Abstract
The gene cluster gspCDEFGHIJKLM codes for various structural components of the type II secretion pathway which is responsible for the secretion of heat-labile enterotoxin by enterotoxigenic Escherichia coli (ETEC). In this work, we used a variety of molecular approaches to elucidate the transcriptional organization of the ETEC type II secretion system and to unravel the mechanisms by which the expression of these genes is controlled. We showed that the gspCDEFGHIJKLM cluster and three other upstream genes, yghJ, pppA, and yghG, are cotranscribed and that a promoter located in the upstream region of yghJ plays a major role in the expression of this 14-gene transcriptional unit. Transcription of the yghJ promoter was repressed 168-fold upon a temperature downshift from 37°C to 22°C. This temperature-induced repression was mediated by the global regulatory proteins H-NS and StpA. Deletion mutagenesis showed that the promoter region encompassing positions −321 to +301 relative to the start site of transcription of yghJ was required for full repression. The yghJ promoter region is predicted to be highly curved and bound H-NS or StpA directly. The binding of H-NS or StpA blocked transcription initiation by inhibiting promoter open complex formation. Unraveling the mechanisms of regulation of type II secretion by ETEC enhances our understanding of the pathogenesis of ETEC and other pathogenic varieties of E. coli.
Enterotoxigenic Escherichia coli (ETEC) is a causative agent of severe watery diarrhea. It is a leading cause of mortality in children in less developed countries and is the most common cause of travelers' diarrhea worldwide. The virulence of ETEC strains is directly associated with the ability to express and deliver one or both of two toxic proteins, heat-labile enterotoxin (LT) and heat-stable enterotoxin (17). LT is structurally and functionally homologous to cholera toxin, which, upon entering epithelial cells, elevates cyclic AMP in the host cell and disrupts the functions of ion channels across the intestinal lumen, leading to diarrhea and dehydration (17, 29). The eltAB operon encoding LT is carried on a virulence plasmid (Ent), and the expression of this operon is regulated by the nucleoid structural protein H-NS in response to temperature changes (31, 33).
LT is secreted by the type II protein secretion pathway, which is encoded by the gspCDEFGHIJKLM gene cluster on the chromosome of ETEC strain H10407 (30). This ETEC gene cluster exhibits a high degree of homology with the gene cluster epsCDEFGHIJKLM of Vibrio cholerae, which codes for various structural components of the type II secretion pathway responsible for secretion of cholera toxin (26). E. coli K-12 strains do not produce LT and cannot secrete endogenous proteins. Sequence analysis of E. coli K-12 strains MG1655 and W3110 has shown that the genetic region at min 67, which corresponds to the region containing the ETEC gspCDEFGHIJKLM cluster, carries a large deletion which encompasses eight open reading frames (gspDEFGHIJK) (30). The DNA sequences of the remnants of the type II secretion pathway and the flanking regions in E. coli K-12 are almost identical to those in ETEC (30). In addition, E. coli K-12 carries a cryptic general secretory pathway at min 74.5 (9), but expression of this group of genes is permanently silenced by H-NS (7).
The ETEC gspCDEFGHIJKLM cluster is located downstream of the glc operon encoding proteins involved in the utilization and transport of glycolate (19). Three open reading frames, yghJ, pppA, and yghG, are positioned between the two gene clusters (Fig. 1A). While the function of the proteins encoded by yghJ and yghG is unknown, the pppA gene codes for a protease (prepilin peptidase). The PppA protein from E. coli K-12 is able to process the precursors of the Neisseria gonorrhoeae type IV pilin PilE and the Klebsiella oxytoca type IV pseudopilin PulG (8). The yghJ-pppA-yghG-gspCDEFGHIJKLM gene cluster is present in other pathogenic E. coli strains, including enteropathogenic E. coli (EPEC), enteroinvasive E. coli, enteroaggregative E. coli, and uropathogenic E. coli strains, but the contribution of the type II secretion pathway to the virulence of these strains, if any, is not known.
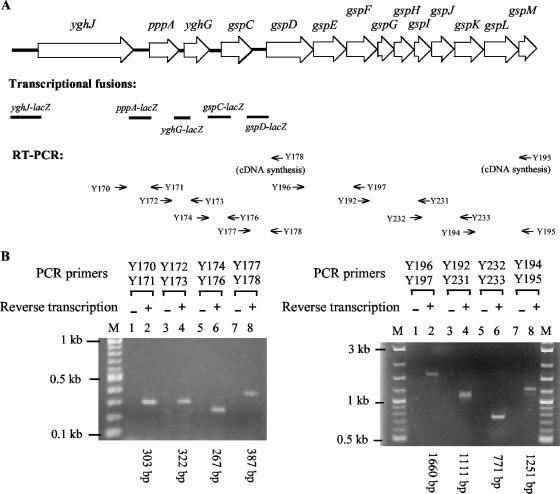
FIG. 1.
Analysis of transcriptional organization of the yghJ-pppA-yghG-gspCDEFGHIJKLM gene cluster of ETEC by transcriptional fusion (A) and by RT-PCR (B). (A) The 14 genes of the cluster are indicated by open arrows (not to scale). The DNA sequence between the downstream region of the yghJ gene and the gspM gene is available in the GenBank database (accession no. AY056599). The sequence of the yghJ upstream region (see Fig. 2B) was determined in this study. The DNA fragments used for constructing different lacZ fusions are indicated by solid lines. The locations of various oligonucleotide primers used for RT-PCR analysis are indicated by thin arrows below the map. cDNA molecules obtained using primer Y178 were used as a template in PCRs involving primer pairs Y170-Y171, Y172-Y173, Y174-Y176, and Y177-Y178, and cDNA molecules obtained using primer Y195 were used as a template in PCRs involving primer pairs Y196-Y197, Y192-Y231, Y232-Y233, and Y194-Y195. (B) RT-PCR analysis was carried out using total RNA from ETEC strain H10407 and primers illustrated in panel A. The paired PCR primers and the sizes of the PCR products are indicated. Control experiments (−) were carried out using the total RNA as the template. A 2-log DNA ladder (Biolabs) was used for DNA size standards (M).
Although the type II secretion pathway plays a key role in pathogenesis of ETEC and V. cholerae, little is known about how the pathway is expressed and regulated in these pathogens. In this study, we carried out a series of experiments to determine the transcriptional organization of this gene cluster, to locate the promoter which governs its expression, and to identify proteins that are involved in regulating its transcription.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides, and reagents.
The E. coli K-12 strains used in this study were as follows. E. coli DH5α [φ80d lacZΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1] (Zymo Research Corp., Glasgow, Scotland) was used for cloning. E. coli MC4100 [Δ(argF-lac)U169 rpsL150 relA araD139 flb5301 deoC1 ptsF25] (1), E. coli PD145 (MC4100 hns-205::Tn10) (3), and E. coli BSN29 (MC4100 trp::Tn10 Δhns stpA60::Kmr) (14) were used as hosts to analyze the expression of various lacZ fusions. The prototypical ETEC strain H10407 (5) was used as a source of DNA for PCR amplification of yghJ, pppA, yghG, gspC, and gspD fragments. Plasmid pMU2385 (32) was used to construct various lacZ fusions. Oligonucleotides used in this study are listed in Table 1. Trimethoprim was used at a final concentration of 40 μg/ml in Luria broth. Restriction enzymes and chemicals were purchased commercially. Purified RNA polymerase holoenzyme was purchased from USB Corp. The purified H-NS and StpA proteins used in the in vitro experiments were provided by A. Ishihama (Nippon Institute for Biological Science, Tokyo, Japan).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Purposea |
|---|---|---|
| Y006 | 5′TTTGGATCCCAGCGGAAACTGGATCGTACC3′ | PCR, gspC fragment |
| Y008 | 5′TTTGGATCCCAGGAGAATCATTCATCGTG3′ | PCR, gspD fragment |
| Y009 | 5′TTTAAGCTTATTGAGCGGGGTCATGGTGCG3′ | PCR, gspD fragment |
| Y119 | 5′TTTGGATCCGTTTTACGTTCAGATAACCTGC3′ | PCR, yghJ fragment (171-622) |
| Y121 | 5′TTTAAGCTTGCTGGCTGATTAATTGCAC3′ | PCR, gspC fragment |
| Y146 | 5′TTTAAGCTTCAACCGGCTAACAGGGTTGC3′ | PCR, yghJ fragment (1-622) |
| Y147 | 5′TTTGGATCCGCCAGCGTTATCGGCATTAT3′ | PCR, yghJ fragment (1-622) |
| Y152 | 5′TTGGATCCACCTGTTTTCTATCAATGATGTC3′ | PCR, yghJ fragment (427-622) |
| Y153 | 5′TTTAAGCTTCCCCTATACGATGTCTATC3′ | PCR, yghJ fragment (171-410) |
| Y170 | 5′GGTTACCGAGCATAAGATGTCTG3′ | RT-PCR |
| Y171 | 5′GCCGATGATCAATCCTCCGAC3′ | RT-PCR |
| Y172 | 5′ATCAACCACACTGCCTTTTGGAC3′ | RT-PCR |
| Y173 | 5′TGCCAGCACTAAGGTATATCCC3′ | RT-PCR |
| Y174 | 5′CACGTACAGGTAACCAGTATCAG3′ | RT-PCR |
| Y176 | 5′CGCCAGAGTGAATGCGCCA3′ | RT-PCR |
| Y177 | 5′GCTCTGATGCGGCAGTTACC3′ | RT-PCR |
| Y178 | 5′CATTGAGCGGGGTCATGGTG3′ | RT-PCR |
| Y182 | 5′GGCATTATCACCCTGCTGCAG3′ | PCR, yghJ fragment |
| Y184 | 5′GTTTTGCGTTCAGATAACCTGCTAA3′ | PCR, yghJ fragment |
| Y185 | 5′ACGACATCATTGATAGAAAACAGGTG3′ | PCR, yghJ fragment |
| Y187 | 5′GCGCTCAAAATAGCCGCTAAAAGC3′ | PCR, yghJ fragment |
| Y192 | 5′GAGCCGATGCTGCAACTGAA3′ | RT-PCR |
| Y194 | 5′GGCATCAGACAGGAGAAAGTG3′ | RT-PCR |
| Y195 | 5′GGCGTTCACGCCATTGCTG3′ | RT-PCR |
| Y196 | 5′GATGCCGCACACTGCACAGCC3′ | RT-PCR |
| Y197 | 5′GCGGAATCCGCCTCAATCATACC3′ | RT-PCR |
| Y203 | 5′TGGATCCGGTTACCGAGCATAAGATGTCTG3′ | PCR, pppA fragment |
| Y205 | 5′GAAGCTTGGGCATCGCCGCAGGGTATTG3′ | PCR, pppA fragment |
| Y208 | 5′TGGATCCAACCACACTGCCTTTTGGACCG3′ | PCR, yghG fragment |
| Y209 | 5′GAAGCTTGTCGTGCCGCTACTTTGTGC3′ | PCR, yghG fragment |
| Y231 | 5′CGAGCATCACTTCCAGCAAGGT3′ | RT-PCR |
| Y232 | 5′GGATATTGAAGTCAGCCTTCACG3′ | RT-PCR |
| Y233 | 5′GCGTGATGGTTACCAGCATCAT3′ | RT-PCR |
Numbers in parentheses signify positions of fragments (for numbering, see Fig. 2B).
DNA manipulation techniques.
Standard recombinant DNA procedures were used as described by Sambrook and Russell (24). Plasmids were purified using a Wizard Plus SV miniprep DNA purification system (Promega Corp., Madison, WI). DNA was sequenced with a model 377 DNA sequencer and ABI Big Dye terminators (Perkin-Elmer Corp., Wellesley, MA). PCRs were carried out using PCR Mastermix from Promega. A TOPO TA cloning kit (Invitrogen, Carlsbad, CA) was routinely used for the cloning of various PCR fragments.
Reverse transcription-PCR (RT-PCR).
RNA was isolated from ETEC strain H10407 grown in Luria broth at 37°C by use of an RNeasy purification kit (QIAGEN GmbH, Hilden, Germany). RNA samples (10 μg) were treated with 8 U of DNase I (New England Biolabs, Ipswich, MA) for 2 h at 37°C in the presence of RNasin (Promega) to remove contaminating DNA. cDNA was synthesized as follows. Total RNA (1 μg) was mixed with specific primer Y178 or Y195. The samples were denatured at 65°C for 5 min and then chilled on ice. Following the addition of deoxynucleoside triphosphates, dithiothreitol, RNasin, and Superscript III reverse transcriptase (Invitrogen), reactions were carried out at 54°C for 2 h. PCRs were performed using the cDNA samples as templates in the presence of the specific primer pairs shown in Fig. 1A and Table 1. Control experiments were done using the total RNA as the template.
Construction of lacZ transcriptional fusions.
The various lacZ transcriptional fusions were constructed by PCR amplification of desirable DNA fragments using ETEC (H10407) chromosomal DNA as the template and the primers described in Table 1. Each of the PCR fragments, which were flanked by a BamHI site and a HindIII site, was cloned into TOPO TA cloning vector and sequenced. The fragments were then excised from the TOPO derivatives and cloned into the BamHI and HindIII sites of the single-copy plasmid pMU2385 to create lacZ transcriptional fusions.
β-Galactosidase assay.
β-Galactosidase activity was assayed as described by Miller (16). Specific activity was expressed in units described therein. The data are the results of at least three independent assays.
In vitro transcription.
Runoff transcription assays were performed by using a method based on the standard single-round conditions described by Igarashi and Ishihama (13). The reaction mixtures contained a linear DNA template (approximately 300 ng) and 1 U of RNA polymerase. The samples were incubated at 37°C for 25 min in a total volume of 35 μl of in vitro transcription buffer (50 mM Tris-HCl [pH 7.8], 50 mM NaCl, 3 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM dithiothreitol, and 25 μg/ml bovine serum albumin). Following the incubation, 15 μl of start solution (containing 1× transcription buffer with 0.67 mg/ml heparin, 0.53 mM each of ATP, CTP, and GTP, 0.053 mM UTP, and 3 μCi of [α-32P]UTP) was added to initiate RNA synthesis. Transcription was allowed to proceed for 5 min before the reaction was terminated by phenol extraction. A portion of each sample (15 μl) was mixed with sequencing dye mix and analyzed on a 6% sequencing gel.
EMSA.
The 32P-labeled yghJ fragment used in an electrophoretic mobility shift assay (EMSA) was generated as follows. The oligonucleotide primer Y182 (Table 1) was labeled with 32P at its 5′ end by using [γ-32P]ATP and T4 polynucleotide kinase. The yghJ fragment (−308 to +282) was amplified by PCR using the 32P-labeled primer 32P-Y182, unlabeled primer Y187 (Table 1), and TOPO-yghJ (1 to 622). The EMSA was carried out as described by Dole et al. (4). Following the incubation of the end-labeled yghJ fragment with various amounts of purified H-NS or StpA protein at 22°C for 20 min, the samples were analyzed by electrophoresis on 5% native polyacrylamide gels (37.5:1). The electrophoresis was carried out at 4°C for approximately 12 h at 10 V/cm.
KMnO4 footprinting.
A portion (0.5 μg) of the linear yghJ fragment (−308 to +282) was incubated in 20 μl of in vitro transcription buffer in the presence or absence of H-NS and StpA protein at 22°C for 10 min. RNA polymerase (1 U) was then added to reaction mixtures. Following a further incubation at 30°C for 30 min to allow open complex formation, the samples were treated with 2.5 μl of KMnO4 (80 mM) for 3 min at room temperature. The reaction was then quenched with 2 μl of β-mercaptoethanol (14.7 M). Following purification of the samples on a MicroSpin column S-200 (GE Healthcare, Buckinghamshire, England), the DNA was cleaved with piperidine at 90°C for 20 min. Primer extension reactions were carried out in a thermal cycler for 25 cycles, using PCR Mastermix (Promega) and 32P-labeled primer (Y184). The extension products were analyzed on a 6% denaturing polyacrylamide gel next to a GA ladder. The latter was made by a Maxam and Gilbert sequencing method using the yghJ fragment, which was generated by PCR using primers 32P-Y184 and Y187.
Nucleotide sequence accession number.
The novel sequence reported here has been assigned GenBank accession number 826429.
RESULTS
Identification of a promoter region and analysis of transcriptional organization.
In the ETEC yghJ-pppA-yghG-gspCDEFGHIJKLM gene cluster, the coding sequences of the last 10 genes (from gspD to gspM) are closely linked, with no intervening gaps between the 10 structural genes. However, intergenic sequences of various lengths are present upstream of yghJ (484 bp), pppA (233 bp), yghG (65 bp), gspC (146 bp), and gspD (239 bp) (Fig. 1A). To determine if these regions harbor promoters, we amplified five DNA fragments, which cover the upstream regions as well as some coding sequences of the yghJ, pppA, yghG, gspC, and gspD genes, respectively, by PCR using total cellular DNA from the prototypical ETEC strain H10407 as the template. These four fragments were each cloned into the single-copy vector pMU2385 (32) to create yghJ-, pppA-, yghG-, gspC-, and gspD-lacZ transcriptional fusions (Fig. 1A). The resulting pMU2385 derivatives and the control plasmid (pMU2385) were each transformed into E. coli K-12 strain MC4100, and β-galactosidase levels in the transformants were assessed following growth in Luria broth at 37°C. Moderately strong promoter activity was detected in the upstream region of yghJ (135 Miller units), whereas relatively weak promoter activity was observed for the pppA (34 Miller units) and yghG (16 Miller units) fragments. No significant promoter activity was found for the gspC and gspD fragments (Table 2).
TABLE 2.
Promoter activities of various lacZ transcriptional fusionsa
| Vector or lacZ fusion | β-Galactosidase activity (Miller units) |
|---|---|
| Vector | 0.3 ± 0.1 |
| yghJ-lacZ | 135 ± 15 |
| pppA-lacZ | 34 ± 5 |
| yghG-lacZ | 16 ± 4 |
| gspC-lacZ | 0.8 ± 0.1 |
| gspD-lacZ | 1.1 ± 0.3 |
E. coli MC4100 derivatives carrying different lacZ fusions were grown in Luria broth at 37°C. β-Galactosidase activity is the mean ± standard deviation of three independent experiments.
To test if the 14-gene cluster (yghJ-pppA-yghG-gspCDEFGHIJKLM) forms one transcription unit, we carried out an RT-PCR analysis. Total RNA was isolated from ETEC strain H10407 grown in Luria broth at 37°C. Following reverse transcription, PCRs were performed using eight sets of primers that cover the entire gene cluster (Fig. 1A). Amplification products of appropriate lengths were obtained only after reverse transcription (Fig. 1B), demonstrating that the yghJ-pppA-yghG-gspCDEFGHIJKLM genes are cotranscribed. Taken together, these results indicate that one or more promoters in the upstream region of the yghJ gene play a major role in the expression of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster.
Transcription of the yghJ promoter is repressed by H-NS and StpA proteins in response to a temperature downshift.
In ETEC, expression of the eltAB operon, which codes for the A and B subunits of LT, is repressed by the regulatory protein H-NS when cells are grown at low temperature, e.g., 22°C (31, 33). To determine if transcription of the yghJ promoter, which is responsible for the expression of the LT-specific type II secretion pathway, is also regulated by H-NS and its paralog, StpA, the pMU2385 derivative which carried the yghJ-lacZ transcriptional fusion was transformed into isogenic E. coli K-12 strains MC4100 (wild type), PD145 (hns), and BSN29 (hns stpA). β-Galactosidase levels were assessed for each of the transformants grown in Luria broth at 37°C or 22°C.
The results showed that at 37°C the yghJ-lacZ fusion expressed 135 units of β-galactosidase activity in the wild-type host, MC4100 (Table 3). Although the level of expression from the yghJ promoter was unchanged (127 units) in the hns background (PD145), there was a twofold increase in yghJ transcription (269 units) in the hns stpA background (BSN29) (Table 3).
TABLE 3.
Temperature-mediated repression of yghJ-lacZ fusion by H-NS and StpA
| Temp (°C) | Sp act of β-galactosidase (Miller units)a produced by strain:
|
||
|---|---|---|---|
| MC4100 (wild type) | PD145 (hns) | BSB29 (hns stpA) | |
| 37 | 135 ± 15 | 127 ± 25 | 269 ± 41 |
| 22 | 0.8 ± 0.2 | 17 ± 3 | 88 ± 11 |
β-Galactosidase activity is the mean ± standard deviation of three independent experiments.
At 22°C, the levels of β-galactosidase activity dropped from 135 (at 37°C) to 0.8 units in the wild-type strain (MC4100), which represents a 168-fold repression of yghJ transcription. In strain PD145 (hns), transcription from the yghJ promoter was 21-fold higher than in MC4100, and in strain BSN29 (hns stpA), the levels of yghJ expression were even higher, at 88 units of β-galactosidase activity, giving a 110-fold derepression compared with levels for MC4100.
The results of this analysis show that transcriptional expression of the yghJ promoter is strongly repressed at 22°C and that the regulatory proteins H-NS and StpA play a key role in the temperature-dependent repression of yghJ. In addition, these results indicate that the 622-bp yghJ fragment which was used for constructing the yghJ-lacZ fusion contains a cis-acting element(s) involved in the repression of yghJ by H-NS and StpA.
Deletion analysis of the yghJ regulatory region.
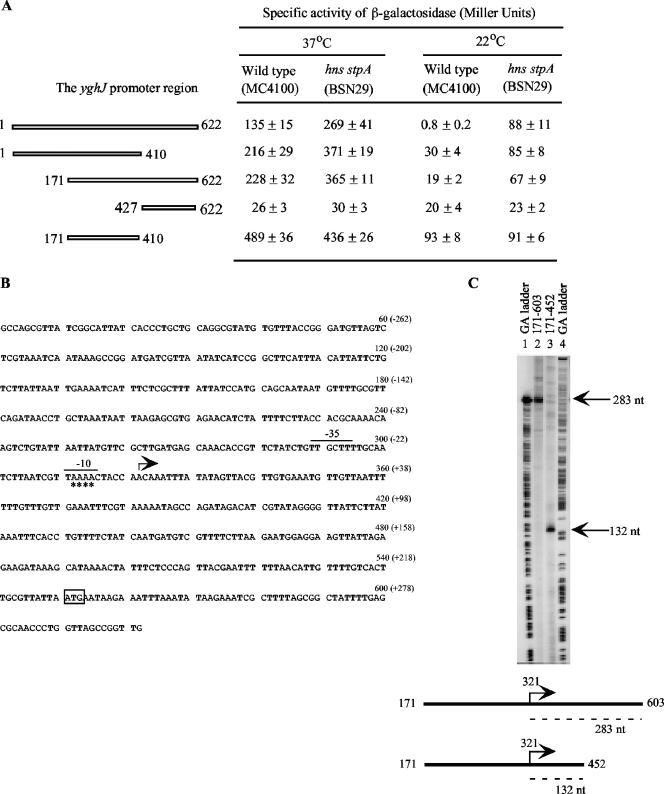
In order to characterize the regulatory region of the yghJ gene, four PCR fragments which contained various deletions were generated (Fig. 2A). Each of the mutant DNA fragments was transcriptionally fused with the lacZ structural gene, and the effects of the deletions on yghJ transcription and regulation were examined by β-galactosidase analysis.
FIG. 2.
Deletion analysis of the yghJ regulatory region and identification of the yghJ promoter. (A) Strains were grown in Luria broth at 22°C or 37°C. The results are the means ± standard deviations of three independent experiments. Numbering of various yghJ fragments is in accordance with that in panel B. (B) The start site of transcription of the yghJ promoter is marked with an angled arrow. The −35 and −10 regions of the yghJ promoter are indicated by lines above the sequence. The numbers in parentheses represent positions relative to the start site of transcription. The positions that were sensitive to KMnO4 treatment during promoter opening are marked with asterisks (see also Fig. 4). The putative translational start codon (ATG) for YghJ is boxed. (C) Runoff in vitro transcription was performed using the two linear yghJ templates 171-603 and 171-452. The 283-nucleotide (nt) and 132-nt transcripts produced by the two templates are indicated. The GA DNA ladders were generated by Maxam-Gilbert sequencing of the yghJ fragments 171-603 and 171-452.
The results showed that mutants 1-410 and 171-622, which had lost 212 bp from the 3′ end and 170 bp from the 5′ end, respectively, exhibited similar phenotypes. Compared to that of the full-length yghJ fragment (bp 1 to 622), the promoter activities of these two mutants showed a minor increase (approximately 1.5-fold) from those of the the wild-type and the hns stpA hosts at 37°C and a major enhancement in β-galactosidase expression in the wild-type background at 22°C, which approximated 38- and 24-fold derepression of yghJ transcription. However, the levels of transcription of the full-length fragment and the 1-410 and 171-622 mutants were not significantly different from each other in the hns stpA background.
Deletion of 426 bp from the 5′ end (mutant 427-622) brought about a severe defect in promoter activity. Moreover, the low levels of transcription from this mutant fragment were no longer significantly repressed by H-NS and StpA at 22°C.
Mutant 171-410, which lacks 170 and 212 bp from the 5′ and 3′ ends, exhibited a sharp increase in transcription at 37°C. The levels of expression by this mutant were increased to 489 and 436 units in the wild-type and hns stpA strains, respectively (Fig. 2A). At 22°C, transcription of this mutant construct was derepressed in the wild-type strain to the same levels as in the hns stpA background.
These results indicate the presence of a major promoter between positions 171 and 410. The finding that deletions from either the 5′ end or the 3′ end of the yghJ promoter region caused significant derepression at 22°C in the wild-type host suggested that the entire yghJ regulatory region is required for efficient repression by the H-NS and StpA proteins.
Mapping the start site of transcription by in vitro transcription.
A single-round in vitro transcription experiment was performed to map the start site of yghJ transcription. In this assay, two linear yghJ fragments encompassing positions 171 and 603 and 171 and 452, respectively, were used as DNA templates (Fig. 2B and C). In the presence of E. coli σ70 RNA polymerase, each template synthesized a single transcript, indicating the presence of only one promoter in the DNA fragments used (Fig. 2C). Transcription from templates 171-452 and 171-603 produced 132-base and 283-base transcripts, respectively. The size difference of the two transcripts matched the difference in length of the two DNA templates at the 3′ ends. Based on these data, the start site of transcription for yghJ was mapped to the adenine residue at position 321 that is 229 bp upstream of the putative start codon for the YghJ protein (Fig. 2B). Inspection of the sequence immediately upstream of the start site of transcription (here designated position +1) revealed the presence of putative −10 (TAAAAC) and −35 (TTGCTT) hexamers (separated by 16 bp) for a σ70 promoter (Fig. 2B).
Binding of H-NS and StpA to the yghJ promoter region.
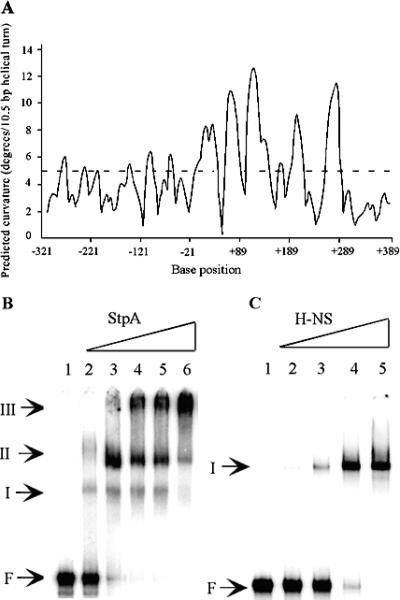
Both the H-NS and StpA proteins are known to have a strong preference for binding to curved DNA structures. To examine if there was any intrinsic DNA curvature in the yghJ promoter region, we analyzed the yghJ upstream sequence by using the program BEND-IT (http://hydra.icgeb.trieste.it/∼kristian/dna/bend_it.html). The yghJ promoter region is predicted to be highly curved (Fig. 3A).
FIG. 3.
In silico prediction of intrinsic curvature of the yghJ regulatory region and analysis of the binding of H-NS and StpA to the yghJ regulatory region by EMSA. (A) The BEND-IT program (http://hydra.icgeb.trieste.it/∼kristian/dna/bend_it.html) was used for DNA curvature analysis. Numbering of base positions is relative to the start site of transcription from the yghJ promoter. Regions with >5 degrees per helical turn of DNA (dashed line) represent curved sequences. (B) Samples in lanes 1 to 6 contained 0, 15.6, 31.3, 62.5, 125, and 250 nM StpA protein, respectively. (C) Samples in lanes 1 to 5 contained 0, 31.3, 62.5, 125, and 250 nM H-NS protein, respectively. F, free DNA.
An EMSA was carried out to test if the H-NS and StpA proteins can bind directly to the yghJ promoter region. A 32P-labeled yghJ fragment spanning the region between positions −308 and +282 was generated by PCR. Following incubation of the DNA fragment with various amounts of purified H-NS or StpA protein for 20 min at 22°C, the reaction mixtures were analyzed on native polyacrylamide gels. Retarded bands representing H-NS-DNA and StpA-DNA complexes were formed at low concentrations of H-NS (31.3 to 250 nM) and StpA (15.6 to 250 nM) (Fig. 3B and C). In contrast, at the same range of concentrations, neither H-NS nor StpA could form a stable protein-DNA complex with an internal noncurved fragment (+129 to +330) from the ETEC eltA gene (33) (data not shown).
In the presence of StpA, three protein-DNA complexes (I, II, and III in Fig. 3B) were formed. At the lowest StpA concentration (15.6 nM), only a very small proportion of the DNA fragment was shifted to form complexes I and II. As the amounts of StpA increased, more DNA was shifted and the slower-migrating complexes (II and III) became predominant. Only a single retarded band (I) was seen in the presence of H-NS (Fig. 3C). Increased amounts of the complex were formed as the concentration of H-NS was raised from 31.3 to 250 nM. Judging by the concentrations of the StpA and H-NS proteins at which the same amounts of DNA were shifted, it appeared that the DNA-binding affinity of StpA was about four times higher than that of H-NS.
H-NS and StpA inhibit open complex formation at the yghJ promoter.
To study the molecular details of the H-NS- and StpA-mediated regulation of the yghJ promoter, we performed a KMnO4 footprinting experiment. The linear yghJ fragment (−308 to +282) was incubated with either the H-NS or the StpA protein (500 nM) at 22°C for 15 min. This was followed by the addition of E. coli RNA polymerase into the reaction. The samples were incubated for a further 25 min at 30°C to allow the formation of open complexes. After the treatment with KMnO4, primer extension was carried out to measure the presence of open complexes. In the presence of RNA polymerase, the primer extension products representing cleavages at positions −7, −8, −9, and −10 of the template strand were observed (Fig. 4). The presence of either H-NS or StpA was able to block the formation of the transcribing bubble.
FIG. 4.
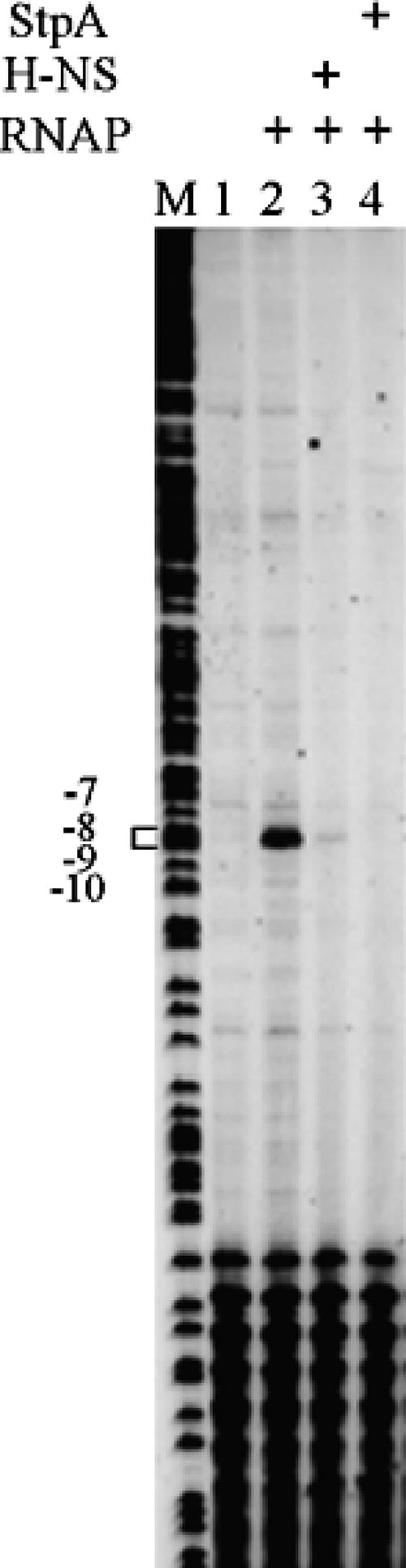
KMnO4 footprinting analysis of the effect of H-NS and StpA on open complex formation of the yghJ promoter. The linear yghJ fragment (−308 to +282) was incubated with 1 U of E. coli RNA polymerase (RNAP) (USB Corp.) in the absence or presence of StpA (500 nM) or H-NS (500 nM). The residues sensitive to oxidation by KMnO4 are indicated. M, GA DNA ladder.
DISCUSSION
In this study, we investigated the expression and regulation of the genes responsible for the production of the type II secretion pathway from the prototypical ETEC strain H10407. We found that the gsp cluster together with the three open reading frames located immediately upstream constitute one transcriptional unit, with a σ70 promoter identified at the start of the transcriptional unit playing a major role in the expression of this pathway. Expression of this operon was tightly controlled by the global regulatory proteins H-NS and StpA in response to low temperature. The temperature-dependent repression was achieved by a specific interaction between the regulatory proteins and their DNA target in the regulatory region of the yghJ promoter.
The first open reading frame (yghJ) in the greater gsp cluster encodes a putative lipoprotein of 1,520 amino acid residues. YghJ shares extensive homology (64%) with an accessory colonization factor, AcfD, of V. cholerae, which is associated with bacterial virulence (21). A proteomic analysis has indicated that the YghJ protein is required for normal functionality of the type II secretion pathway in E. coli (D. Baldi et al., unpublished data). Downstream of yghJ is the pppA gene coding for the prepilin peptidase. Homologues of the PppA protein from the bacterial pathogens Pseudomonas aeruginosa and V. cholerae are required for processing of the type IV prepilin subunits and modification of the prepilin-like components of the type II secretion pathway, including GspG, GspH, GspI, GspJ, and GspK (25). Given the functional relationship of YghJ and PppA to the type II secretion pathway, it is not surprising that the yghJ and pppA genes and the gsp genes have evolved to form a single operon and that their transcription is coordinately regulated. Studies of pppA expression in E. coli K-12 have suggested that a promoter upstream of yghJ drives at least part of pppA transcription and that genes of the cluster yghJ-pppA-yghG-gspCLM are cotranscribed (8).
The identification of weak promoter activity in the upstream region of the ETEC pppA gene is consistent with findings made with the pppA gene of E. coli K-12 (8). However, the weak activity of this promoter suggests that it plays only a relatively minor role in the expression of pppA and the type II secretion pathway in ETEC. Peptidase activity encoded by the pppA gene of E. coli K-12 was detectable in strains grown only at or above 37°C but not at 30°C. Analysis using a translational pppA-lacZ fusion demonstrated fourfold thermoregulation at the level of translation (8). The identification of a much stronger upstream promoter (the yghJ promoter) whose expression is temperature dependent suggests that the thermoinduction of pppA expression occurs not only at the level of translation but also at the level of transcription in both E. coli K-12 and ETEC.
The E. coli K-12 counterpart of the ETEC gspC gene has been designated ecfA, following the identification of a putative σE promoter near the start of the open reading frame (2). The −35 and −10 regions of the proposed σE promoter of ecfA contain the sequences AAAATT and GCTGA, respectively, and are separated by a 17-bp spacer (2). However, this putative E. coli K-12 σE promoter was not among the members of the σE regulon identified in a separate genetic study (22). Recently, Rhodius et al. (23) reported the composition of the σE regulons in E. coli K-12, as well as those in other gram-negative bacteria, but were unable to confirm the function of the putative ecfA promoter in E. coli K-12. The corresponding region of the ETEC gspC gene contains the same sequence as the proposed ecfA promoter of E. coli K-12, except for a C-to-T change at the second position of the spacer. Using a gspC-lacZ transcriptional fusion, we were unable to detect any basal level of transcription from the gspC upstream region at 37°C (Table 2) or 42°C (results not shown). This suggests that either the putative σE promoter is also not functional in ETEC or its expression requires stimulation by a transcriptional regulator(s). Nevertheless, even if this putative promoter is active under certain conditions in ETEC, it will contribute to the expression of genes only downstream of gspC as the proposed start site of transcription is positioned downstream of the putative start codon for GspC.
Expression of the yghJ promoter was strongly repressed at 22°C in the wild-type E. coli K-12 strain MC4100, but temperature-mediated repression was significantly relieved in an hns-deficient host and was further alleviated in an hns stpA mutant strain (Table 3). Similarly, deleting cis-acting elements located upstream and downstream of the yghJ core promoter also caused derepression (Fig. 2A). At 37°C, however, only two- to threefold repression of yghJ transcription by H-NS and StpA was observed (Fig. 2A). In an hns stpA host, levels of expression from the full-length yghJ fragment, as well as from the three truncated yghJ fragments, were three to four times lower at 22°C than at 37°C. It is possible that E. coli RNA polymerase functions at a suboptimal capacity at this promoter at low temperatures. Alternatively, an additional regulatory protein(s), which binds closer to the promoter core sequence, may also be involved in the temperature-mediated repression, as in the case of Salmonella enterica, where the Hha protein represses transcription of the hilA gene in concert with H-NS by binding to similar target DNA sequences under low osmolarity (6, 20).
The StpA protein is susceptible to proteolysis by Lon protease (15) and is present at 1/10 the level of H-NS in the E. coli cell (28). However, the binding affinity of StpA for the proU downstream regulatory element as well as for the yghJ promoter region is four times higher than that of H-NS (reference 28 and this study). H-NS can form heteromeric complexes with StpA, which enhances the stability of StpA (15). The mutant hns allele hns-205 codes for a C-terminally truncated H-NS molecule lacking the DNA binding domain (12). Free et al. (10, 11) have shown that StpA can function as a molecular adaptor for this truncated H-NS protein in the repression of the bgl operon. Based on this model, the partial repression of the yghJ promoter observed to occur in strain PD145 (hns-205) (Table 3) may be due to the presence of H-NS(truncated)-StpA hetero-oligomers.
The yghJ promoter region is AT rich (G+C content of 34%). A recent study has shown that H-NS specifically targets AT-rich sequences, resulting in the silencing of virulence genes in Salmonella enterica (18). Our findings indicate that H-NS also binds to the AT-rich and curved sequences within and around the yghJ promoter, leading to the inhibition of open complex formation during transcription initiation. This is in contrast to the situation with the eltAB operon where H-NS binds to silencer sites located within the first structural gene, blocking chain elongation by RNA polymerase (33). The bgl operon contains two separate silencer sites, one upstream of the promoter and the other 600 to 700 bp downstream (27). The binding of H-NS to these two sites inhibits transcription initiation and elongation, resulting in the complete silencing of bgl expression (4). Obviously, by binding to different positions and by interacting with other regulatory proteins, H-NS can generate a wide range of regulatory outcomes that reflect the physiological or pathogenic context of the proteins that it controls.
The type II secretion pathway which is required for the secretion of LT is an important virulence determinant of ETEC (30). However, the yghJ-pppA-yghG-gspCDEFGHIJKLM operon is also present and highly conserved in the genomes of many other pathogenic E. coli strains. A recent study in our laboratory has shown that the type II secretion pathway is expressed by EPEC strain E2348/69 at 37°C and is fully functional in EPEC and required for virulence (D. Baldi et al., unpublished data). As EPEC and other pathogenic E. coli strains do not synthesize LT, elucidation of the role of this operon in these strains would enhance our general understanding of E. coli pathogenicity.
Acknowledgments
We thank A. Ishihama and J. Pittard for strains, plasmids, and proteins used in this study.
This work was supported by program grant no. 284214 and project grant no. 400029 from the Australian National Health and Medical Council. D.L.B. is supported by Peter Doherty fellowship no. 251760 from the Australian National Health and Medical Council.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 2.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 3.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245:255-259. [DOI] [PubMed] [Google Scholar]
- 4.Dole, S., V. Nagarajavel, and K. Schnetz. 2004. The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol. Microbiol. 52:589-600. [DOI] [PubMed] [Google Scholar]
- 5.Evans, D. G., D. J. Evans, Jr., and N. F. Pierce. 1973. Differences in the response of rabbit small intestine to heat-labile and heat-stable enterotoxins of Escherichia coli. Infect. Immun. 7:873-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francetic, O., D. Belin, C. Badaut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francetic, O., S. Lory, and A. P. Pugsley. 1998. A second prepilin peptidase gene in Escherichia coli K-12. Mol. Microbiol. 27:763-775. [DOI] [PubMed] [Google Scholar]
- 9.Francetic, O., and A. P. Pugsley. 1996. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J. Bacteriol. 178:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-917. [DOI] [PubMed] [Google Scholar]
- 11.Free, A., R. M. Williams, and C. J. Dorman. 1998. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J. Bacteriol. 180:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulton, C. S., A. Seirafi, J. C. Hinton, J. M. Sidebotham, L. Waddell, G. D. Pavitt, T. Owen-Hughes, A. Spassky, H. Buc, and C. F. Higgins. 1990. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell 63:631-642. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi, K., and A. Ishihama. 1991. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 65:1015-1022. [DOI] [PubMed] [Google Scholar]
- 14.Johansson, J., B. Dagberg, E. Richet, and B. E. Uhlin. 1998. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J. Bacteriol. 180:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1974. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 19.Nunez, M. F., M. T. Pellicer, J. Badia, J. Aguilar, and L. Baldoma. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147:1069-1077. [DOI] [PubMed] [Google Scholar]
- 20.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357:373-386. [DOI] [PubMed] [Google Scholar]
- 21.Peterson, K. M., and J. J. Mekalanos. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezuchova, B., H. Miticka, D. Homerova, M. Roberts, and J. Kormanec. 2003. New members of the Escherichia coli sigma E regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the sigma E stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 25.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, and V. J. DiRita. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnetz, K. 1995. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 14:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenfield, J. M., C. M. Burns, C. F. Higgins, and J. C. Hinton. 2001. The nucleoid-associated protein StpA binds curved DNA, has a greater DNA-binding affinity than H-NS and is present in significant levels in hns mutants. Biochimie 83:243-249. [DOI] [PubMed] [Google Scholar]
- 29.Spangler, B. D. 1992. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol. Rev. 56:622-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tauschek, M., R. J. Gorrell, R. A. Strugnell, and R. M. Robins-Browne. 2002. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7066-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trachman, J. D., and W. K. Maas. 1998. Temperature regulation of heat-labile enterotoxin (LT) synthesis in Escherichia coli is mediated by an interaction of H-NS protein with the LT A-subunit DNA. J. Bacteriol. 180:3715-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, J., J. S. Hwang, H. Camakaris, W. Irawaty, A. Ishihama, and A. J. Pittard. 2004. Mode of action of the TyrR protein: repression and activation of the tyrP promoter of Escherichia coli. Mol. Microbiol. 52:243-256. [DOI] [PubMed] [Google Scholar]
- 33.Yang, J., M. Tauschek, R. Strugnell, and R. M. Robins-Browne. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199-1208. [DOI] [PubMed] [Google Scholar]