded proteins. C. jejuni uses two alternative σ factors, σ28 and σ54, to mediate transcriptional regulation of specific flagellar genes (6, 13, 15, 16, 28). σ28 is involved in the transcription of a small subset of genes, including the major flagellin gene, flaA. σ54 is required for transcription of many genes encoding the flagellar rod, basal body, and hook components and a minor flagellin. Previous studies have determined that activation of transcription of σ54-dependent flagellar genes is dependent on the FlgSR two-component system and the proteins of the flagellar export apparatus, including FlhA, FlhB, FliP, and FliR (15). We have proposed that these two systems may constitute a regulatory cascade where formation or the secretory activity of the flagellar export apparatus may activate the FlgS sensor kinase. Through a phosphorelay system, FlgS would be predicted to then phosphorylate the FlgR σ54-dependent response regulator (28), activating it for transcription of σ54-dependent flagellar genes.
In a previous screen to identify genes of C. jejuni strain 81-176 that are required for flagellar motility, we used the mariner-based solo transposon to randomly mutagenize the bacterium and then isolated 28 mutants defective for motility (13). One nonmotile mutant was found to have a solo insertion in Cj1026c/Cjj81176_1045 (gene designations are based on the previously annotated C. jejuni 11168 genome [22] and the more recently sequenced C. jejuni 81-176 genome [GenBank accession number AANY01000004], respectively) but this mutant was not investigated further. Because this gene is located two genes upstream of flgR, which is required for motility and transcription of σ54-dependent flagellar genes, the possibility existed that the solo insertion in Cj1026c may have a polar effect on expression of flgR.
In this study, we have characterized Cj1026c and the downstream gene Cj1025c/Cjj81176_1044 and have found that both genes are required for motility. (Due to simplicity in communicating our findings, we propose to annotate Cj1026c and Cj1025c as flgP and flgQ, respectively, and will refer to these genes as such hereafter.) Mutants lacking either of these genes are nonmotile but possess flagella of normal appearance. In addition, the ΔflgP and ΔflgQ mutants are not defective for transcription of σ28- or σ54-dependent flagellar genes. Localization studies revealed that a large proportion of FlgP in C. jejuni is associated with the outer membrane. Furthermore, association of FlgP with the outer membrane is dependent on FlgQ, suggesting that FlgQ may be necessary for localization and subsequent stabilization of FlgP to contribute to the larger macromolecular flagellar complex to promote motility.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. jejuni 81-176 is a clinical isolate from a patient with gastroenteritis that has since been shown to promote gastroenteritis in humans and commensal colonization of the chick gastrointestinal tract (3, 14, 17). C. jejuni was routinely grown on Mueller-Hinton (MH) agar containing 10 μg/ml trimethoprim (TMP) in microaerobic conditions (10% CO2, 5% O2, and 85% N2) at 37°C. Antibiotics for C. jejuni growth were added to MH agar when necessary at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 15 μg/ml; cefoperazone, 30 μg/ml; or streptomycin, 0.5, 1, 2, or 5 mg/ml. All C. jejuni strains were stored at −80°C in a 85% MH broth-15% glycerol solution. Escherichia coli DH5α and XL1-Blue were grown in Luria-Bertani (LB) agar or broth. Antibiotics were used in the following concentrations: kanamycin, 100 μg/ml; chloramphenicol, 15 μg/ml; or ampicillin, 100 μg/ml. All E. coli strains were stored at −80°C in a 80% LB-20% glycerol solution.
Construction of bacterial strains.
PCR was used to amplify a 2.4-kb fragment from the chromosome of C. jejuni 81-176 containing the flgPQ locus with approximately 700 nucleotides of upstream and downstream DNA sequence. Primers were used with BamHI restriction sites to facilitate cloning into BamHI-digested pUC19 to create pDRH1954.
Defined deletion mutagenesis was used to create in-frame deletions of flgP and flgQ from the chromosome of C. jejuni (13). To generate a construct with insertion of a cat-rpsL cassette in flgP, pDRH1954 was digested with NheI, which cleaves the DNA within the coding sequence of flgP. Blunt-end fragments were generated with T4 DNA polymerase and the DNA was then ligated to SmaI-digested cat-rpsL from pDRH265. The resulting plasmid was designated pDRH1955. Because of the lack of a convenient restriction site in flgQ, PCR-mediated mutagenesis was used to generate an MscI site in flgQ by making A-to-G and T-to-C mutations at nucleotides 188 and 189 of the flgQ coding sequence to generate pDRH1981 (19). This DNA was then digested with MscI and ligated to SmaI-digested cat-rpsL to create the plasmids pDRH2028 and pDRH2029, which differed by the orientation of the cat-rpsL cassette. PCR-mediated deletion mutagenesis was also used to create in-frame deletions of flgP or flgQ from pDRH1954 (19). For flgP, primers were designed and used in a reaction with pDRH1954 to fuse in frame the start codon to the last 23 codons of the gene, deleting the intervening 148 codons to create pDRH1957. Similarly, the first 10 codons of flgQ were fused to the stop codon of flgQ to delete the intervening 139 codons to create pDRH1958.
Strains DRH212 (81-176 Smr) and DRH461 (81-176 Smr ΔastA) were electorporated with pDRH1955, pDRH2028, or pDRH2029 and then grown on MH agar with chloramphenicol to select for transformants with cat-rpsL insertions in flgP or flgQ (25). The resulting mutants were then electroporated with the appropriate plasmid, pDRH1957 or pDRH1981, and grown on MH agar with streptomycin to select for transformants that replaced the flgP::cat-rpsL or flgQ::cat-rpsL mutations with the in-frame deletion constructs on the chromosome of C. jejuni. The resulting mutants were designated DRH2064 (81-176 Smr ΔastA ΔflgP), DRH2070 (81-176 Smr ΔflgP), DRH2071 (81-176 Smr ΔflgQ), and DRH2118 (81-176 Smr ΔastA ΔflgQ).
For generation of flgP::astA or flgQ::astA transcriptional fusions in C. jejuni, NheI-digested pDRH1954 that had been filled in by T4 DNA polymerase to create blunt ends and MscI-digested pDRH1981 were ligated to a SmaI-digested astA-kan cassette from pDRH580. The resulting plasmids, pDRH2047 containing flgQ::astA-kan and pDRH2080 containing flgP::astA-kan, were electroporated into the appropriate ΔastA deletion strains to replace native flgP or flgQ with the transcriptional fusions on the chromosome of C. jejuni strains. For creation of DRH2316 (81-176 Smr ΔastA ΔflgQ flgP::astA-kan), pDRH1958 containing the flgPQ locus with the ΔflgQ deletion was digested with NheI, treated with T4 DNA polymerase, and ligated to the SmaI-digested astA-kan cassette to create a transcriptional fusion of flgP to astA. The resulting plasmid, pDRH2308, was then electroporated into DRH2118 to generate DRH2316.
To create astA transcriptional fusions to flgD or motA, PCR was used with chromosomal DNA from C. jejuni 81-176 to amplify a 2.1-kb fragment containing flgD and a 2.7-kb fragment containing the motAB locus. These DNA fragments were then digested with BamHI to facilitate cloning into pUC19 to generate pDRH325 (containing flgD) and pDRH2312 (containing motAB). MscI-digested pDRH325 and SpeI-digested pDRH2312 were then ligated to the SmaI-digested astA-kan cassette to create pDRH669 and pDRH2322, respectively.
For generation of flaA-, flaB-, flgD-, flgE2-, and motA-astA transcriptional fusions on the chromosomes of C. jejuni strains, appropriate ΔastA mutants were electroporated with pDRH608, pDRH610, pDRH669, pDRH532, or pDRH2322, respectively. These constructs replaced the native loci with the respective astA transcriptional fusions.
To complement ΔflgP or ΔflgQ mutants with plasmids expressing flgP or flgQ, primers with in-frame 5′ BamHI restriction sites were designed to amplify the coding sequence of flgP or flgQ from the second codon to the stop codon. After PCR, the amplified DNA was digested with BamHI and ligated to BamHI-digested pECO102 to generate plasmids expressing flgP (pSMS265) or flgQ (pSMS269) from the chloramphenicol acetyltransferase (cat) promoter (27). These plasmids were then transferred to DH5α/RK212.1 to facilitate conjugation into wild-type C. jejuni 81-176 Smr or the ΔflgP or ΔflgQ mutants (10).
Protein homology and domain analyses.
Homology searches were performed with BLASTP and PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST). Analyses of N-terminal signal sequences and lipoprotein sorting signals were performed with the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) and the LipoP 1.0 server (http://www.cbs.dtu.dk/services/LipoP/), respectively.
Preparation of antisera.
The flgP coding sequence from the second codon to the stop codon was cloned into the BamHI site of pQE30 (QIAGEN) to generate pSMS511 in E. coli XL1-Blue for induction of His6-tagged FlgP. For induction of a His6-tagged FlgQ protein, the coding sequence from codon 30 to the stop codon of flgQ was amplified and inserted into the BamHI site of pQE30 to generate pSMS506. For purification of proteins, XL1-Blue containing pSMS511 or pSMS506 was grown in 500 ml LB broth with ampicillin to mid-log phase, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and then incubated for another 4 h at 37°C. Bacteria were harvested and disrupted by passage twice through an EmulsiFlex-C5 cell disruptor (Avesin) at 15,000 to 20,000 lb/in2. Insoluble matter was separated from lysates by centrifugation for 2 h at 20,000 × g at 4°C. Protein was purified with Ni-nitrilotriacetic acid agarose under denaturing conditions in the presence of 8 M urea. After purification, the urea concentration was gradually reduced to below 100 mM urea through dialysis. Anti-FlgP (α-FlgP) and α-FlgQ antisera were generated by immunizing five and eight mice, respectively.
α-RpoA antiserum was generated by synthesizing a peptide with an N-terminal cysteine followed by the sequence of amino acids representing residues 207 to 229 of the C. jejuni RpoA protein (N′-CITPNEAFQNALEAMYKQLSVFDK-C′) at the Protein Synthesis Core at the University of Texas Southwestern Medical Center. This peptide was conjugated to keyhole limpet hemocyanin and then used with TiterMax to immunize BALB/c mice for production of antisera.
Fractionation of C. jejuni strains.
C. jejuni strains were grown from frozen stocks on MH agar containing 10 μg/ml TMP or 15 μg/ml chloramphenicol for 48 h at 37°C under microaerobic conditions. Strains were then restreaked on MH agar containing TMP or chloramphenicol and grown at 37°C under microaerobic conditions for 16 h. Strains were suspended from agar plates and diluted to an optical density at 600 nm (OD600) of 0.8. These bacterial cultures were used for all fractionation procedures described below.
For whole-cell lysates, 1 ml of bacteria was pelleted, washed once with phosphate-buffered saline (PBS), and then resuspended in 50 μl 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. Outer and inner membrane proteins were prepared and separated based on Sarkosyl insolubility by the method of Carlone et al. (5). Briefly, 5-ml aliquots of bacteria were pelleted, washed once in 10 mM HEPES (pH 7.4), and resuspended in 1 ml of the same buffer. Sonication to disrupt bacteria was achieved with a Branson Sonifier 450 with a microtip attachment. Unbroken bacteria were removed by brief centrifugation at 16,000 × g for 2 min. To recover total membranes, bacteria were then centrifuged for 16,000 × g for 30 min at 4°C. Membrane pellets were resuspended in 400 μl 1% sodium lauryl sulfate in 10 mM HEPES (pH 7.4) and incubated at room temperature for 30 min. Sarkosyl-insoluble outer membrane proteins were then removed by centrifugation for 30 min at 16,000 × g at 4°C. The supernatant contained Sarkosyl-soluble inner membrane proteins.
Periplasmic and cytoplasmic proteins were collected by pelleting 20-ml aliquots of bacteria at 4,200 × g for 10 min. To isolate periplasmic proteins, we modified the procedure of Donohue-Rolf and Keusch (8). The bacterial pellets were washed twice with 2 ml of PBS containing 0.1% gelatin by centrifugation at 4,200 × g at 4°C. The pellets were then suspended in 200 μl of PBS containing 0.1% gelatin, and polymixin B sulfate was added to a final concentration of 2 mg/ml. The bacteria were agitated for 30 min at 4°C. The preparation was then centrifuged at 16,000 × g for 30 min at 4°C to separate the spheroplasts in the pellet from the periplasmic proteins in the supernatant. The supernatants were removed and centrifuged again at 16,000 × g for 30 min at 4°C to remove any contaminating spheroplasts.
The spheroplasts obtained from the periplasmic preparations were suspended in 1 ml of 10 mM HEPES (pH 7.4) and then disrupted by sonication as described above. Insoluble matter and unbroken bacteria were removed by centrifugation at 16,000 × g for 2 min at 4°C. The supernatant was removed and centrifuged again at 16,000 × g for 30 min at 4°C. The resulting supernatants represented the proteins from the cytoplasm of the bacteria.
Immunoblot analyses.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 10% acrylamide gels. For whole-cell lysates, protein samples from each bacterial preparation were loaded to represent the proteins recovered from 200 μl of bacterial cultures that had been equilibrated to the same density by OD600 readings. For outer membrane proteins and cytoplasmic proteins, protein samples from each bacterial preparation were loaded to represent those obtained from 750 μl of culture. To detect FlgP or RpoA, a 1:1,500 dilution of murine α-FlgP M1 antiserum or murine α-RpoA M1 antiserum was used, respectively. Anti-major outer membrane protein (α-MOMP) rabbit antiserum diluted 1:1000 was used to detect MOMP in outer membrane preparations (30). For immunoblot analysis of FlgR production, 5 μl of whole-cell lysates from C. jejuni strains (representing 100 μl of bacterial culture) were used. Rabbit α-FlgR antiserum Rab13 was used at a dilution of 1:3,500 as the primary antibody for detection of FlgR (12). Horseradish peroxidase-conjugated goat α-mouse or α-rabbit immunoglobulin G antiserum was used as the secondary antibody.
Motility assays.
C. jejuni strains from frozen stocks were grown on MH agar with TMP for 48 h in microaerobic conditions at 37°C. After growth, strains were streaked on MH agar with TMP or chloramphenicol and grown for an additional 16 h in microaerobic conditions at 37°C. Strains were suspended from agar plates and diluted to an OD600 of 1.0. For agar-based assays for motility, bacterial strains were stabbed into semisolid MH agar plates containing 0.4% agar as previously described (13). Motility phenotypes were assessed approximately 32 h after inoculation and incubation at 37°C in microaerobic conditions. For direct observation of motile bacteria, bacterial cultures were grown as described above but diluted to an OD600 of 0.75. The bacteria were then diluted 1:10 in MH broth, and 10 μl of each sample was placed on a glass slide and topped with a glass coverslip. Bacteria were then visualized under a magnification of ×400 using an Olympus BH2 dark-field microscope.
Transmission electron microscopy.
C. jejuni strains were grown from frozen stocks on MH agar with TMP for 48 h in microaerobic conditions at 37°C. After growth, strains were streaked on MH agar with TMP and grown for an additional 16 h at 37°C in microaerobic conditions. Strains were suspended from agar plates and diluted to an OD600 of 0.4. Bacteria were pelleted and then resuspended in 2% glutaraldehyde for 1 hour at room temperature. Samples were stained with 1% uranyl acetate and visualized with a JOEL 1200 EX transmission electron microscope at 80 kV.
Arylsulfatase reporter assays.
Arylsulfatase assays were performed as previously described (15), which was based on previously published procedures (11, 29). C. jejuni strains were grown from frozen stocks on MH agar with TMP or chloramphenicol in microaerobic conditions at 37°C for 48 h. Strains were then restreaked on MH agar and grown again for 16 h in microaerobic conditions at 37°C. Bacteria were suspended from agar plates to an appropriate dilution, washed, and then incubated with 10 mM nitrophenylsulfate and 1 mM tyramine at 37°C for 1 h. Reactions were terminated with NaOH, and the amount of nitrophenol released in each sample was determined by reading the OD410 of the sample with a spectrophotometer. Values were compared to a standard curve of OD410 readings of known nitrophenol concentrations to determine the number of arylsulfatase units produced by each strain. One arylsulfatase unit is defined as the amount of enzyme catalyzing the release of 1 nmol of nitrophenol per hour per OD600 unit. Each strain was tested in triplicate, and each assay was performed three times.
RESULTS
Deletion of flgP or flgQ causes defects in flagellar motility.
We previously identified a nonmotile mutant of C. jejuni 81-176 with an insertion of the solo transposon in flgP (DRH241) (13). However, this mutant was not further characterized to verify that the nonmotile phenotype is actually due to interruption of flgP by the transposon. The 3′ end of flgP overlaps the 5′ end of the uncharacterized downstream gene flgQ by 27 nucleotides. These genes are positioned on the C. jejuni chromosome immediately upstream of flgR (Fig. 1A), which we and others have also shown is required for flagellar motility and transcription of σ54-dependent flagellar genes (15, 16, 28). The solo transposon in the original transposon mutant is located 34 bases upstream of the stop codon of flgP and 10 bases upstream of the start codon of flgQ.
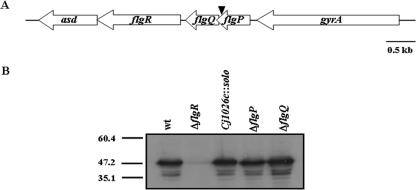
FIG. 1.
Deletion of flgP or flgQ does not affect FlgR production in C. jejuni. (A) Organization of the flgPQ locus and surrounding genes. flgP and flgQ are immediately upstream of flgR, and their coding sequences overlap by 27 nucleotides. The arrowhead indicates the location of the solo transposon in strain 81-176 flgP::solo (DRH241). The annotations of flgP and flgQ in the genomic sequences of C. jejuni 11168 and 81-176, respectively, are Cj1026c and Cjj81176_1045 for flgP and Cj1025c and Cjj81176_1044 for flgQ. (B) Immunoblot analysis of FlgR production in C. jejuni flgP and flgQ mutants. Whole-cell lysates of wild-type (wt) 81-176 Smr (DRH212), 81-176 Smr ΔflgR (DRH737), 81-176 flgP::solo (DRH241), 81-176 Smr ΔflgP (DRH2070), and 81-176 Smr ΔflgQ (DRH2071) were used for analysis.
Due to the location of the solo transposon and the organization of flgP, flgQ, and flgR on the C. jejuni chromosome, it is possible that the motility defect of the solo transposon mutant is due to either interruption of flgP or polar effects on expression of downstream genes such as flgQ or flgR. Immunoblot analysis with antiserum against FlgR revealed that the original transposon mutant (81-176 flgP::solo) produces normal levels of FlgR compared to the wild-type strain 81-176 Smr (DRH212), demonstrating that the solo insertion in flgP does not cause a defect in FlgR production (Fig. 1B).
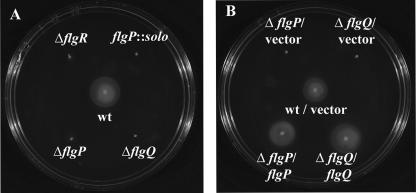
We then constructed mutants of 81-176 Smr with in-frame deletions of large portions of flgP or flgQ to determine if either one of these genes is required for motility. To construct a ΔflgP mutant, we deleted from the chromosome of 81-176 Smr codons 2 to 149, fusing the start codon of flgP to the last 23 codons. For the ΔflgQ mutant, we fused the first 10 codons of flgQ to the stop codon, deleting the intervening 139 codons. Deletion of either of these genes does not affect levels of FlgR (Fig. 1B), demonstrating that the deletions in flgP and flgQ that we created do not have polar effects on FlgR production. Analysis of the ΔflgP and ΔflgQ mutants in semisolid motility agar revealed that both mutants display a nonmotile phenotype (Fig. 2A), similar to that of a ΔflgR mutant. Complementation of each mutant with a plasmid expressing only the respective gene deleted from each strain restores the motility phenotype to wild-type levels (Fig. 2B), confirming that flgP and flgQ are both required for motility. Since each flgP or flgQ mutant could be complemented with just the respective gene supplied in trans, these findings indicate that the flgP deletion construct is nonpolar with respect to flgQ and vice versa. Dark-field microscopy was also used to examine the motile phenotypes of individual bacterial mutants. Wild-type bacteria were observed to migrate rapidly across the field, whereas mutants lacking flgP or flgQ were unable to move (data not shown), suggesting that the flagella of these mutants may have a paralyzed phenotype. These results combined indicate that both flgP and flgQ are required for motility of C. jejuni.
FIG. 2.
FlgP and FlgQ are required for flagellar motility of C. jejuni. Strains were inoculated into MH motility agar. Motility was assessed 32 h after inoculation. Strains are (A) wild-type (wt) 81-176 Smr (DRH212), 81-176 Smr ΔflgR (DRH737), 81-176 flgP::solo (DRH241), 81-176 Smr ΔflgP (DRH2070), and 81-176 Smr ΔflgQ (DRH2071) and (B) wild-type 81-176 Smr/pECO102 (DRH837), 81-176 Smr ΔflgP/pECO102 (SMS304), 81-176 Smr ΔflgQ/pECO102 (SMS306), 81-176 Smr ΔflgP/pSMS265 (SMS308), and 81-176 Smr ΔflgQ/pSMS269 (SMS312). pECO102 is the empty vector control, and pSMS265 and pSMS269 are pECO102 derivatives that express flgP and flgQ, respectively.
Domain predictions and homologues of FlgP and FlgQ proteins.
flgP is predicted to encode a precursor protein of 171 amino acids with a size of 18.5 kDa containing an N-terminal signal peptide cleaved after amino acid 23. Alternatively, a lipoprotein sorting signal peptide contained in the first 16 residues is also predicted. PSI-BLAST analysis suggests that FlgP is encoded in the genomes of other Campylobacter species, with 59 to 84% identity and 73 to 91% similarity across the lengths of the homologous proteins of C. coli (CCO1093), C. lari (CLA1520), C. upsaliensis (CUP0109), and C. fetus (CFF8240_1669). FlgP is also homologous to proteins of Helicobacter pylori (JHP_0775 and HPAG1_0822) (1, 21) and Helicobacter hepaticus (HH0979) (24), with 36 to 38% identity and 77% similarity across approximately 77% of the lengths of the proteins. In addition, Wolinella succinogenes encodes a protein (WS1756) (2) with 42% identity and 66% similarity to FlgP. The only predicted motif found in FlgP by BLAST analysis is a DUF400 domain contained within amino acids 73 to 171 of the protein. However, the DUF400 domain is undefined, with no known function. This domain is present in a series of functionally uncharacterized proteins of many other bacteria. As such, these bacterial proteins, which include the ones of pathogenic species of Vibrio, contain approximately 25% identity and 50% similarity to FlgP, but this homology is primarily limited to the DUF400 domains with little to no homology to the first 72 amino acids of FlgP.
flgQ is predicted to encode a precursor protein of 149 amino acids and 17.4 kDa with an N-terminal signal peptide predicted to be cleaved after the first 17 amino acids. PSI-BLAST analysis predicts only FlgQ homologues with 56 to 79% identity and 75 to 91% similarity to proteins of C. coli (CCO1092), C. lari (CLA1519), and C. upsaliensis (CUP0110). C. fetus possesses a FlgQ homologue (CFF8240_1668) with less homology (32% identity and 51% similarity). No other bacteria are predicted to have a protein with similarity to FlgQ.
FlgP and FlgQ are not required for flagellar biosynthesis or expression of σ54- or σ28-dependent flagellar genes.
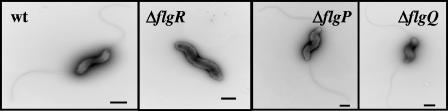
To determine the possible roles of FlgP and FlgQ in motility, we analyzed the ΔflgP and ΔflgQ mutants for defects in flagellar biosynthesis and transcription of known regulated flagellar genes. As could be determined from visualization of strains by transmission electron microscopy, ΔflgP and ΔflgQ mutants construct a single flagellum at one or both bacterial poles that is similar in length and appearance to those produced by a wild-type strain (Fig. 3). These phenotypes are different than those of a ΔflgR mutant, which fails to produce any flagella (Fig. 3).
FIG. 3.
FlgP and FlgQ are not required for flagellar biosynthesis. Bacteria were stained with 1% uranyl acetate. All electron micrographs are at a magnification of ×20,000. The bar for each micrograph represents 0.5 μm. Strains shown are wild-type (wt) 81-176 Smr (DRH212), 81-176 Smr ΔflgR (DRH737), 81-176 Smr ΔflgP (DRH2070), and 81-176 Smr ΔflgQ (DRH2071).
C. jejuni uses two alternative σ factors, σ28 and σ54, to transcribe many flagellar genes. Whereas σ28 is involved in the transcription of flaA, encoding the major flagellin, σ54 and FlgR are required for transcription of many genes encoding the basal body, rod, hook, and minor flagellin proteins (6, 13, 15, 16, 28). Through analysis of expression of flaA- and the σ54-dependent flagellar gene flgD-, flgE2-, and flaB-astA transcriptional fusions, we found that expression of these genes was not reduced in the ΔflgP or ΔflgQ mutants (Table 1). These results are in contrast to the defects in transcription of flaA in the σ28 mutant (ΔfliA) or of flgD, flgE2, and flaB in the σ54 (ΔrpoN) or flgR mutants. In fact, we observed a slight elevation of expression (less than two- to threefold) of flaA and flgE2 in mutants lacking flgP or flgQ.
TABLE 1.
Arylsulfatase activities of flaA-, flgD-, flgE2-, flaB-, and motA-astA transcriptional fusions in ΔflgP and ΔflgQ mutants
| Strain | Arylsulfatase unitsa (mean ± SD)
|
||||
|---|---|---|---|---|---|
| flaA::astAb | flgD::astAc | flgE2::astAd | flaB::astAe | motA::astAf | |
| Wild type | 133.2 ± 2.7 | 196.4 ± 7.3 | 33.5 ± 1.5 | 175.5 ± 6.8 | 174.6 ± 6.7 |
| ΔfliA | 64.5 ± 7.0 | NDg | ND | ND | 179.9 ± 13.6 |
| ΔrpoN | ND | 0.6 ± 0.1 | <0.1 ± 0.1h | <0.2 ± 0.1h | 185.9 ± 10.6 |
| ΔflgR | 139.1 ± 4.7 | <0.4 ± 0.3h | <0.1 ± 0.1h | 0.2 ± 0.1 | 181.5 ± 11.7 |
| ΔflgP | 340.6 ± 8.5 | 275.2 ± 20.5 | 78.5 ± 5.0 | 225.7 ± 4.1 | 189.7 ± 16.9 |
| ΔflgQ | 341.8 ± 15.3 | 275.5 ± 18.9 | 78.0 ± 1.0 | 245.5 ± 9.1 | 190.4 ± 7.7 |
Results are from a typical assay with determinations for each sample performed in triplicate. One unit equals the amount of arylsulfatase required to generate 1 nmol of nitrophenol per hour per OD600 unit.
Strains used were DRH655, DRH1070, DRH1122, SMS360, and SMS407.
Strains used were SMS354, SMS401, SNJ230, SNJ316, and SNJ322.
Strains used were DRH533, DRN536, DRH830, SMS357, and SMS404.
Strains used were DRH665, DRH667, DRH842, SMS364, SMS410.
Strains used were DRH2329, DRH2331, DRH2333, DRH2335, DRH2337, and DRH2342.
ND, not determined.
At least one of the three tested samples had a value below the limit of detection of the assay, which was 0.06 arylsulfatase unit.
Since the ΔflgP and ΔflgQ mutants are nonmotile but produce flagella, we examined the possibility that these mutants may be defective for expression of one or more of the genes required for production of the flagellar motor, such as motA and motB. These two genes are separated by two nucleotides, with motA 5′ to motB (22). This type of organization suggests that motA and motB may be in an operon and cotranscribed. We constructed a motA::astA transcriptional fusion to replace native motA on the chromosomes of ΔflgP and ΔflgQ mutants. Via arylsulfatase assays, we were unable to detect a defect in expression of motA in these mutants or any other mutants lacking σ28, σ54, or FlgR (Table 1). Thus, the flagellated, nonmotile phenotype of the ΔflgP or ΔflgQ mutants cannot be explained by a defect in expression of the motor component encoded by motA.
Considering that many flagellar genes in C. jejuni are under regulation by the σ54-dependent or σ28-dependent regulatory cascades, we explored whether expression of flgP or flgQ is a target of one of these cascades. flgP::astA and flgQ::astA transcriptional fusions were used to replace the native flgP or flgQ loci on the chromosomes of mutants lacking σ28 or σ54. Expression of flgP or flgQ is reduced only approximately two- to threefold, respectively, in the σ54 mutant (ΔrpoN) compared to wild-type strains (Table 2). Expression levels could be partially restored upon complementation of the ΔrpoN mutants with a plasmid expressing rpoN, suggesting that flgP and flgQ may be partially dependent either directly or indirectly on σ54 for expression.
TABLE 2.
Arylsulfatase activities of flgP- and flgQ-astA transcriptional fusions in σ28 and σ54 mutants
| Strain | Arylsulfatase unitsa (mean ± SD)
|
|
|---|---|---|
| flgP::astAb | flgQ::astAc | |
| Wild type | 282.7 ± 40.0 | 29.3 ± 1.1 |
| ΔfliA | 308.1 ± 22.9 | 29.0 ± 0.7 |
| ΔrpoN | 163.9 ± 1.5 | 10.6 ± 0.9 |
| ΔrpoN/vector | 155.6 ± 6.3 | 11.4 ± 0.8 |
| ΔrpoN/rpoN | 215.4 ± 4.2 | 19.1 ± 1.2 |
Results are from a typical assay with determinations for each sample performed in triplicate. One unit equals the amount of arylsulfatase required to generate 1 nmol of nitrophenol per hour per OD600 unit.
Strains used were SMS341, SMS350, SMS420, SMS524, and SMS526.
Strains used were SMS281, SMS323, SMS335, SMS532, and SMS534.
FlgP requires FlgQ for localization to or stability in the outer membrane of C. jejuni.
To better understand the roles of FlgP and FlgQ in flagellar motility, we constructed His6-tagged fusion proteins of FlgP and FlgQ for generation of polyclonal murine antisera to determine the locations of the proteins in C. jejuni. Whereas the generated antisera were reactive to the respective purified recombinant proteins, the α-FlgQ antiserum was not reactive to protein preparations from wild-type C. jejuni with normal FlgQ production (data not shown); thus, analysis of FlgQ localization in C. jejuni could not be done.
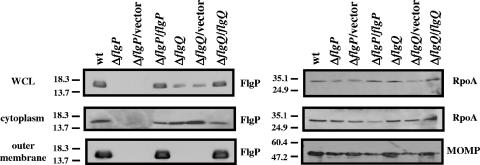
To determine the location of FlgP in C. jejuni, we separated wild-type and mutant strains of C. jejuni into fractions containing outer membrane, periplasmic, inner membrane, or cytoplasmic proteins. Immunoblot analysis revealed the presence of a protein of approximately 16 kDa in whole-cell lysates (total protein) of wild-type C. jejuni (Fig. 4). Examination of the cytoplasmic and outer membrane proteins derived from an equivalent number of wild-type bacteria revealed that more FlgP is located in the outer membrane than in the cytoplasm (Fig. 4). Thus, the outer membrane appears to represent the terminal destination for FlgP. The band corresponding to FlgP was absent in fractions from a ΔflgP mutant but could again be observed when the ΔflgP mutant was complemented with a plasmid expressing flgP in trans. No bands representing FlgP were found in the periplasm or inner membrane (data not shown). To ensure that our fractionation procedures were appropriate and represented the proper subcellular compartments, immunoblot analyses were performed with antiserum against RpoA, a component of the cytoplasmic RNA polymerase, or MOMP of C. jejuni (30). As expected, RpoA was found in the whole-cell lysates and cytoplasm, whereas the outer membrane contained a protein of 48 kDa representing MOMP. In addition, these markers served as loading controls to ensure that proteins from equivalent amounts of bacteria were analyzed for each sample.
FIG. 4.
FlgP requires FlgQ for localization to or stability in the outer membrane. Proteins from whole-cell lysates representing 200 μl of bacterial culture or proteins from outer membrane preparations or cytoplasmic preparations representing 750 μl of bacterial culture were used for immunoblotting. Blots were developed with α-FlgP mouse antiserum M1, α-RpoA mouse antiserum M1, or α-MOMP rabbit antiserum (30). Blots on the left are detecting FlgP. The top and middle blots on the right are detecting RpoA, whereas the bottom blot is detecting MOMP. Strains are wild-type (wt) 81-176 Smr (DRH212), 81-176 Smr ΔflgP (DRH2070), 81-176 Smr ΔflgP/pECO102 (SMS304), 81-176 Smr ΔflgP/pSMS265 (SMS308), 81-176 Smr ΔflgQ (DRH2071), 81-176 Smr ΔflgQ/pECO102 (SMS306), and 81-176 Smr ΔflgQ/pSMS269 (SMS312). pECO102 is the empty vector control, and pSMS265 and pSMS269 are pECO102 derivatives that express flgP and flgQ, respectively. WCL, whole-cell lysates.
We then determined if the amount or location of FlgP was affected in the ΔflgQ mutant. Analysis of the ΔflgQ mutant revealed that the amount of FlgP from whole-cell lysates was noticeably decreased compared to that in wild-type bacteria (Fig. 4). Further analysis revealed that roughly equivalent levels of FlgP are present in the cytoplasms of the ΔflgQ mutant and the wild-type strain. However, no FlgP is present in the outer membrane of the ΔflgQ mutant. Wild-type levels of FlgP in the whole-cell lysates and outer membrane are restored when the ΔflgQ mutant is complemented with a plasmid expressing flgQ. These results suggest that FlgQ may be required for localizing FlgP to the outer membrane and perhaps maintaining its stability at this location. We also investigated whether FlgP accumulates in the periplasm of the ΔflgQ mutant, but we could not detect FlgP in this compartment in wild-type C. jejuni or any mutant (data not shown). Since the ΔflgQ mutants have less FlgP in the whole-cell lysates, a reasonable hypothesis is that FlgQ may be required for transcription or translation of FlgP. However, the cytoplasms of the ΔflgQ mutants have levels of FlgP similar to those of the wild-type strain, but the outer membranes of the flgQ mutants lack FlgP. The most likely possibility for the decrease in total FlgP in the whole-cell lysates is due to the failure of FlgP to localize to and be stable in the outer membrane in the absence of FlgQ. Thus, FlgQ is unlikely to influence the transcription of flgP or its initial production. To ensure that FlgQ is not required for expression of flgP, flgP::astA transcriptional fusions were analyzed in the wild-type and ΔflgQ strains. No differences in the levels of arylsulfatase were observed (data not shown), indicating that flgQ is not required for expression of flgP.
DISCUSSION
In this work, we have identified two additional proteins of C. jejuni required for flagellar motility. Deletion of either flgP or flgQ from C. jejuni resulted in mutants that produce a single flagellum of normal appearance at the poles of the bacterium. These mutants, however, are nonmotile, as they are unable to migrate in motility agar or swim as visualized by dark-field microscopy, indicating that the lack of FlgP or FlgQ in C. jejuni may create paralyzed flagella. Localization studies revealed that the terminal location of FlgP is the outer membrane and that FlgQ is required for localizing FlgP to the outer membrane and possibly stabilizing FlgP at this location. These results suggest that FlgP may play a direct role in motility, whereas the requirement of FlgQ may be indirect in aiding FlgP to localize or be stable at or near the flagellar structure so that it can function in motility. Considering our findings, our leading hypothesis for the role of FlgP in motility is that it may be an outer membrane protein required for the actual process of flagellar rotation.
It is currently unclear how FlgP may contribute to flagellar rotation if the lack of these proteins does indeed cause a paralyzed flagellar phenotype. Visualization of individual bacteria by dark-field microscopy gives credence to this phenotype, since individual bacteria lacking flgP are unable to move. One possible role for FlgP is that it could be required for stabilization of parts of the flagellar motor complex, such as the MotA or MotB stators. Whereas MotA is an integral membrane protein, MotB is localized to the inner membrane with a large periplasmic domain (4), which could conceivably interact with an outer membrane-localized FlgP protein if it is oriented appropriately at this location. Future experiments will be directed at analyzing the fate of MotA and MotB in the presence and absence of FlgP.
Since our immunoblot analyses were performed by examining cytoplasmic or outer membrane proteins extracted from the same amount of bacteria, we were able to determine that more FlgP is located in the outer membrane than in the cytoplasm in wild-type bacteria. In addition, we were also able to determine that the levels of FlgP in the cytoplasms of the strains are similar in wild-type and ΔflgQ mutant strains, but no FlgP was found associated with the outer membrane of the ΔflgQ mutant. Thus, we hypothesize that the outer membrane is the terminal destination of FlgP and that FlgQ is required to localize FlgP to this compartment. For this to occur, FlgQ may interact with FlgP at some point in the secretion, localization, or stabilization of FlgP. Thus, FlgQ may have some type of chaperone function for FlgP. In the absence of FlgQ and proper localization, FlgP may be highly unstable and degraded in the periplasm; we were unable to detect FlgP in the periplasms of the wild-type and ΔflgQ mutant strains. Like FlgP, FlgQ is also predicted to have an N-terminal signal sequence, suggesting that it could be secreted across the inner membrane as well. However, we were unable to monitor the localization of FlgQ in C. jejuni since the antisera we generated did not recognize the native protein. Once in the periplasm, FlgQ may be able to interact with FlgP to target it to the outer membrane, possibly close to the flagellar basal body and rod complex. If FlgQ possesses a chaperone function for FlgP, FlgQ may form a physical interaction with FlgP and we may be able to capture interacting complexes. Thus, future experiments will be focused on developing methods to detect FlgQ and performing experiments with C. jejuni or with a heterologous system such as E. coli to attempt to detect a complex of FlgP and FlgQ by a combination of cross-linking and immunoprecipitation experiments or two-hybrid experiments.
We do not favor the hypothesis that FlgQ is required for the actual secretion of FlgP out of the cytoplasm for two reasons. First, FlgP is predicted to possess an N-terminal signal sequence or lipoprotein sorting signal; each would presumably function to transport the protein via the Sec system across the inner membrane, independent of FlgQ. Second, if FlgQ is required for secretion of FlgP, we would have expected to see an accumulation of FlgP in the cytoplasm of a ΔflgQ mutant. Instead, similar levels of FlgP were found in the cytoplasms of wild-type and ΔflgQ mutant strains. However, we cannot entirely exclude the possibility that FlgQ could be required for secretion of FlgP. A series of pulse-chase experiments in combination with induction of FlgP and FlgQ proteins would allow us to follow the fate of FlgP in the presence and absence of FlgQ to better analyze a possible role for FlgQ in FlgP transport, localization, and stability. However, these types of experiments require induction strategies that are currently unavailable for C. jejuni; we are actively pursuing the development of such technologies.
Further analysis will also allow us to more precisely determine whether FlgP is located on the surface of C. jejuni or associated with the periplasmic face of the outer membrane. Currently, we know only that FlgP is associated with the outer membrane. FlgP is predicted to have an N-terminal lipoprotein sorting signal, which may indicate that the protein could be associated with the periplasmic face of the outer membrane. We attempted to localize FlgP to the outer surface of C. jejuni by using whole-cell dot immunoblotting and immunogold labeling with transmission electron microscopy, but the results were inconclusive. These methods could have been limited by the α-FlgP antiserum that is currently available. Since Kyte-Doolittle analysis predicts a strong hydrophobic region only at the N terminus of FlgP where the predicted N-terminal signal sequence or lipoprotein sorting sequence is located, we do not expect that FlgP is an integral outer membrane protein. Additional studies with site-directed mutagenesis of flgP and improved reagents and methods will be required to better determine the nature of the association of FlgP with the outer membrane.
Considering our findings with C. jejuni, the role of FlgP in motility of other bacteria is unclear. Both FlgP and FlgQ are required for motility in C. jejuni, and we suspect that the same will be true in other Campylobacter species, as they contain genes to encode homologues of both proteins. However, other bacteria such as Helicobacter species and W. succinogenes contain a highly similar FlgP homologue but lack a gene encoding FlgQ. Meanwhile, other bacteria appear to encode a protein homologous only to the portion of FlgP that contains the uncharacterized DUF400 domain (composing the last 60% of FlgP), while also lacking a FlgQ homologue. Since FlgQ is required for motility in C. jejuni and appears to be necessary for proper levels and localization of FlgP, it is unclear whether FlgP-like proteins of other bacteria would function in motility without a cognate FlgQ protein. Alternatively, these other bacteria may have FlgP-like proteins that are independent of FlgQ or are partnered with an unknown protein that can substitute for FlgQ to promote proper localization and function of FlgP. Determining if these FlgP-like proteins of other bacteria function in motility warrants further investigation.
In summary, we have identified a pair of proteins required for flagellar motility that may be unique to C. jejuni and other campylobacters. Further characterization of these proteins may reveal a better understanding of the role of FlgQ in the localization and stability of FlgP and the role of FlgP in motility. In addition, analysis of these proteins may reveal new insights regarding membrane proteins of bacteria that are possibly involved in the physical rotation of flagella. Due to the uniqueness of these two proteins in Campylobacter species, further analysis of these proteins may reveal new understandings in protein-protein interactions required for the process of motility in campylobacters.
Acknowledgments
We thank Kayla Hagman for assistance with dark-field microscopy and Laurie Mueller of the University of Texas Southwestern Medical Center Molecular and Cellular Imaging Facility for assistance with transmission electron microscopy. We thank Stephanie Joslin for construction of C. jejuni derivatives SNJ230, SNJ316, and SNJ322. The MOMP antibody was kindly provided by Qijing Zhang.
This work was supported by the U.S. Department of Agriculture National Research Initiative Grants Program, CSREES project 2006-35201-17382. This work was also supported by a Distinguished Young Investigator Award from the President's Research Council of the University of Texas Southwestern Medical Center and start-up funds from the University of Texas Southwestern Medical Center.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 4.Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. [DOI] [PubMed] [Google Scholar]
- 5.Carlone, G. M., M. L. Thomas, H. S. Rumschlag, and F. O. Sottnek. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—ten States, United States, 2005. Morb. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 8.Donohue-Rolfe, A., and G. T. Keusch. 1983. Shigella dysenteriae 1 cytotoxin: periplasmic protein releasable by polymyxin B and osmotic shock. Infect. Immun. 39:270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman, C. R., R. M. Hoekstra, M. Samuel, R. Marcus, J. Bender, B. Shiferaw, S. Reddy, S. D. Ahuja, D. L. Helfrick, F. Hardnett, M. Carter, B. Anderson, and R. V. Tauxe. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin. Infect. Dis. 38(Suppl. 3):S285-S296. [DOI] [PubMed] [Google Scholar]
- 10.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, M. J., and F. H. Milazzo. 1979. Arylsulfatase in Salmonella typhimurium: detection and influence of carbon source and tyramine on its synthesis. J. Bacteriol. 139:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 13.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 14.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 16.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 18.Lindblom, G.-B., E. Sjörgren, and B. Kaijser. 1986. Natural campylobacter colonization in chickens raised under different environmental conditions. J. Hyg. (London) 96:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29:970-972. [DOI] [PubMed] [Google Scholar]
- 20.Nachamkin, I., X.-H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA 103:9999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 23.Pokamunski, S., N. Kass, E. Borochovich, B. Marantz, and M. Rogol. 1986. Incidence of Campylobacter spp. in broiler flocks monitored from hatching to slaughter. Avian Pathol. 15:83-92. [DOI] [PubMed] [Google Scholar]
- 24.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Dröge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. König, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Vliet, A. H. M., A. C. Wood, J. Henderson, K. Wooldridge, and J. M. Ketley. 1997. Genetic manipulation of enteric Campylobacter species. Methods Microbiol. 27:407-419. [Google Scholar]
- 26.Wassenaar, T. M., B. A. M. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 27.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wösten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 29.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]