Abstract
Neotrehalosadiamine (3,3′-diamino-3,3′-dideoxy-α,β-trehalose; NTD) is an amino-sugar antibiotic produced by several Bacillus species that functions as an autoinducer by activating its own biosynthetic operon, ntdABC. We previously reported that the introduction of a certain rpoB mutation (rpoB5) into Bacillus subtilis enables the cells to overproduce NTD. B. subtilis mini-Tn10 transposant libraries have been screened for genes that affect NTD production. Inactivation of ccpA, which encodes a major transcriptional regulator of carbon catabolite regulation, markedly reduced NTD production. By contrast, inactivation of glcP, which is situated just downstream of ntdABC and encodes a glucose/mannose:H+ symport permease, stimulated NTD production. Overexpression of glcP led to the repression of ntdABC expression (and thus NTD production) in response to GlcP-mediated glucose uptake. These results suggest that CcpA-mediated catabolite activation of ntdABC expression occurs in response to the increase of the in vivo concentration of fructose-1,6-bisphosphate via glucose-6-phosphate and that GlcP-mediated glucose repression of ntdABC expression occurs in association with the increase of the in vivo concentration of unphosphorylated glucose. In addition, Northern analysis showed that glcP is transcribed from the ntdABC promoter through transcription readthrough at the ntdABC transcription terminator site, which enables NTD to function as a modulator of glucose uptake through the stimulation of ntdABC-glcP transcription, even in wild-type (rpoB+) cells. A trace amount (0.5 to 3 μg/ml) of NTD was sufficient to ensure expression of glcP, thus demonstrating the physiological role of “antibiotic” in the producing bacteria by functioning as an autoinducer for glucose uptake modulation.
Carbon catabolite regulation (involving repression and activation) is a ubiquitous phenomenon whereby bacteria alter the expression of a large number of genes in response to the availability of carbon sources in their surroundings and is dependent upon the organism's ability to take up sugar. Bacillus subtilis has at least three different pathways for glucose uptake. The first is the glucose-specific phosphoenolpyruvate-dependent phosphotransferase system (glucose-PTS), which is widespread among bacteria and plays an essential role in both the transport and the phosphorylation of glucose (31, 32). Encoded by the ptsGHI operon, glucose-PTS is the predominant glucose uptake pathway of B. subtilis and promotes carbon catabolite regulation mediated by a master transcriptional regulator, CcpA (4). The remaining two pathways are the non-PTS-type transporters GlcP (30) and GlcU (7). GlcP is a proton-dependent glucose/mannose symport permease and a member of the major facilitator superfamily (29). Paulsen et al. (30) showed that cells lacking functional GlcP import 30% less glucose than wild-type cells and that GlcP contributes partially to glucose-promoted catabolite repression. The gene encoding GlcU and a downstream gdh gene encoding glucose dehydrogenase form an operon that is transcribed in forespores after the onset of sporulation (26). Thus, GlcU probably functions in forespores during sporulation or in germinating spores rather than in vegetative cells. In contrast to PTS-mediated sugar uptake, which has been extensively studied, little is known about the regulation of GlcP and GlcU.
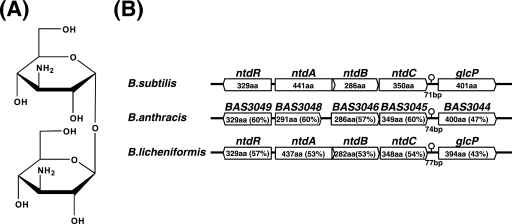
glcP, the gene encoding GlcP, is located just downstream of an operon consisting of three genes, ntdA, ntdB, and ntdC (formerly yhjL, yhjK, and yhjJ, respectively). We previously used heterologous expression in Escherichia coli to establish that ntdABC encodes all the enzymes required for the biosynthesis of an antibiotic, neotrehalosadiamine (NTD; 3,3′-diamino-3,3′-dideoxy-α,β-trehalose) (Fig. 1A) (18). Although B. subtilis normally produces lower-than-detectable levels of NTD, we were able to induce overproduction of NTD in B. subtilis by introducing a mutation into RNA polymerase (RNAP) and selecting for the rifampin (Rif)-resistant phenotype (18). Interestingly, transcriptional analysis using the ntdABC promoter (PntdABC) fused to lacZ revealed that the inactivation of the NTD biosynthesis operon shuts off its own promoter. Conversely, PntdABC was dose-dependently activated by the addition of purified NTD. A transcriptional regulator, NtdR, whose gene is situated upstream of the ntdABC operon in the opposite orientation, mediates the NTD-dependent activation of PntdABC. Thus, NTD appears to function as an autoinducer, at least for its own biosynthetic operon. Still, the mechanism by which ntdABC expression is regulated is not fully understood.
FIG. 1.
Structure of NTD and organization of the NTD biosynthesis operon in Bacillus species. (A) Chemical structure of NTD. (B) The ntdABC operon region of B. subtilis is compared with those of B. anthracis and B. licheniformis. Amino acid lengths or amino acid identities (indicated as percentages in parentheses) with the corresponding B. subtilis proteins (for B. anthracis and B. licheniformis) are shown. The BAS3048 gene of B. anthracis has a frameshift mutation (possibly by 1 base deletion), which resulted in a protein product smaller than the B. subtilis NtdA.
Autoinduction has been extensively studied in several bacteria in relation to quorum-sensing systems, which are important for various physiological processes, including acquisition of competence, sporulation, antibiotic production, motility, biofilm formation, bioluminescence, and virulence (3, 6, 9, 10, 19, 20, 22, 33). Whereas in gram-positive bacteria, most of these signaling molecules are peptides or modified peptides (6, 19, 20), NTD is unique in that it is an amino-sugar. NTD was originally identified as an antibiotic produced by Bacillus pumilus (36) and Bacillus circulans (27) that inhibits growth of Staphylococcus aureus and Klebsiella pneumoniae. In addition, genome sequencing revealed that Bacillus anthracis and Bacillus licheniformis contain orthologues for a complete set of NTD biosynthetic enzymes and its transcriptional regulator, NtdR (Fig. 1B). This suggests that NTD is used as a common “language” between B. subtilis and its close relatives. Here we present evidence that glucose can exert alternative effects on ntdABC gene expression in B. subtilis, depending upon the organism's uptake pathway. The significance of NTD as a novel signaling molecule that modulates glucose metabolism is discussed.
MATERIALS AND METHODS
Bacterial strains and their construction.
B. subtilis strains used in this study are listed in Table 1. All strains except JCM1465 and 168 were derived from strain 61884 (trpC2 aspB66) (28). Strain GM273 (ΔptsGHI::erm) was provided by J. Deutscher, and QB7103 (crh::aphA3 ptsH1) was kindly provided by I. Martin-Verstraete via J. Deutscher.
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Construction, reference, or sourcea |
|---|---|---|
| JCM1465 | Prototroph (type species for Bacillus subtilis) | JCMb |
| 168 | trpC2 (Marburg strain) | Laboratory stock |
| QB7103 | trpC2 crh::aphA3 ptsH1 amyE::(PΔB levD′-lacZ cat) | 11 |
| GM273 | trpC2ΔptsGHI::erm | 5 |
| 61884 | trpC2 aspB66 | 28 |
| 84R5 | trpC2 aspB66 rpoB5 | 18 |
| 84R2 | trpC2 aspB66 rpoB2 | 18 |
| 84R6 | trpC2 aspB66 rpoB6 | 18 |
| 84R32 | trpC2 aspB66 rpoB32 | 18 |
| 84R5-UP1 | trpC2 aspB66 rpoB5 glcP::Tn10 | This study |
| 84R5-DN1 | trpC2 aspB66 rpoB5 ymfI::Tn10 | This study |
| 84R5-DN2 | trpC2 aspB66 rpoB5 ccpA::Tn10 | This study |
| TI122 | trpC2 aspB66 amyE::(PntdABC-lacZ cat) | 18 |
| TI122-UP1 | trpC2 aspB66 glcP::Tn10 amyE::(PntdABC-lacZ cat) | 84R5-UP1→TI122 |
| TI122R5 | trpC2 aspB66 rpoB5 amyE::(PntdABC-lacZ cat) | 18 |
| TI122R5-UP1 | trpC2 aspB66 rpoB5 glcP::Tn10 amyE::(PntdABC-lacZ cat) | 84R5-UP1→TI122R5 |
| TI122R5-DN1 | trpC2 aspB66 rpoB5 ymfI::Tn10 amyE::(PntdABC-lacZ cat) | 84R5-DN1→TI122R5 |
| TI122R5-DN2 | trpC2 aspB66 rpoB5 ccpA::Tn10 amyE::(PntdABC-lacZ cat) | 84R5-DN2→TI122R5 |
| TI127R5 | trpC2 aspB66 rpoB5 ntdR::neo amyE::(PntdABC-lacZ cat) | 18 |
| TI130 | trpC2 aspB66 ntdA::Tn10 amyE::(PntdABC-lacZ cat) | TI130R5→TI122 |
| TI130R5 | trpC2 aspB66 rpoB5 ntdA::Tn10 amyE::(PntdABC-lacZ cat) | 18 |
| TI138 | trpC2 aspB66 ΔntdABC::cat | pUC18-ΔntdABC::cat →61884 |
| TI167 | trpC2 aspB66 ntdC-pMutinT3-TntdABC | pMutinT3-ntdC→61884 |
| TI167R5 | trpC2 aspB66 rpoB5 ntdC-pMutinT3-TntdABC | TI167→84R5 |
| TI168 | trpC2 aspB66 TntdABC-pMutinT3-glcP | pMutinT3-glcP→61884 |
| TI168R5 | trpC2 aspB66 rpoB5 TntdABC-pMutinT3-glcP | TI168→84R5 |
| TI168Δ4 | trpC2 aspB66 TntdABC-pMutinT3-ΔglcP | pMutinT3-ΔglcP→61884 |
| TI168R5Δ4 | trpC2 aspB66 rpoB5 TntdABC-pMutinT3-ΔglcP | TI168Δ4→84R5 |
| TI171 | trpC2 aspB66 ΔntdABC::cat-pMutinT3-TntdABC | pMutinT3-ntdC→TI138 |
| TI171R5 | trpC2 aspB66 rpoB5 ΔntdABC::cat-pMutinT3-TntdABC | 84R5→TI171 |
| TI171R2 | trpC2 aspB66 rpoB2 ΔntdABC::cat-pMutinT3-TntdABC | 84R2→TI171 |
| TI171R6 | trpC2 aspB66 rpoB6 ΔntdABC::cat-pMutinT3-TntdABC | 84R6→TI171 |
| TI171R32 | trpC2 aspB66 rpoB32 ΔntdABC::cat-pMutinT3-TntdABC | 84R32→TI171 |
| TI172 | trpC2 aspB66 ΔntdABC::cat-TntdABC-pMutinT3-glcP | pMutinT3-glcP→TI138 |
| TI172R5 | trpC2 aspB66 rpoB5 ΔntdABC::cat-TntdABC-pMutinT3-glcP | 84R5→TI172 |
| TI172R2 | trpC2 aspB66 rpoB2 ΔntdABC::cat-TntdABC-pMutinT3-glcP | 84R2→TI172 |
| TI172R6 | trpC2 aspB66 rpoB6 ΔntdABC::cat-TntdABC-pMutinT3-glcP | 84R6→TI172 |
| TI172R32 | trpC2 aspB66 rpoB32 ΔntdABC::cat-TntdABC-pMutinT3-glcP | 84R32→TI172 |
| TI179 | trpC2 aspB66 ΔptsGHI::erm amyE::(PntdABC-lacZ cat) | GM273→TI122 |
| TI179R5 | trpC2 aspB66 rpoB5 ΔptsGHI::erm amyE::(PntdABC-lacZ cat) | TI179→TI122R5 |
| TI184R5 | trpC2 aspB66 rpoB5 crh::aphA3 amyE::(PntdABC-lacZ cat) | QB7103→TI122R5 |
| TI185R5 | trpC2 aspB66 rpoB5 crh::aphA3 ptsH1 amyE::(PntdABC-lacZ cat) | QB7103→TI179R5 |
| TI191R5 | trpC2 aspB66 rpoB5 ntdR::neo glcP::Tn10 amyE::(PntdABC-lacZ cat) | TI127R5→TI122R5-UP1 |
| TI196R5 | trpC2 aspB66 rpoB5 ptsH1 amyE::(PntdABC-lacZ cat) | TI185R5→TI179 |
Arrows indicate construction by transformation (with congression in some cases).
JCM, Japan Collection of Microorganisms.
Strain TI138 (ΔntdABC::cat) was constructed as follows. The 5′ fragment of ntdA (501 bp) and the 3′ fragment of ntdC (554 bp) were amplified by PCR with the primers HindIII-ntdAF (5′-ccaagcttattggaggtactgttcATGCA-3′) and Sse8387I-ntdAR (5′-cctgcaggTAATAGCATCGGTTCCACTG-3′) for ntdA, and Sse8387I-ntdCF (5′-cctgcagGTAAGAAGACGAGTGGATCAT-3′) and EcoRI-ntdCR (5′-ggaattcCTAGTTGACTGCTGAAACAGACAT-3′) for ntdC. Capital letters indicate the coding region of each gene, while the underlined sequences are the restriction sites for HindIII, Sse8387I, or EcoRI, used for cloning the resulting PCR product; lowercase letters indicate the noncoding region. The amplified DNAs were cloned into pCR2.1 (Invitrogen), generating pCR2.1-ΔntdA and pCR2.1-ΔntdC. The HindIII-EcoRI fragment derived from pCR2.1-ΔntdA was subcloned into the corresponding region of pUC18, resulting in pUC18-ΔntdA. The Sse8387I-EcoRI fragment derived from pCR2.1-ΔntdC was inserted into the corresponding region of pUC18-ΔntdA, generating pUC18-ΔntdABC. An Sse8387I fragment of the cat gene from pCR2.1-cat (15) was inserted at the Sse8387I site of pUC18-ΔntdABC. The resulting plasmid, pUC18-ΔntdABC::cat, was linearized with EcoRI and used in the transformation of B. subtilis 61884.
Strains TI167 (ntdC-pMutinT3-TntdABC), TI168 (TntdABC-pMutinT3-glcP) and TI168Δ4 (TntdABC-pMutinT3-ΔglcP) were constructed by integrating plasmid pMutinT3 (25) upstream or downstream of the ntdABC transcriptional terminator (TntdABC). A DNA fragment containing the C-terminal coding region of ntdC or the N-terminal coding region of glcP was amplified by PCR with the following specific primer pairs: HindIII-ntdCF (5′-ctctaagcttCTGTGGTTTTAGCATTGAGG-3′) and BamHI-ntdCR (5′-cgggatcctaGTTGACTGCTGAAACAGAC-3′) for ntdC, and HindIII-glcPF (5′-ctctaagcttataggggtgtaatgaATGTTAA-3′) and BamHI-glcPR (5′-cgggatccAACAAATGAACCAACTGTCG-3′) for glcP. For construction of strain TI168Δ4, HindIII-ΔglcPF (5′-ctctaagcttataggggtgtaatgaATGTAA-3′) was used instead of HindIII-glcPF for PCR. The capital letters indicate the coding region of each gene, while the underlined sequences are the restriction sites for HindIII or BamHI used for cloning the resulting PCR product. The amplified DNAs were cloned into pCR2.1, generating pCR2.1-ntdC, pCR2.1-glcP, and pCR2.1-ΔglcP, respectively, after which the cloned fragments were fully sequenced to confirm their correctness. The HindIII-BamHI fragments were inserted into the corresponding region of pMutinT3, generating pMutinT3-ntdC, pMutinT3-glcP, and pMutinT3-ΔglcP, respectively, which were used to transform B. subtilis 61884, after which strains TI167, TI168, and TI168Δ4 were selected on L agar plates based on erythromycin (Erm) resistance. To generate strains TI171 and TI172, strain TI138 (ΔntdABC::cat) was transformed with the plasmids pMutinT3-ntdC and pMutinT3-glcP, respectively.
Growth conditions.
B. subtilis strains were grown overnight on L agar plates (17) at 30°C, after which they were inoculated into S7N medium (17) and incubated at 37°C. Both media were supplemented as required with tryptophan (Trp; 50 μg/ml) and aspartate (Asp; 2 mM for L medium or 20 mM for S7N medium). S7N medium contained 1% glucose, if otherwise not stated. Purified NTD (prepared in our laboratory) was used at appropriate concentrations for the induction of ntdABC. Isopropyl-β-d-thiogalactopyranoside (IPTG; 2 mM) was used for the activation of the spac promoter (Pspac). Rif (1 μg/ml), Erm (0.5 μg/ml), spectinomycin (Spc; 100 μg/ml), kanamycin (5 μg/ml), and chloramphenicol (Cm; 5 μg/ml) were used for the selection of B. subtilis transformants. Ampicillin (100 μg/ml) was used for the selection of E. coli transformants.
NTD assay.
Cells were grown in S7N medium for 24 h, as described above, after which the cultures were centrifuged and the resultant supernatants were appropriately diluted, and a 50-μl sample was applied to a paper disk (8-mm diameter; Advantec). The paper disk was then placed on an NTD assay plate (S7N soft agar [0.7%] supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactoside [X-Gal], Spc, Cm, Trp, and Asp) inoculated with the B. subtilis indicator strain TI130R5 (rpoB5 ntdA::Tn10 amyE::PntdABC-lacZ), which lacks the ability to synthesize NTD because of the presence of a transposon inserted within ntdA. After incubation at 37°C for 24 h, the diameter of the blue zone (representing the NTD activity) was measured. Purified NTD (>95% purity) was used as the standard, where linearity of the standard curve was found in the concentration range of 5 to 100 μg/ml NTD.
Transposon mutagenesis.
For transposon mutagenesis, the temperature-sensitive mini-Tn10-containing plasmid pIC333 was used as described by Steinmetz and Richter (35). Plasmid pIC333 was introduced into B. subtilis 84R5 at 28°C, after which transformants were selected for Erm resistance. The transformant colonies were then used to inoculate 20 independent cultures (1 ml each) of L medium containing Spc. During the exponential phase of growth (optical density at 650 nm [OD650], 0.5), the temperature was shifted from 28°C to 42°C, and the incubation was continued for an additional 4 h. Appropriate dilutions of these cultures were then spread onto L agar plates containing Spc. We selected colonies that are resistant to Spc but sensitive to Erm. The mutants with altered NTD production were identified using the NTD assay plate (see above) after the isolates were cultivated for 24 h in S7N medium containing 1% glucose. The chromosomal region targeted by the transposon was rescued as follows. Chromosomal DNA from each transposant was prepared and digested with EcoRI or HindIII. The mini-Tn10 transposon-inserted gene, along with its flanking region, was cloned into E. coli JM109 by self-religation and sequenced using primers Tn10-F (5′-GCCGCGTTGGCCGATTC-3′) and Tn10-R (5′-GATATTCACGGTTTAC-3′).
Northern blot analysis.
Cells grown on L agar plate were inoculated into S7N medium, cultured at 37°C with vigorous shaking, and harvested at the indicated times, and total cellular RNA was prepared using Isogen reagent (Nippon Gene) according to the protocols provided by the manufacturer. The RNAs were subjected to electrophoresis in the presence of formamide, transferred onto a membrane (Hybond-N+; Amersham), and hybridized with a digoxigenin-labeled RNA probe. For RNA dot blotting, RNAs were spotted directly onto a membrane. Transcription of the RNA probes was driven by the T7 promoter in the pCR2.1 vector. To prepare the template for the ntdB probe, the DNA fragment containing the full-length ntdB coding region was amplified by PCR using primers ntdBF (5′-ATGTTATTAAGCAAGAAATCGGAG-3′) and ntdBR (5′-TTATTTCCTCCTCATGAATCCA-3′). The amplified DNA was then cloned into pCR2.1 to generate pCR2.1-ntdB. Plasmid pCR2.1-ntdB together with pCR2.1-ntdC and pCR2.1-glcP (see above) served as a template for in vitro transcription. A digoxigenin RNA labeling kit (SP6/T7) was purchased from Roche Diagnostics and used for RNA labeling.
Assay of β-Gal activity.
Strains were grown aerobically in S7N medium, after which β-galactosidase (β-Gal) activity was measured as described previously (16). One unit is equivalent to 1,000 × A420/OD650/min, where A420 is the absorbance at 420 nm.
RESULTS
NTD biosynthesis is regulated by CcpA-mediated catabolite activation.
We previously showed that a single Ser487→Leu substitution (rpoB5 mutation) within the β subunit of RNAP causes a dramatic activation of NTD production in B. subtilis (18). We then carried out transposon mutagenesis of the B. subtilis rpoB5 mutant (strain 84R5) using the mini-Tn10 delivery plasmid pIC333 and identified the NTD biosynthesis operon. During the course of that transposon mutagenesis, three other insertion mutants were also found to be affected on NTD production. One mutant represented by strain 84R5-UP1 produced 3.5-fold more NTD than the 84R5 strain, while the other two mutants represented by strains 84R5-DN1 and 84R5-DN2, respectively, displayed dramatically reduced NTD production (Table 2). A backcross transformation revealed that the insertion mutation was directly responsible for the observed phenotype (data not shown), and the genes inactivated by the transposon insertion in each strain were identified through target rescue and sequencing (Table 2). The 84R5-UP1 mutant carried a mini-Tn10 insertion within the N-terminal coding region of glcP. The 84R5-DN1 mutant carried the insertion within ymfI, which encodes an uncharacterized protein with similarity to 3-oxoacyl-acyl carrier protein reductase, and the 84R5-DN2 mutant carried the insertion within ccpA, which encodes a major transcription factor mediating carbon catabolite regulation (12).
TABLE 2.
Characterization of genes that affect NTD production
| Strain | Tn10 inserted gene | Function or similarity | NTD titer (μg/ml)a |
|---|---|---|---|
| 61884 | NAb | NA | <5 |
| 84R5 | NA | NA | 200 |
| 84R5-UP1 | glcP | Glucose/mannose:H+ symporter | 700 |
| 84R5-DN1 | ymfI | Similar to 3-oxoacyl-acyl carrier protein reductase | <5 |
| 84R5-DN2 | ccpA | Catabolite control protein A | 5 |
B. subtilis strains were grown overnight on L agar plates at 30°C, after which they were inoculated into S7N medium containing excess (1%) glucose and incubated for 24 h at 37°C. NTD titers were determined by using paper disk agar diffusion assays with B. subtilis TI130R5 (rpoB5 ntdA::Tn10 amyE::PntdABC-lacZ) as the indicator strain (see Materials and Methods). NTD titers for strains 84R5 and 84R5-UP1 were determined after appropriate dilution of their cultured broth. Purified NTD (>95% purity) was used as the standard. At least two independent experiments were performed.
NA, not applicable.
To further analyze the effects of these mutations, each insertion was introduced into reporter strain TI122R5 carrying the ntdABC promoter (PntdABC) fused to lacZ at the amyE locus, after which the β-Gal activities in resultant strains were measured. In this way, we were able to study the possible effect of the mutations on the promoter activity for ntdABC expression. As shown in Table 3, the β-Gal activity in each mutant was in good agreement with the mutants' abilities to produce NTD. The CcpA-mediated catabolite regulation is widely believed to depend on either of two Ser46-phosphorylated proteins: the histidine-containing protein HPr, which is a component of PTS (1, 5, 8), or its paralogue, Crh (11). These Ser46-phosphorylated proteins (HPr and Crh) stimulate CcpA to bind catabolite-responsive element (cre). We therefore tested the effect of these mutations (ptsH and crh, which encode HPr and Crh proteins, respectively) on the expression of PntdABC-lacZ. Similar to the results obtained with a ccpA mutation, the double mutation of crh and ptsH (but not crh or ptsH mutation alone) diminished the expression of PntdABC-lacZ (Table 3), supporting the intrinsic role of CcpA in NTD production. Unlike the glcP mutant, a mutant lacking glucose-PTS (ΔptsGHI) also exhibited a lower expression level. The reduced expression of PntdABC-lacZ observed in all of these mutants was restored extensively by adding purified NTD (Table 3), which can be taken as an indication that ccpA and ymfI play a role, directly or indirectly, in the supply of substrate(s) for NTD biosynthesis, the synthesis of which is dependent on a glucose activation mechanism. In fact, as shown in Fig. 2, the expression of PntdABC-lacZ was induced at the stationary phase in a glucose-dependent manner; almost no induction was observed in wild-type strain TI122 under the glucose-limited (0.1%) condition. Further evidence supporting the above notion came from the fact that the addition of NTD (200 μg/ml) markedly stimulated ntdABC expression (reaching 103 U/OD650 in the rpoB+ strain), even in cells cultured with 0.1% glucose, demonstrating a bypass of the requirement of glucose for ntdABC expression in the presence of exogenous NTD (data not shown). On the other hand, the enhanced ntdABC expression observed in the glcP mutant appears to contradict the positive effect of glucose on NTD production, as mentioned above. We therefore further investigated the regulatory function of GlcP on ntdABC expression.
TABLE 3.
Expression of ntdABC-lacZ in various B. subtilis mutants
| Strain | Relevant genotype | PntdABC-lacZ (U/OD650)a
|
|
|---|---|---|---|
| −NTD | +NTD | ||
| TI122R5 | 330 ± 47 | 410 ± 74 | |
| TI122R5-UP1 | glcP::Tn10 | 440 ± 98 | 490 ± 130 |
| TI122R5-DN1 | ymfI::Tn10 | 23 ± 7.2 | 300 ± 8.5 |
| TI122R5-DN2 | ccpA::Tn10 | 48 ± 33 | 200 ± 92 |
| TI179R5 | ΔptsGHI | 150 ± 79 | 220 ± 18 |
| TI184R5 | crh::aphA3 | 270 ± 62 | 470 ± 14 |
| TI196R5 | ptsH1 | 170 ± 18 | 430 ± 33 |
| TI185R5 | crh::aphA3 ptsH1 | 26 ± 12 | 180 ± 82 |
Cells were grown for 15 h at 37°C in S7N medium containing excess (1%) glucose with or without 500 μg/ml NTD. β-Gal activity is expressed in units/cell optical density at 650 nm. The values are the means ± standard deviations of three or more experiments conducted independently.
FIG. 2.
Growth and transcription of PntdABC-lacZ in the B. subtilis wild type and rpoB5 mutant strains. Strains TI122 (wild type) (A) and TI122R5 (rpoB5 mutant) (B) were grown in the presence of limited (0.1%, open symbols) or excess (1%, closed symbols) glucose. Culture samples were withdrawn at the indicated times, and culture densities (OD650, circles) and β-Gal activities (squares) were measured. At least two independent experiments were performed, and one of them is shown.
glcP is cotranscribed with the NTD biosynthesis operon via transcriptional readthrough.
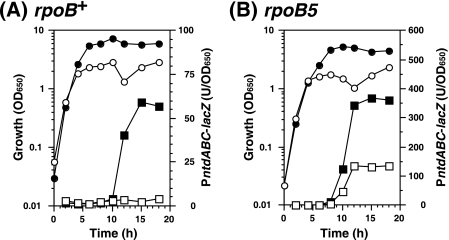
To analyze the role of GlcP during NTD production, we first compared the growth and expression of PntdABC-lacZ in the wild type with those in the glcP mutant strains using S7N medium which contained an excess amount (1%) of glucose (Fig. 3). We found that the glcP mutant grew somewhat slower than the parent strain, presumably due to the impaired glucose uptake. The glcP mutation resulted in a dramatic elevation of β-Gal activity in both the rpoB+ wild-type and the rpoB5 mutant strains. It is noteworthy that the elevation of β-Gal activity was detected even at the exponential growth phase, indicating that GlcP protein exists and functions throughout the entire growth phase. In addition, because NtdR is a transcriptional activator of ntdABC (18), we then measured the ntdR promoter (PntdR)-lacZ expression in the glcP mutant. We found that whereas the glcP mutation induced expression of PntdABC-lacZ, the expression of PntdR-lacZ in the glcP mutant was rather less than that in the glcP+ parental strain, especially during the late growth phase (data not shown). However, in a mutant (strain TI191R5) lacking functional NtdR, the effect of the glcP mutation on PntdABC-lacZ expression was no longer observed (data not shown). From these results, it is concluded that, although NtdR is an essential element for ntdABC expression even in the rpoB5 mutant or glcP mutant strain, a basal level of ntdR expression is sufficient to fully activate PntdABC.
FIG. 3.
Effect of glcP mutation on the expression of PntdABC-lacZ in B. subtilis wild-type and rpoB5 mutant strains. The expression of PntdABC-lacZ in B. subtilis rpoB+ wild-type (A) and rpoB5 mutant (B) strains is shown. Strains TI122 (rpoB+, squares), TI122-UP1 (rpoB+ glcP::Tn10, circles), TI122R5 (rpoB5, squares), and TI122R5-UP1 (rpoB5 glcP::Tn10, circles) were grown in S7N medium containing excess (1%) glucose. Culture samples were withdrawn at the indicated times, and culture densities (OD650, open symbols) and β-Gal activities (closed symbols) were measured. At least three independent experiments were performed, and one of them is shown.
Since no obvious promoters exist downstream of the ntdABC transcriptional terminator (TntdABC), we hypothesized that glcP may be cotranscribed along with ntdABC via transcription readthrough. To test this idea, we subjected the transcripts to Northern blotting using probes for ntdC and glcP (Fig. 4A). As expected, in rpoB5 mutant cells grown to mid-exponential phase (OD650, 0.5), the ntdABC transcript (3.2 kb) and a transcription readthrough product (4.5 kb) were both clearly detected using an ntdC probe (Fig. 4B, left panel). By contrast, when the glcP probe was used, only the readthrough product was detected (Fig. 4B, right panel). Likewise, these transcripts (ntdABC and ntdABC-glcP) were observed in transition (OD650, 1.5) and stationary growth phase (OD650, 8.0) (data not shown). These results indicate that PntdABC is the sole promoter of glcP expression. Although it was difficult to detect the ntdABC-glcP readthrough transcript in rpoB+ wild-type cells, the ntdABC transcript (though slight) was readily detectable, even during the exponential growth phase (Fig. 4B), a time when only basal levels of PntdABC activity were seen (see Fig. 3). These results suggest that a basal level of ntdABC expression (as seen during the exponential growth phase [see Fig. 2]) is sufficient for cells to express glcP, at least under the experimental conditions used here.
FIG. 4.
Northern analysis of ntdABC transcription. (A) Structure of the ntdABC operon. Promoters and transcriptional terminators are indicated. The two transcripts of the ntdABC operon and the probes used for Northern analysis are shown. (B) Northern analysis of the ntdABC transcript using probes for ntdC and glcP. B. subtilis strains 61884 (wild-type) and 84R5 (rpoB5 mutant) were grown in S7N medium containing excess (1%) glucose until OD650 reached 0.5, after which total cellular RNA was prepared from each strain using Isogen reagent. RNA samples (2 or 10 μg) were subjected to electrophoresis, transferred to a membrane, and then hybridized with the RNA probe for ntdC (left panel) or glcP (right panel) shown in panel A. Lane wt, wild type.
Expression of the NTD biosynthesis operon is repressed by GlcP-mediated glucose uptake.
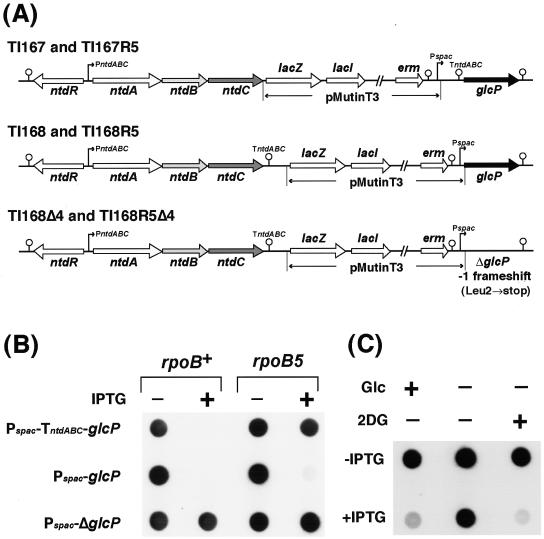
To further analyze the role of GlcP in NTD biosynthesis, we next inserted the pMutinT3 plasmid immediately upstream or downstream of the TntdABC of the rpoB+ wild-type and rpoB5 mutant strains, yielding glcP-interrupted strains carrying ntdC-pMutinT3-TntdABC and TntdABC-pMutinT3-glcP, respectively (Fig. 5A). These strains enabled us to control the expression of glcP via the IPTG-dependent promoter Pspac. In addition, a glcP-disrupted strain carrying TntdABC-pMutinT3-ΔglcP was also constructed. In this strain, T nucleotide in the second codon of glcP was deleted, resulting in a −1 frameshift and a nonsense mutation at Leu2 (TTA)→ochre (TAA). Initially, we directly detected the ntdABC transcript in the presence or absence of IPTG using RNA dot blotting with an ntdB probe (Fig. 5B). With the rpoB+ wild-type genetic background, induction of glcP (in the presence of IPTG) apparently repressed ntdABC expression in both the TI167 and TI168 strains (Fig. 5B). With the rpoB5 mutant genetic background, however, this repressive effect was no longer observed in strain TI167R5 (but not in TI168R5), possibly due to the blockade of glcP transcription by TntdABC in TI167R5. An alternative explanation for the absence of the repressive effect is that glcP is expressed in TI167 and TI167R5 less than in TI168 and TI168R5, and the lower concentration of GlcP is sufficient to repress transcription in wild-type rpoB+ cells but not in rpoB5 mutant cells. Given that the addition of IPTG had no effect on ntdABC expression in the ΔglcP strains (TI168Δ4 and TI168R5Δ4) as a control experiment, we concluded that expression of ntdABC is repressed by a GlcP protein-mediated mechanism.
FIG. 5.
Repression of ntdABC transcription by GlcP-mediated glucose transport. (A) Construction of glcP-interrupted strains. B. subtilis glcP-interrupted strains carrying ntdC::pMutinT3 or glcP::pMutinT3 were constructed by integrating a plasmid, pMutinT3, immediately upstream or downstream of TntdABC, respectively. These strains enabled us to control the expression of the glcP gene via the IPTG-dependent Pspac. A glcP-disrupted strain carrying ΔglcP::pMutinT3 also was constructed. In this strain, T nucleotide in the second codon of glcP was deleted, resulting in a −1 frameshift and a nonsense mutation at Leu-2 (TTA) (→ochre [TAA]). (B) Repression of ntdABC by induction of glcP. Total cellular RNAs were prepared from cells of strains TI167 (rpoB+ ntdC::pMutinT3), TI168 (rpoB+ glcP::pMutinT3), TI168Δ4 (rpoB+ ΔglcP::pMutinT3), TI167R5 (rpoB5 ntdC::pMutinT3), TI168R5 (rpoB5 glcP::pMutinT3), and TI168R5Δ4 (rpoB5 ΔglcP::pMutinT3), which were grown for 10 h in S7N medium containing excess (1%) glucose with or without 2 mM IPTG. RNA samples (10 μg each) were spotted directly onto a membrane and hybridized with the RNA probe for ntdB. (C) Repression of ntdABC by GlcP-mediated sugar transport. B. subtilis strain TI168 (rpoB+ glcP::pMutinT3) was grown in S7N medium containing excess (1%) glucose (Glc) until OD650 reached 1.0, after which the cells were harvested by centrifugation and transferred into the same S7N medium (left lane), S7N medium without glucose (center lane), or S7N medium supplemented with 1% 2-deoxy-glucose (2DG) instead of glucose (right lane). Where indicated, IPTG was added to final a concentration of 2 mM just after the cells were transferred. After incubation for an additional 4 h, cells were harvested and RNA samples (4 μg each) were spotted directly onto a membrane and hybridized with the RNA probe for ntdC.
Using glcP-interrupted strain TI168, we tested whether or not GlcP-dependent glucose uptake is responsible for the down-regulation of ntdABC transcription (Fig. 5C). When glcP was expressed in the absence of glucose (Fig. 5C), the expression of ntdABC was derepressible. However, the addition of 2-deoxy-glucose (a nonmetabolizable carbon source) instead of glucose entirely repressed ntdABC expression in cells expressing glcP, indicating that GlcP protein alone does not function as a repressor for ntdABC. The repression of ntdABC by glucose was also observed in a ccpA mutant genetic background (data not shown). Thus, GlcP-mediated glucose uptake appears to negatively regulate expression of the NTD biosynthesis operon in a CcpA-independent manner, which led us to conclude that glucose is able to act both as an inducer and as a repressor of ntdABC expression in B. subtilis, depending upon its uptake pathway, although we cannot completely eliminate the possibility that GlcP together with glucose regulates expression of the operon.
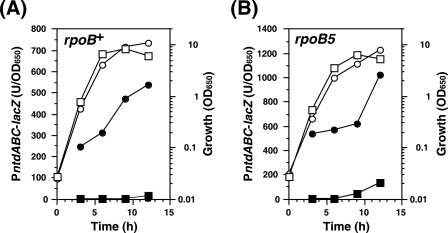
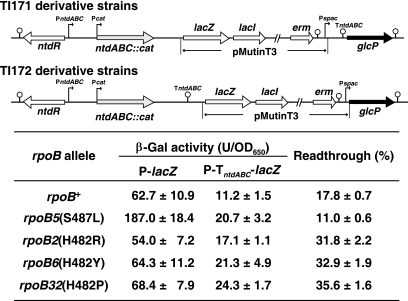
Mutant RNAP efficiently recognizes the ntdABC transcription termination site.
The results shown in Fig. 5B imply that the readthrough frequency at TntdABC in the rpoB5 mutant strain is less than that in the rpoB+ wild-type strain. Accordingly, we compared the readthrough frequency of the rpoB+ wild-type strain with those of various rpoB mutant strains at TntdABC. To avoid the effect of endogenously synthesized NTD, ntdABC in the strains TI167 and TI168 was deleted and replaced with the cat gene, generating strains TI171 and TI172, respectively (Fig. 6). The cat gene used here contains a weak promoter (Pcat). In the TI171 derivative strains carrying ΔntdABC::cat-pMutinT3-TntdABC, β-Gal activity represents the total promoter activity from PntdABC and Pcat, whereas in the TI172 derivative strains carrying ΔntdABC::cat-TntdABC-pMutinT3-glcP, β-Gal activity represents only the activity resulting from transcription readthrough at TntdABC. Therefore, the transcription readthrough level can be expressed as the β-Gal activity resulting from transcription readthrough divided by the activity representing the total promoter activity. In this way, the readthrough frequency at TntdABC was measured using various rpoB mutants. Expectedly, the rpoB5 mutant revealed a decreased readthrough frequency compared to that in the rpoB+ wild-type strain (Fig. 6). In contrast, the readthrough frequencies for other rpoB mutants harboring a His482→Arg (rpoB2), a His482→Tyr (rpoB6), or a His482→Pro (rpoB32) substitution within the RNAP β subunit were even higher than that in the rpoB+ wild-type strain. The rpoB5 RNAP mutant, which carries a Ser487→Leu substitution within its β subunit, thus appears to have a greater ability to recognize TntdABC than the wild-type RNAP or the His482 mutants described above.
FIG. 6.
Readthrough frequency at the ntdABC terminator site for various rpoB mutants. (Upper panel) Construction of strains for the TntdABC transcription readthrough assay. To avoid the effect of endogenously synthesized NTD, the ntdABC operons in strains TI167 and TI168 were replaced with cat genes. The cat gene used here contains a weak promoter. In a TI171 derivative strain carrying ΔntdABC::cat-pMutinT3-TntdABC, β-Gal activity represents the total promoter activity, whereas in a TI172 derivative strain carrying ΔntdABC::cat-TntdABC-pMutinT3-glcP, β-Gal activity represents the activity resulting only from transcription readthrough at TntdABC. Therefore, the transcription readthrough level can be expressed as the β-Gal activity resulting from transcription readthrough divided by the activity representing the total promoter activity. (Lower panel) TI171 and TI172 derivative strains containing each rpoB allele were cultivated for 15 h in S7N medium (containing 1% glucose) supplemented with purified NTD (500 μg/ml) and then harvested, after which measurements of β-Gal activities were made. Readthrough frequencies are expressed as β-Gal activity in TI172 derivative strains divided by the activity in a TI171 derivative strain. β-Gal activities are shown as the means ± standard deviations of three or more experiments.
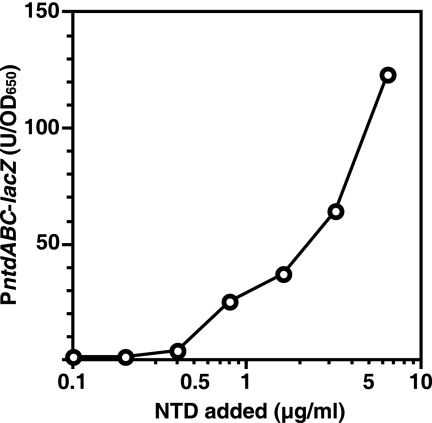
NTD functions in rpoB+ wild-type cells at concentrations below the detectable level.
Although NTD activity normally could not be detected (with a bioassay) in culture broth conditioned by the B. subtilis wild-type strain (rpoB+) (Table 2), an insertional mutation of glcP led to increased ntdABC expression (Fig. 3A) and, in turn, detectable NTD production (data not shown). In addition, we previously reported that the insertional mutation of ntdA causes a loss of promoter activity (18). Taking these facts together, it is possible that NTD can function for ntdABC expression at a concentration level below that which is detectable (5 μg/ml). We therefore determined the minimum concentration of NTD required for autoinduction using the reporter strain TI130, which contains a transposon inserted within ntdA. As shown in Fig. 7, B. subtilis cells sensed the exogenously added NTD at concentrations higher than 0.5 μg/ml. The amount of NTD produced by the B. subtilis rpoB+ 61884 strain was 1 to 3 μg/ml, as calculated based on the expression level of PntdABC-lacZ in the rpoB+ strain. Two other B. subtilis standard strains (JCM1465 and 168) and a number of B. subtilis strains isolated from soil also produced levels of NTD that were low but detectable with a bioassay (about 10 μg/ml). It is therefore concluded that the NTD produced in a wide variety of B. subtilis spp., though in small amounts, is able to function as a glucose uptake modulation factor in this bacterial group.
FIG. 7.
Minimum concentration of NTD needed for ntdABC transcriptional activation. B. subtilis strain TI130 [ntdA::Tn10 amyE::(PntdABC-lacZ cat)] was grown for 10 h in S7N medium (containing 1% glucose) supplemented with various concentrations of NTD, after which culture samples were withdrawn and their β-Gal activities were measured. At least four independent experiments were performed, and one of them is shown.
DISCUSSION
Bacteria are able to monitor their surroundings by releasing and detecting signaling molecules called autoinducers. This process enables bacteria to control gene expression so they can adapt to changes within their environment. Among the different classes of autoinducers, the best studied is LuxI, an acyl-homoserine lactone autoinducer produced by Vibrio fischeri (reviewed by Schauder and Bassler [33]). Moreover, B. subtilis is known to express two oligopeptide autoinducers, ComX and CSF (competence and sporulation factors), but many gram-positive autoinducer systems are currently under study (reviewed by Dunny and Leonard [6]). We previously reported that NTD is a novel autoinducer needed for transcription of its own biosynthesis operon in B. subtilis (18). This notion is confirmed in the present work, making NTD the third known autoinducer produced by B. subtilis. Our work also demonstrates that the downstream glcP gene is coexpressed with the ntdABC operon through transcription readthrough (Fig. 4). In other words, GlcP-mediated glucose uptake is under the control of NTD. Although the B. subtilis rpoB+ wild-type strain produces less-than-detectable amounts of NTD, our results indicate that a trace amount (0.5 to 3 μg/ml) of NTD is sufficient to ensure expression of glcP. Thus, by stimulating the transcription of ntdABC-glcP, NTD functions as a glucose uptake modulator in B. subtilis and probably in other NTD-producing bacteria, with a gene arrangement identical to that in B. subtilis. This conclusion is important because glucose metabolism takes a central place in a wide variety of metabolic systems. Another significant aspect of our finding is based on the fact that in addition to its function as an autoinducer, NTD has an antimicrobial activity against certain bacteria, possibly by inhibiting the cell wall synthesis, as demonstrated by the decline of antimicrobial activity in the presence of 1 mM d-glucosamine (T. Inaoka and K. Ochi, unpublished data). This is particularly intriguing, since the intrinsic biological role of antibiotics (if any) in the producing bacteria has not been verified (or has been largely overlooked) so far. Many bacteria, including B. subtilis (34), often produce antibiotics. NTD therefore may offer a feasible system for the study of possible physiological roles of “antibiotics” in the producing bacteria.
The effect of glucose on NTD production is enigmatic. The expression of ntdABC is apparently regulated, perhaps indirectly, by a mechanism related to CcpA-mediated carbon catabolite activation. Consistent with this idea are the observations that expression of PntdABC-lacZ was induced during the stationary phase in a glucose-dependent manner (Fig. 2) and that NTD production was severely impaired in a ccpA mutant (Tables 2 and 3). That the addition of purified NTD to the growth medium stimulated PntdABC-lacZ expression in ccpA and ymfI mutants suggests that these genes are involved in de novo synthesis of NTD, perhaps playing a role for the supply of a precursor(s) for NTD biosynthesis. At present, however, we cannot exclude the possibility that CcpA directly regulates ntdABC expression. Although there is no cre CcpA-binding motif in the ntdABC promoter region, a number of other genes subject to catabolite activation also lack a cre motif (2, 23, 24). It was somewhat surprising that GlcP-mediated glucose uptake repressed ntdABC expression in a CcpA-independent manner (Fig. 5C). This finding appears to contradict the positive effect of glucose on NTD production, but one possible explanation is that glucose can exert alternative (positive and negative) effects on ntdABC transcription, depending upon the uptake pathway. As summarized in Fig. 8, glucose transported through the predominant uptake pathway (glucose-PTS) is phosphorylated to glucose-6-phosphate during the permeation process and is then rapidly metabolized in the glycolytic pathway. In response to high cellular glycolytic activity, a glycolytic intermediate fructose-1,6-bisphosphate stimulates phosphorylation of HPr and Crh at Ser46 by activating the HPr kinase/phosphorylase (HprK), eventually resulting in CcpA-dependent catabolite activation (11). However, PntdABC remained repressed until the cells entered the stationary phase.
FIG. 8.
Proposed model of NTD production in B. subtilis. Glucose transported through glucose-PTS (EIICBA) positively regulates NTD production. This activation mechanism is based on the CcpA-dependent catabolite activation mechanism. Under conditions of high glycolytic activity, fructose-1,6-bisphosphate stimulates the phosphorylation of HPr and Crh at Ser46 by activating HprK, leading to CcpA-dependent catabolite activation (11). CcpA and YmfI proteins appear to be involved, directly or indirectly, in de novo synthesis of a precursor(s) for NTD, rather than in ntdABC promoter regulation. By contrast, ntdABC is repressed by GlcP-dependent glucose uptake. Possibly, a presumptive transcriptional regulator (designated X) linking to GlcP-dependent glucose uptake participates in this regulation. The transcriptional activator NtdR binds directly to NTD and activates the ntdABC promoter (18). NTD transporters (Y and Z), which are required for export or import of NTD, have not yet been identified. Positive effects are represented as arrows (→), while the wavy line represents a regulation of the ntdABC promoter by a presumptive transcriptional regulator, X. The glucose-PTS is comprised of enzyme I (EI), enzyme II (EIICBA), and HPr. G6P, glucose-6-phosphate; FBP, fructose-1,6-bisphosphate.
The repression seen during the exponential growth phase appears to depend solely on the glucose transport mediated by GlcP (Fig. 5C). Although details of the mechanism remain unclear, it is likely that a presumptive transcription regulator of ntdABC expression linked to GlcP-dependent glucose uptake (designated X in Fig. 8) might be involved. Within the framework of this notion, NtdR per se can be one of the candidates for X. Another hypothesis is that glucose itself transported by GlcP might interfere with the synthesis of a precursor of NTD. Investigation of the molecular mechanism underlying this glucose repression mediated by GlcP is now in progress. Such a study may provide a clue for uncovering the regulation mechanism of gene expression for NTD synthesis and also may help to further our understanding of the intrinsic mechanism of catabolite regulation in B. subtilis. In that regard, the glcP mutation partially but specifically relieves glucose- and sucrose-promoted catabolite repression of the gluconate operon (30).
Another point of interest is the mechanism underlying the gene activation induced by a certain Rif-resistant rpoB mutation. The rpoB mutations also reportedly activate antibiotic production in several Streptomyces spp. (13, 14, 21, 37). These earlier studies suggest that the mechanism of gene activation involves the alteration of the ternary structure of RNAP. We previously showed that a mutant RNAP (Ser487→Leu, corresponding to the rpoB5 mutation) efficiently recognizes house-keeping sigma factor (σA)-dependent promoters, including PntdABC (18). In agreement with that work, a quantitative analysis using surface plasmon resonance also revealed that this mutated RNAP binds to PntdABC more efficiently than the wild-type RNAP (H. Aoki, T. Inaoka, and K. Ochi, unpublished results). In addition to those earlier works, the present work shows that the readthrough frequency at TntdABC of the rpoB5 mutant strain was less than that of the rpoB+ wild-type strain (Fig. 6B), indicating that the RNAP mutant acquired a superior ability to recognize not only the promoter but also the transcription terminator, thus leading to a synergistic activation of NTD production. We do not know at present whether or not the enhancement of the transcription termination caused by the mutant RNAP is specific to the ntdABC-glcP operon. Moreover, very marked differences in ntdABC expression eventuated between rpoB+ and rpoB5 mutant cells (Fig. 5B), despite rather small differences (at most twofold) in readthrough frequencies (Fig. 6). Although further work is needed for more-convincing evidence of the mutant RNAP effect on transcription readthrough, these novel findings would be useful not only for basic microbiology but also for constructing new systems for screening secondary metabolites and enhancing the ability of strains to produce useful compounds.
Acknowledgments
We thank Chie Inaoka for performing several experiments and J. Deutscher (CNRS/INRA-PG) and I. Martin-Verstraete (Institut Pasteur) for providing the B. subtilis strains GM273 and QB7103.
This work was supported by grants (to T.I.) from the Kurata Memorial Hitachi Science and Technology Foundation and (to K.O.) from the Organized Research Combination System and the Effective Promotion of Joint Research of Special Coordination Funds (the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government).
Footnotes
Published ahead of print on 20 October 2006.
REFERENCES
- 1.Aung-Hilbrich, L. M., G. Seidel, A. Wagner, and W. Hillen. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319:77-85. [DOI] [PubMed] [Google Scholar]
- 2.Blencke, H. M., G. Homuth, H. Ludwig, U. Mader, M. Hecker, and J. Stulke. 2003. Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab. Eng. 5:133-149. [DOI] [PubMed] [Google Scholar]
- 3.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher, J., A. Galinier, and I. Martin-Verstraete. 2002. Carbohydrate uptake and metabolism. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 5.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 7.Fiegler, H., J. Bassias, I. Jankovic, and R. Bruckner. 1999. Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J. Bacteriol. 181:4929-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., and E. P. Greenberg. 1998. Cell-to-cell communication in Escherichia coli and Salmonella typhimurium: they may be talking, but who's listening? Proc. Natl. Acad. Sci. USA 95:6571-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 11.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stulke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94:8439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 13.Hu, H., and K. Ochi. 2001. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 67:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, H., Q. Zhang, and K. Ochi. 2002. Activation of antibiotic biosynthesis by specified mutations in the rpoB gene (encoding the RNA polymerase β subunit) of Streptomyces lividans. J. Bacteriol. 184:3984-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inaoka, T., K. Kasai, and K. Ochi. 2001. Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis. J. Bacteriol. 183:4958-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 18.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 279:3885-3892. [DOI] [PubMed] [Google Scholar]
- 19.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 20.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 21.Lai, C., J. Xu, Y. Tozawa, Y. Okamoto-Hosoya, X. Yao, and K. Ochi. 2002. Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology 148:3365-3373. [DOI] [PubMed] [Google Scholar]
- 22.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 23.Lorca, G. L., Y. J. Chung, R. D. Barabote, W. Weyler, C. H. Schilling, and M. H. Saier, Jr. 2005. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J. Bacteriol. 187:7826-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 25.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29:179-187. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani, Y., W. L. Nicholson, K. D. Neitzke, P. Setlow, and E. Freese. 1989. Sigma-G RNA polymerase controls forespore-specific expression of the glucose dehydrogenase operon in Bacillus subtilis. Nucleic Acids Res. 17:999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Numata, K., F. Satoh, M. Hatori, T. Miyaki, and H. Kawaguchi. 1986. Isolation of 3,3′-neotrehalosadiamine (BMY-28251) from a butirosin-producing organism. J. Antibiot. (Tokyo) 39:1346-1348. [DOI] [PubMed] [Google Scholar]
- 28.Ochi, K., and S. Ohsawa. 1984. Initiation of antibiotic production by the stringent response of Bacillus subtilis Marburg. J. Gen. Microbiol. 130:2473-2482. [DOI] [PubMed] [Google Scholar]
- 29.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen, I. T., S. Chauvaux, P. Choi, and M. H. Saier, Jr. 1998. Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel hexose:H+ symporter. J. Bacteriol. 180:498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saier, M. H., Jr., S. Chauvaux, G. M. Cook, J. Deutscher, I. T. Paulsen, J. Reizer, and J. J. Ye. 1996. Catabolite repression and inducer control in Gram-positive bacteria. Microbiology 142:217-230. [DOI] [PubMed] [Google Scholar]
- 33.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 34.Stein, T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56:845-857. [DOI] [PubMed] [Google Scholar]
- 35.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuno, T., C. Ikeda, K. Numata, K. Tomita, M. Konishi, and H. Kawaguchi. 1986. 3,3′-Neotrehalosadiamine (BMY-28251), a new aminosugar antibiotic. J. Antibiot. (Tokyo) 39:1001-1003. [DOI] [PubMed] [Google Scholar]
- 37.Xu, J., Y. Tozawa, C. Lai, H. Hayashi, and K. Ochi. 2002. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Genet. Genomics 268:179-189. [DOI] [PubMed] [Google Scholar]