Abstract
The Bacillus subtilis TnrA transcription factor belongs to the MerR family of proteins and regulates gene expression during nitrogen-limited growth. When B. subtilis cells are grown with excess nitrogen, feedback-inhibited glutamine synthetase forms a protein-protein complex with TnrA that prevents TnrA from binding to DNA. The C-terminal region of TnrA is required for the interaction with glutamine synthetase. Alanine scanning mutagenesis of the C-terminal region of TnrA identified three classes of mutants that altered the regulation by glutamine synthetase. While expression of the TnrA-regulated amtB gene was expressed constitutively in the class I (M96A, Q100A, and A103G) and class II (L97A, L101A, and F105A) mutants, the class II mutants were unable to grow on minimal medium unless a complex mixture of amino acids was present. The class III tnrA mutants (R93A, G99A, N102A, H104A, and Y107A mutants) were partially defective in the regulation of TnrA activity. In vitro experiments showed that feedback-inhibited glutamine synthetase had a significantly reduced ability to inhibit the DNA-binding activity of several class I and class II mutant TnrA proteins. A coiled-coil homology model of the C-terminal region of TnrA is used to explain the properties of the class I and II mutant proteins. The C-terminal region of TnrA corresponds to a dimerization domain in other MerR family proteins. Surprisingly, gel filtration and cross-linking analysis showed that a truncated TnrA protein which contained only the N-terminal DNA binding domain was dimeric. The implications of these results for the structure of TnrA are discussed.
The MerR family of transcription factors regulates gene expression in response to a wide variety of physiological signals (3, 25). Bacillus subtilis TnrA is a member of the MerR family that controls transcription in response to nitrogen availability (45). During nitrogen-limited growth, TnrA regulates the expression of many genes, including genes involved in the transport and catabolism of nitrogen-containing compounds (2, 45, 50). The DNA-binding activity of TnrA is controlled by glutamine synthetase (GS), a key enzyme in nitrogen metabolism (45). B. subtilis GS is subject to feedback inhibition by glutamine and AMP (11). Under excess-nitrogen growth conditions, feedback-inhibited GS forms a complex with TnrA that prevents TnrA from binding DNA (48).
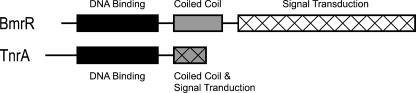
MerR family proteins are dimers that typically contain three different domains (Fig. 1). Members of the MerR family are defined by a conserved N-terminal DNA-binding domain that is approximately 70 amino acids in length (3, 25, 28). Structural studies have shown that this DNA-binding domain contains a helix-turn-helix motif, a β-loop wing, and a second wing formed by helices 3 and 4 (Fig. 2) (7, 19, 21, 34). The C-terminal domain is involved in signal transduction, and the sequences of these regions are conserved only among orthologs (Fig. 1) (3, 23, 25). The N- and C-terminal domains are linked together by a long α-helix that forms an antiparallel coiled-coil structure that is involved in dimerization (Fig. 1) (6, 7, 9, 19, 21, 53).
FIG. 1.
Domain structures of the MerR family proteins BmrR and TnrA. Black rectangles denote DNA-binding domains. Gray rectangles denote protein regions capable of forming coiled-coil structures. Cross-hatched rectangles denote signal transduction domains. The size of each rectangle is proportional to the size of each protein domain. The domain organization of BmrR was derived from its crystal structure (21).
FIG. 2.
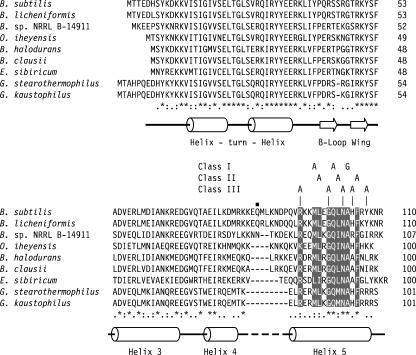
Alignment of TnrA orthologs. The level of amino acid residue conservation is shown below the sequences, where an asterisk denotes positions with identical amino acid residues, a colon indicates positions where all of the residues are members of the same substitution group (49), and a period represents positions where the majority of the residues are members of the same substitution group. Positions with identical or highly conserved amino acid residues within the C-terminal region are shown in reverse text. The locations of the amino acid substitutions for the three classes of tnrA mutants are shown above the alignment. Residue Gln-84, which is the location of the nonsense mutation in TnrAC213, is denoted with a small black square. A graphical representation of the secondary structures is shown below the aligned sequences, where α-helices and β-strands are depicted as cylinders and arrows, respectively. These secondary structural assignments are based on the crystal structures of other MerR family proteins (7, 19, 21) and analysis of the B. subtilis TnrA protein sequence with the secondary structural prediction algorithms GOR V (26) and PSIPRED (22).
The B. subtilis TnrA protein, which contains only 110 amino acids, is smaller than most MerR family members and contains only two domains (Fig. 1). The conserved N-terminal DNA binding domain is located between residues 10 and 81 in the B. subtilis TnrA protein (Fig. 1 and 2). Comparison of the amino acid sequences of TnrA orthologs reveals the presence of a conserved 15-amino-acid C-terminal region that is connected to the N-terminal DNA binding domain by a linker region of variable length (Fig. 2). This conserved C-terminal region functions as the TnrA signal transduction domain, because several previously isolated missense mutations in this region of the B. subtilis TnrA protein resulted in constitutive regulation of gene expression (TnrAC) due to impaired interaction with feedback-inhibited GS (48). The region of TnrA involved in dimerization has not been identified.
In this work, alanine substitution mutagenesis was used to identify amino acid residues in the C-terminal region of TnrA that are required for regulation by glutamine synthetase. Although computational analysis indicates that the TnrA C-terminal signal transduction domain may form a coiled-coil structure (Fig. 1), this region of TnrA was not required for dimerization.
MATERIALS AND METHODS
Bacterial strains and growth.
Mutant tnrA genes generated by oligonucleotide mutagenesis were integrated as single copies into the amyE locus by transforming plasmid DNA containing each tnrA allele into B. subtilis 168 strain SF29TN (amtB::Tn917-lacZ tnrA111::neo trpC2) with selection for chloramphenicol resistance and screening for loss of plasmid-encoded spectinomycin resistance. Previously reported methods were used to grow bacteria in liquid cultures (1). The minimal medium described by Neidhardt et al. (33) was used for the growth of liquid cultures. The carbon source was 0.5% glucose, while the final concentration of all nitrogen sources was 0.2% Casamino Acids, an acid hydrolysate of casein, was obtained from Difco Laboratories. Luria-Bertani (LB) medium (30) containing glutamine at a final concentration of 0.2% was used as an excess nitrogen growth medium in some experiments. Minimal medium agar plates were prepared as previously described (8). The expression of amtB in the tnrA mutants was screened on glucose minimal agar plates supplemented with 40 μg/ml of 5-bromo-4-chloro-3-indoyl-β-d-galactoside and various nitrogen sources.
Enzyme assays.
β-Galactosidase assays were performed as previously described (1). Cell extracts for β-galactosidase assays were prepared from cultures harvested during exponential growth. The reported β-galactosidase levels were corrected for the endogenous activity present in B. subtilis 168 cells lacking a Tn917-lacZ transposon insertion.
Oligonucleotide mutagenesis.
A two-step PCR-based method was used to generate mutations in the tnrA gene (24). The first amplification utilized a primer containing the desired mutation and the oligonucleotide TNRH3 (5′-TAC GAA GCT TGC ACA AAC TGA AAG TAG ACC). The TNRH3 primer is complementary to sequences downstream of the tnrA gene and contains a HindIII site. The reaction product from the first amplification and oligonucleotide TNRECO (5′-ATG CGA ATT CTC CAT GAT TAT CCT TCC TCC) were used as primers for the second amplification. Primer TNRECO is complementary to sequences upstream of the tnrA gene and contains an EcoRI site. The proofreading Tgo DNA polymerase (Roche Applied Science) was used for all amplifications. The final PCR products were cloned into the chromosomal integration vector pDG1662 (20) as EcoRI-HindIII DNA fragments and subsequently sequenced to confirm the presence of the desired mutation.
DNA and protein methods.
Construction of TnrA overexpression plasmids was performed as previously described (15). Overexpression and purification of the GS and TnrA proteins were performed as previously described (47, 48). The concentrations of TnrA and GS were determined by measuring their absorbances at 280 nm. The molar absorption coefficients of the proteins were calculated from their amino acid sequences (36). Gel mobility shift experiments to examine the ability of GS to inhibit DNA binding by wild-type and mutant TnrA proteins were performed as previously described (46, 47).
The molecular sizes of the purified wild-type and mutant TnrA proteins were determined by size exclusion chromatography on a 1.6-cm-by-60-cm HiPrep Sephacryl S-200 HR column (Amersham Pharmacia Biotech). Buffer containing 50 mM Bicine (pH 8.3), 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol was used in the gel filtration analysis. The protein protomer concentrations of the wild-type and mutant TnrA proteins in the peak column fractions were 4 to 8 μM. Ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa) were used as size standards.
Cross-linking was performed with buffer containing 20 mM sodium phosphate (pH 8) and 100 mM sodium chloride with protein protomer concentrations of 8 μM. Reactions were initiated by adding disuccinimidyl suberate (DSS) dissolved in dimethyl sulfoxide to a final concentration of 0.5 mM. After incubation at 30°C for various time periods, the reactions were quenched by the addition of 0.15 volumes of 0.5 M glycine. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue.
Bioinformatics.
A multiple-sequence alignment that included all known TnrA orthologs was prepared with ClustalW (42) and manually edited with GeneDoc (http://www.psc.edu/biomed/genedoc/). The accession numbers for the TnrA protein sequences are as follows: NP_389214 (B. subtilis), YP_078674 (Bacillus licheniformis), ZP_01170844 (Bacillus sp. strain NRRL B-14911), NP_242360 (Bacillus halodurans), YP_175256 (Bacillus clausii), NP_691871 (Oceanobacillus iheyensis), EAM88231 (Exiguobacterium sibiricum), and YP_147713 (Geobacillus kaustophilus). The sequence for Geobacillus stearothermophilus TnrA was obtained from the web site of the G. stearothermophilus genome sequencing project (http://www.genome.ou.edu/bstearo.html). Secondary structural predictions of the B. subtilis TnrA protein sequence utilized the GOR V (39) and PSIPRED (29) web servers. Coiled-coil evaluation of the B. subtilis TnrA protein sequence was carried out using the COILS web server (http://www.ch.embnet.org/software/COILS_form.html) with a 14-residue scanning window and the weighting option enabled (27). Homology modeling of the C-terminal region of TnrA was performed with Swiss-PDB Viewer software and the SWISS-MODEL web server (38). The structural model for the C-terminal region of TnrA was constructed by using MtaN residues 87 to 103 (19) as a template for TnrA residues 90 to 106.
RESULTS AND DISCUSSION
Mutagenesis of the TnrA C-terminal domain.
To identify individual amino acid residues that are essential for the interaction between TnrA and the feedback-inhibited form of GS, alanine substitutions in the C-terminal region of TnrA (residues 83 to 110) were generated. Residue Ala-103 was replaced with a glycine. The resulting mutant tnrA genes were integrated as single copies into the amyE locus of a tnrA mutant strain that contained an amtB::Tn917-lacZ transcriptional fusion. Three of the tnrA mutants (L97A, L101A, and F105A mutants) had a conditional growth phenotype. These three mutants grew like wild-type cells on nutrient-rich LB plates but could not form colonies on glucose minimal medium unless the minimal medium was supplemented with Casamino Acids. Although these mutants grew like wild-type cells when supplemented with Casamino Acids, no single amino acid or group of biosynthetically related amino acids could be identified which restored the wild-type growth phenotype to these mutants. The conditional growth phenotype was also seen for strains that did not contain the amtB::Tn917-lacZ insertion, indicating that this phenotype is not dependent on expression of the amtBglnK operon.
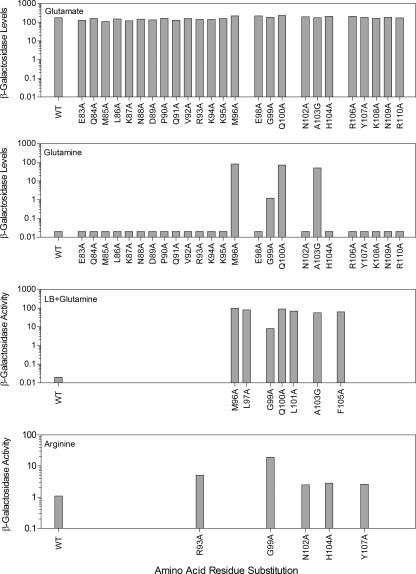
Since activation of amtB transcription requires TnrA (45), β-galactosidase expression from an amtB-lacZ transcriptional fusion was used to examine the ability of GS to regulate the activity of the mutant TnrA proteins in vivo. β-Galactosidase levels were 3,000-fold higher in wild-type cells grown in minimal medium containing the limited-nitrogen source glutamate than in minimal medium with the excess-nitrogen source glutamine (Fig. 3). LB-plus-glutamine medium also contains excess nitrogen, because expression of the amtB-lacZ fusion is not activated in wild-type cells grown in this medium (Fig. 3).
FIG. 3.
β-Galactosidase expression in mutant tnrA strains. β-Galactosidase levels were determined in extracts of cells growing exponentially in minimal media containing the indicated nitrogen sources or in LB-plus-glutamine medium. Data are the averages of 2 to 10 determinations. The sample standard deviation did not vary by more than 15%.
The tnrA mutants were placed in three classes based on their growth phenotypes and their effects on TnrA-dependent gene regulation. The class I tnrA mutants (M96A, Q100A, and A103G mutants) had highly constitutive amtB expression and did not have any growth defect on minimal media. Similar levels of β-galactosidase were present for the three class I tnrA mutants when grown in minimal medium containing either excess (glutamine) or limited (glutamate) nitrogen (Fig. 3). The class II mutants (L97A, L101A, and F105A mutants) had the conditional growth phenotype, and β-galactosidase levels were determined only for cells grown in LB plus glutamine medium, because these mutants were unable to grow on minimal media. Since similar high levels of β-galactosidase were present in class I and class II tnrA mutants grown in LB-plus-glutamine medium (Fig. 3), amtB expression is highly constitutive for the class II mutants. The class I and II substitutions are all located in highly conserved residues (Fig. 2). These results indicate that these six residues are critical for the interaction of TnrA with GS and are consistent with the idea that highly conserved residues are essential for protein function.
The GS-dependent regulation of TnrA was partially defective for the five class III mutants (R93A, G99A, N102A, H104A, and Y107A mutants). Four of the class III tnrA mutants (R93A, N102A, H104A, and Y107A mutants) had only minor defects in the regulation of TnrA activity. This effect was observed only in cultures where arginine was the sole nitrogen source and amtB expression was partially activated. β-Galactosidase levels were 61-fold higher in wild-type cells grown in minimal medium with arginine as the nitrogen source than in those grown in minimal medium with the nitrogen source glutamine (Fig. 3). While these four tnrA mutants contained wild-type levels of β-galactosidase when grown in minimal medium containing the excess-nitrogen source glutamine, β-galactosidase levels were two- to threefold higher for these class III mutants than in wild-type cells grown in minimal medium containing the nitrogen source arginine (Fig. 3). A more significant defect in the regulation of TnrA activity was seen with the G99A mutant. Compared to those in wild-type cells, β-galactosidase levels for this mutant were 30-fold higher with glutamine minimal medium and 180-fold higher with LB-plus-glutamine medium (Fig. 3).
Complementation analysis of the class I and class II mutations was performed by introducing these mutations into a strain containing a wild-type copy of tnrA. Surprisingly, the merodiploid strains with the class II mutations could grow on minimal medium containing glutamate or glutamine as the nitrogen source. Expression of the amtB-lacZ fusion in the complemented strains was partially constitutive when cells were grown on minimal medium with glutamine as the nitrogen source (Table 1). Under these excess-nitrogen growth conditions, strains containing the class II mutations had higher levels of β-galactosidase than strains containing the class I mutations. All of this suggests that the class II mutations cause a more severe defect in GS-dependent regulation than the class I mutations and argues that high-level expression of TnrA-regulated genes is responsible for the growth phenotype of the class II mutants.
TABLE 1.
β-Galactosidase expression of an amtB-lacZ fusion in merodiploid strains
| tnrA allele integrated into the amyE genea | β-Galactosidase sp act (U/mg protein) in cells grown withb:
|
Nitrogen regulation ratioc | |
|---|---|---|---|
| Glutamine | Glutamate | ||
| Wild type | 0.05 | 129 | 2,580 |
| Class I | |||
| tnrA(M96A) | 14 | 128 | 9.1 |
| tnrA(Q100A) | 14 | 133 | 9.5 |
| tnrA(A103G) | 6.6 | 176 | 27 |
| Class II | |||
| tnrA(L97A) | 52 | 164 | 3.2 |
| tnrA(L101A) | 59 | 154 | 2.6 |
| tnrA(F105A) | 54 | 163 | 3.0 |
All strains contained a wild-type tnrA allele and an amtB::Tn917-lacZ insertion.
Cells were grown in glucose minimal medium containing the indicated nitrogen sources. Each value is the average of two or more determinations, and values did not vary by more than 20%.
The nitrogen regulation ratio was calculated by dividing the enzyme activity found in cultures grown with glutamate by the enzyme activity found in glutamine-grown cultures.
Characterization of mutant TnrA proteins in vitro.
To examine their in vitro properties, two class I proteins (M96A and Q100A mutants) and two class II proteins (L97A and L101A mutants) were overexpressed and purified. All four mutant TnrA proteins behaved as dimers according to gel filtration analysis. In gel mobility shift experiments, the mutant TnrA proteins were found to have equilibrium disassociation binding constants for amtB promoter DNA (5.9 ± 0.6 to 13.5 ± 1.8 nM) which were similar to that of wild-type TnrA (8.6 ± 0.8 nM).
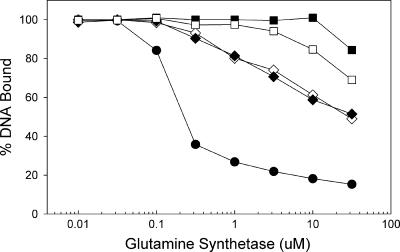
The ability of feedback-inhibited GS to inhibit the binding of the mutant TnrA proteins to amtB promoter DNA was analyzed in vitro using a DNA gel mobility shift assay. In these experiments, a fixed amount of the TnrA protein was incubated with various amounts of GS in the presence of 20 mM glutamine. This concentration of the GS feedback inhibitor closely matches the intracellular glutamine levels of wild-type B. subtilis cells grown in the presence of excess nitrogen (16). The ability of feedback-inhibited GS to inhibit DNA binding by the mutant TnrA proteins was greatly reduced compared to that with wild-type TnrA (Fig. 4). DNA binding by the two class II TnrA proteins (L97A and L101A mutants) was almost completely refractory to inhibition by high concentrations of feedback-inhibited GS (Fig. 4). In contrast, the two class I TnrA proteins (M96A and Q100A mutants) had intermediate levels of sensitivity to inhibition by feedback-inhibited GS (Fig. 4). Since these four mutant TnrA proteins have DNA binding affinities that are similar to that of the wild-type protein, the constitutive transcriptional activation by these proteins observed in vivo most likely results from a reduced ability to interact with feedback-inhibited GS.
FIG. 4.
Effect of feedback-inhibited glutamine synthetase on the DNA-binding activity of TnrAC proteins. A gel mobility shift assay was used to examine the binding of wild-type (•), M96A (⋄), Q100A (⧫), L97A (□), and L101A (▪) TnrA proteins to amtB DNA. The TnrA dimer concentration was 50 nM in all binding reactions. Each data point is the mean of at least two independent determinations and is reproducible to within 10%.
Homology model of TnrA C-terminal region.
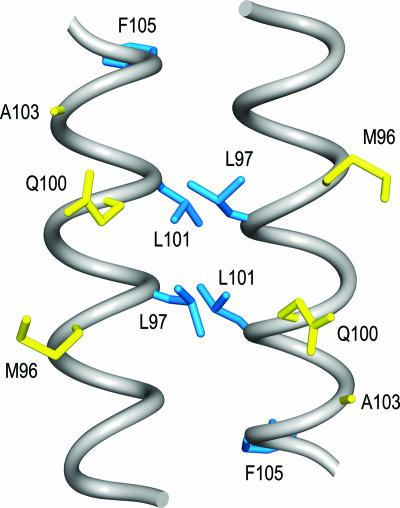
The coiled coil is a protein structural motif which contains two or more α-helices that are coiled around each other (5). The sequences of helices within coiled coils are characterized by heptad repeats, designated abcdefg, where residues in the a and d positions form the interface between interacting helices (5). The crystal structures of the MerR family members BmrR, CueR, MtaN, and ZntR revealed that these proteins contained antiparallel coiled-coil dimerization structures located C terminal to the DNA binding domain (Fig. 1) (7, 19, 21). Since the α-helices that form these coiled-coil structures have the same position relative to the DNA binding domain as helix 5 of TnrA (Fig. 1 and 2.), the ability of helix 5 of TnrA to form a coiled-coil structure was examined computationally. Analysis of TnrA with the coiled coil prediction program COILS indicated that residues 91 to 106 in the C-terminal region of TnrA may form a coiled-coil structure (27). The COILS program predicts that Lys-94, Leu-97, Leu-101, and His-104 would be the interface residues for helix 5 of TnrA. Based on the structures of BmrR, CueR, MtaN, and ZntR (7, 19, 21), helix 5 from one TnrA subunit most likely interacts with helix 5 of the other subunit. This information was used to construct a comparative structural model for the C-terminal region of TnrA (Fig. 5).
FIG. 5.
Structural model of the TnrA C-terminal region. The backbone of residues 90 to 106 is shown as smoothed gray tubes. The side chains of the class I and II residues are colored yellow and blue, respectively. This diagram was produced with UCSF Chimera (37).
In this model of the C-terminal region of TnrA, all of the class I residues (Met-96, Gln-100, and Ala-103) would be located on the same face of the coiled coil and thus could be surface exposed (Fig. 5). By this hypothesis, the class I residues would make direct contact with GS, and alanine substitutions for these residues would remove these contacts. In contrast, the hydrophobic class II residues (Leu-97, Leu-101, and Phe-105) would be buried, and thus, the alanine substitutions of these residues would generate cavities within the interior of TnrA (Fig. 5). Since protein cavities can rearrange the local structure (4, 12), the class II substitutions would indirectly interfere with the binding of TnrA to GS and disrupt the TnrA-GS interaction more severely than class I substitutions. This interpretation is supported by the in vitro studies, which demonstrated that the class II proteins had a more significant defect in their interaction with GS than the class I proteins (Fig. 4). The relatively severe defect in the ability of GS to regulate the activity of the class II mutant proteins may result in highly constitutive activation and/or repression of TnrA-regulated genes. This aberrant gene expression may be the cause of the growth impairment observed on minimal media with the class II mutants. Interestingly, a similar phenomenon has been observed for Escherichia coli, where highly constitutive mutants of the cAMP activator protein have been shown to inhibit cell growth (51). Nonetheless, the growth phenotype of the class II mutants is puzzling in that strains containing a null mutation in glnA, the gene encoding GS, are able to grow on glutamine minimal media even though TnrA is constitutively active (45). Curiously, strains that contain both a class II mutation and a glnA null mutation can grow on glutamine minimal media. This observation indicates that the glnA mutation can suppress the growth phenotype associated with the class II mutations. Since B. subtilis glnA mutants are known to have a variety of pleiotropic effects on gene expression (16), the suppression of the class II mutant growth defect is most likely indirect.
The class III substitutions had relatively weak effects on the regulation of TnrA, and they define a set of residues that appear to make only minor contributions to the interaction of TnrA with GS. Surprisingly, two of these residues, Arg-93 and Asn-102, are highly conserved in the TnrA proteins (Fig. 2). Although the alanine substitutions for these residues did not significantly disrupt the regulation of TnrA activity, it is still possible that these residues are involved in important interactions with GS. For instance, a mutational study of a lysozyme-antibody complex described an Asp-to-Ala substitution of a contact residue that had no significant effect on affinity (10). Structural analysis revealed that the Asp side-chain interactions were replaced by water-mediated contacts (10). The substitution of water molecules for truncated polar side chains has also been observed in the barnase-barstar complex (43). Thus, it is possible that the polar Arg-93 and Asn-102 residues of TnrA interact with GS and that the alanine substitutions have only a weak effect on binding because solvent molecules are able to replace the missing side chains. A definitive explanation of the roles of these residues in the interaction between TnrA and GS will require the structural determination of the TnrA-GS complex.
TnrA C-terminal region is not required for dimerization.
The crystal structures of MerR family proteins revealed that their N-terminal DNA-binding domains do not contact one another and that dimerization is mediated by a coiled-coil structure located between the N-terminal and C-terminal domains (Fig. 1) (7, 19, 21). This raises the possibility that helix 5 of TnrA, which may form a coiled-coil structure, also functions as the TnrA dimerization domain. We have previously described a constitutive mutant of TnrA (TnrAC213) in which the substitution of an amber codon for the Gln-84 codon results in the synthesis of a truncated TnrA protein which lacks the C-terminal 27 amino acid residues (48). If TnrA has the same topology as these other MerR family proteins, then the TnrAC213 protein would lack the coiled-coil dimerization domain and would thus be monomeric.
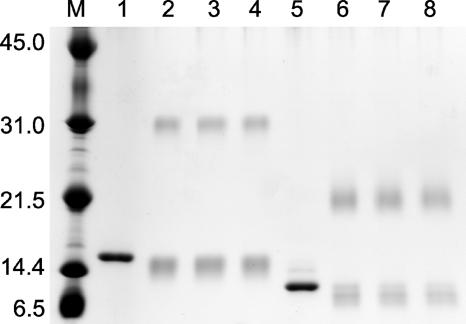
To test this hypothesis, the oligomeric states of the wild-type TnrA and mutant TnrAC213 proteins were determined by size exclusion chromatography. The wild-type TnrA protein has a calculated protomer size of 13 kDa and eluted from the column as a dimer with an apparent molecular mass of 26 kDa. The TnrAC213 protein, which has a calculated protomer size of 10 kDa, eluted as a single peak with an apparent molecular mass of 24 kDa, indicating that this protein is also dimeric. The quaternary structure of these two proteins was also analyzed with the amine-specific cross-linker DSS. In these experiments, cross-linked dimers were seen with both the wild-type TnrA and mutant TnrAC213 proteins (Fig. 6). The bands migrating slightly faster than those of the monomeric unlinked proteins were not converted to cross-linked dimers upon extended incubation and are most likely the result of intrachain cross-linking. Taken together, these results indicate that the 27 C-terminal amino acids of TnrA are not required for dimerization. While we cannot rule out the possibility that the C-terminal region of TnrA contributes to dimerization, these results argue that the N-terminal DNA-binding domains of TnrA interact with one another.
FIG. 6.
Cross-linking of the wild-type TnrA and mutant TnrAC213 proteins. After cross-linking with DSS, samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lanes 1 to 4 contain the wild-type TnrA protein. Lanes 5 to 8 contain the mutant TnrAC213 protein. For lanes 1 and 5, DSS was not present in the reaction mixture. The other samples were incubated with DSS for different time periods: lanes 2 and 6, 15 min; lanes 3 and 7, 30 min; lanes 4 and 8, 45 min. Lane M contains molecular size markers with the masses of the standards indicated at the left in kDa.
These results are supported by two other experimental observations (48). First, because the affinity of the TnrAC213 protein for amtB DNA is only twofold lower than that of the wild-type TnrA protein, the TnrAC213 protein is not significantly impaired in its ability to bind DNA. Second, sigmoidal DNA-binding curves are observed with DNA-binding proteins that have similar equilibrium constants for the protein monomer-dimer and dimer-DNA interactions (14, 40, 41). Since the TnrAC213 protein has a hyperbolic DNA binding curve similar to that seen with the wild-type TnrA protein, no defect in dimerization is apparent in the DNA-binding assay where nanomolar concentrations of TnrA are used. While it is possible that the TnrAC213 protein uses a protein interface for dimerization different from that of the wild-type protein, this seems unlikely, since both proteins have similar affinities for amtB promoter DNA.
Unique properties of TnrA.
Several distinctive attributes of TnrA indicate that it is a unique member of the MerR family of proteins. First, while the activity of a typical MerR family member is regulated by a small molecular coeffector, such as a metal ion or organic compound (3), TnrA is regulated by a protein-protein interaction with feedback-inhibited GS (48). Second, the observation that the DNA-binding domains of TnrA are able to dimerize argues that there is a difference in the quaternary structures of TnrA and those of the structurally characterized MerR family proteins. Finally, the mechanism of transcriptional activation by TnrA is different than that of other well-studied MerR transcription factors. MerR family proteins activate expression of nonoptimal promoters transcribed by RNA polymerase complexed with the general housekeeping sigma factor (3, 47). TnrA activates transcription at promoters that have optimal spacing between the −35 and −10 elements but are suboptimal because of mismatches with the consensus sequences for the −35 and −10 regions (13, 17, 31, 32, 44, 52). The binding of TnrA to an inverted repeat located upstream of the −35 region enhances the binding of RNA polymerase to the promoter (47). In contrast, other MerR family proteins activate transcription at promoters that are suboptimal because the spacing between the −35 and −10 promoter regions is 2 to 3 bp larger than the ideal 17-bp spacer length (35). These MerR proteins bind to inverted repeats which overlap the promoter and cause a distortion in the DNA that spatially realigns the −35 and −10 regions so that RNA polymerase can bind to the promoter (18, 21, 34, 35). While the DNA-binding domains of TnrA appear to be in contact with one another, the DNA-binding domains in other MerR family proteins are spatially separated. This separation of the DNA-binding domains may be required to cause DNA distortion and thus would not be necessary for transcriptional activation by TnrA.
Acknowledgments
This research was supported by Public Health Service research grant GM51127 from the National Institutes of Health.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 182:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 4.Buckle, A. M., P. Cramer, and A. R. Fersht. 1996. Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities. Biochemistry 35:4298-4305. [DOI] [PubMed] [Google Scholar]
- 5.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 6.Caguiat, J. J., A. L. Watson, and A. O. Summers. 1999. Cd(II)-responsive and constitutive mutants implicate a novel domain in MerR. J. Bacteriol. 181:3462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Changela, A., K. Chen, Y. Xue, J. Holschen, C. E. Outten, T. V. O'Halloran, and A. Mondragón. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383-1387. [DOI] [PubMed] [Google Scholar]
- 8.Chasin, L. A., and B. Magasanik. 1968. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J. Bacteriol. 178:4778-4786. [PubMed] [Google Scholar]
- 9.Chiu, M. L., P. H. Viollier, T. Katoh, J. J. Ramsden, and C. J. Thompson. 2001. Ligand-induced changes in the Streptomyces lividans TipAL protein imply an alternative mechanism of transcriptional activation for MerR-like proteins. Biochemistry 40:12950-12958. [DOI] [PubMed] [Google Scholar]
- 10.Dall'Acqua, W., E. R. Goldman, W. Lin, C. Teng, D. Tsuchiya, H. Li, X. Ysern, B. C. Braden, Y. Li, S. J. Smith-Gill, and R. A. Mariuzza. 1998. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry 37:7981-7991. [DOI] [PubMed] [Google Scholar]
- 11.Deul, T. F., and S. Prusiner. 1974. Regulation of glutamine synthetase from Bacillus subtilis by divalent cations, feedback inhibitors, and L-glutamine. J. Biol. Chem. 249:257-264. [PubMed] [Google Scholar]
- 12.Eriksson, A. E., W. A. Baase, X. J. Zhang, D. W. Heinz, M. Blaber, E. P. Baldwin, and B. W. Matthews. 1992. Response of a protein to cavity-creating mutations and its relation to the hydrophobic effect. Science 255:178-183. [DOI] [PubMed] [Google Scholar]
- 13.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693-701. [DOI] [PubMed] [Google Scholar]
- 14.Filée, P., C. Vreuls, R. Herman, I. Thamm, T. Aerts, P. P. De Deyn, J.-M. Frère, and B. Joris. 2003. Dimerization and DNA binding properties of the Bacillus licheniformis 749/I BlaI repressor. J. Biol. Chem. 278:16482-16487. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, S. H., J. L. Brandenburg, and L. V. Wray, Jr. 2002. Mutations in Bacillus subtilis glutamine synthetase that block its interaction with transcription factor TnrA. Mol. Microbiol. 45:627-635. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, S. H., and A. L. Sonenshein. 1984. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J. Bacteriol. 157:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, S. H., and L. V. Wray, Jr. 2002. Bacillus subtilis 168 contains two differentially regulated genes encoding l-asparaginase. J. Bacteriol. 184:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frantz, B., and T. V. O'Halloran. 1990. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry 29:4747-4751. [DOI] [PubMed] [Google Scholar]
- 19.Godsey, M. H., N. N. Baranova, A. A. Neyfakh, and R. G. Brennan. 2001. Crystal structure of MtaN, a global multidrug transporter gene activator. J. Biol. Chem. 276:47178-47184. [DOI] [PubMed] [Google Scholar]
- 20.Guérout-Fleury, A.-M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 21.Heldwein, E. E. Z., and R. G. Brennan. 2001. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409:378-382. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 23.Kahmann, J. D., H. J. Sass, M. G. Allan, H. Seto, C. J. Thompson, and S. Grzesiek. 2003. Structural basis for antibiotic recognition by the TipA class of multidrug-resistance transcriptional regulators. EMBO J. 22:1824-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kammann, M., J. Laufs, J. Schell, and B. Gronenborn. 1989. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR). Nucleic Acids Res. 17:5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidd, S. P., A. J. Potter, M. A. Apicella, M. P. Jennings, and A. G. McEwan. 2005. NmlR of Neisseria gonorrhoeae: a novel redox responsive transcription factor from the MerR family. Mol. Microbiol. 57:1676-1689. [DOI] [PubMed] [Google Scholar]
- 26.Kloczkowski, A., K. L. Ting, R. L. Jernigan, and J. Garnier. 2002. Combining the GOR V algorithm with evolutionary information for protein secondary structure prediction from amino acid sequence. Proteins 49:154-166. [DOI] [PubMed] [Google Scholar]
- 27.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 28.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p. 439. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 31.Nakano, M. M., T. Hoffmann, Y. Zhu, and D. Jahn. 1998. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J. Bacteriol. 180:5344-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano, M. M., F. Yang, P. Hardin, and P. Zuber. 1995. Nitrogen regulation of nasA and nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J. Bacteriol. 177:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newberry, K. J., and R. G. Brennan. 2004. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 279:20356-20362. [DOI] [PubMed] [Google Scholar]
- 35.Outten, C. E., F. W. Outten, and T. V. O'Halloran. 1999. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J. Biol. Chem. 274:37517-37524. [DOI] [PubMed] [Google Scholar]
- 36.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 38.Schwede, T., J. Koop, N. Guex, and M. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen, T. Z., R. L. Jernigan, J. Garnier, and A. Kloczkowski. 2005. GOR V server for protein secondary structure prediction. Bioinformatics 21:2787-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetzer, K., J. Behlke, and S. Brantl. 1998. Plasmid pIP501 encoded transcriptional repressor CopR binds to its target DNA as a dimer. J. Mol. Biol. 283:595-603. [DOI] [PubMed] [Google Scholar]
- 41.Steinmetzer, K., and S. Brantl. 1997. Plasmid pIP501 encoded transcriptional repressor CopR binds asymmetrically at two consecutive major grooves of the DNA. J. Mol. Biol. 269:684-693. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaughan, C. K., A. M. Buckle, and A. R. Fersht. 1999. Structural response to mutation at a protein-protein interface. J. Mol. Biol. 286:1487-1506. [DOI] [PubMed] [Google Scholar]
- 44.Wray, L. V., Jr., M. R. Atkinson, and S. H. Fisher. 1994. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J. Bacteriol. 176:108-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wray, L. V., Jr., A. E. Ferson, K. Rohrer, and S. H. Fisher. 1996. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:8841-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray, L. V., Jr., and S. H. Fisher. 2005. A feedback-resistant mutant of Bacillus subtilis glutamine synthetase with pleiotropic defects in nitrogen-regulated gene expression. J. Biol. Chem. 280:33298-33304. [DOI] [PubMed] [Google Scholar]
- 47.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2000. Purification and in vitro activities of the Bacillus subtilis TnrA transcription factor. J. Mol. Biol. 300:29-40. [DOI] [PubMed] [Google Scholar]
- 48.Wray, L. V., Jr., J. M. Zalieckas, and S. H. Fisher. 2001. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107:427-435. [DOI] [PubMed] [Google Scholar]
- 49.Wu, T. D., and D. L. Brutlag. 1996. Discovering empirically conserved amino acid substitution groups in databases of protein families. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4:230-240. [PubMed] [Google Scholar]
- 50.Yoshida, K., H. Yamaguchi, M. Kinehara, Y. Ohki Y. Nakaura, and Y. Fujita. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol. Microbiol. 49:157-165. [DOI] [PubMed] [Google Scholar]
- 51.Youn, H., R. L. Kerby, M. Conrad, and G. P. Roberts. 2006. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 281:1119-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 2006. Cross-regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA. J. Bacteriol. 188:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng, Q., C. Stålhandske, M. C. Anderson, R. A. Scott, and A. O. Summers. 1998. The core metal-recognition domain of MerR. Biochemistry 37:15885-15895. [DOI] [PubMed] [Google Scholar]