Abstract
The plasmids of the members of the Bacillus cereus sensu lato group of organisms are essential in defining the phenotypic traits associated with pathogenesis and ecology. For example, Bacillus anthracis contains two plasmids, pXO1 and pXO2, encoding toxin production and encapsulation, respectively, that define this species pathogenic potential, whereas the presence of a Bt toxin-encoding plasmid defines Bacillus thuringiensis isolates. In this study the plasmids from B. cereus isolates that produce emetic toxin or are linked to periodontal disease were sequenced and analyzed. Two periodontal isolates examined contained almost identical ∼272-kb plasmids, named pPER272. The emetic toxin-producing isolate contained one ∼270-kb plasmid, named pCER270, encoding the cereulide biosynthesis gene cluster. Comparative sequence analyses of these B. cereus plasmids revealed a high degree of sequence similarity to the B. anthracis pXO1 plasmid, especially in a putative replication region. These plasmids form a newly defined group of pXO1-like plasmids. However, these novel plasmids do not contain the pXO1 pathogenicity island, which in each instance is replaced by plasmid specific DNA. Plasmids pCER270 and pPER272 share regions that are not found in any other pXO1-like plasmids. Evolutionary studies suggest that these plasmids are more closely related to each other than to other identified B. cereus plasmids. Screening of a population of B. cereus group isolates revealed that pXO1-like plasmids are more often found in association with clinical isolates. This study demonstrates that the pXO1-like plasmids may define pathogenic B. cereus isolates in the same way that pXO1 and pXO2 define the B. anthracis species.
The different species comprising the Bacillus cereus sensu lato group are largely defined by differences in plasmid-encoded features, while the chromosomes have been shown to be similar in both gene content and gene order (31). The current species concept defines Bacillus anthracis as containing two plasmids, pXO1 and pXO2, that encode for the tripartite toxin and the poly-d-γ-glutamic acid capsule, respectively (28, 29). Bacillus thuringiensis isolates contain plasmids that encode various isoforms of the Bt toxin (36). The plasmid profile of Bacillus cereus sensu stricto is extremely variable, and no well-defined conserved members have been identified that could delineate the species. These plasmid-based species definitions have resulted in the classification of members of the B. cereus group that are not valid when molecular typing is applied and the suggestion that these three species should be regarded as a single species (18, 19, 31).
B. cereus is the causative agent of a number of human ailments, including two forms of food poisoning and local or systemic diseases such as endophthalmitis, endocarditis, meningitis, osteomyelitis, and periodontitis. Cases of B. cereus associated with wound infection and septicemia are rare; however, an increasing number of infections in immunocompromised patients have been documented (11, 24). The two distinct types of gastrointestinal disease, termed emetic and diarrheal, are based on the clinical presentation (11, 24) and have been attributed to the production of toxins by B. cereus (37). The diarrheal disease has been characterized on a molecular level and shown to be associated with the production of two peptide-based toxins: the tripartite hemolysin BL (HBL) (6) and the nonhemolytic enterotoxin Nhe (15, 26). The emetic disease has been linked to a single toxin known as the emetic toxin or cereulide (1, 2). A number of studies have demonstrated that the strains producing cereulide are restricted to what appears to be a relatively clonal group of B. cereus isolates (1, 7). Nonetheless, recent studies have suggested that there may be more diversity within this group of pathogens than previously thought (4) and a Bacillus weihenstephanensis strain has recently been shown to produce cereulide (41). It has been demonstrated that the emetic toxin biosynthetic gene cluster, containing two nonribosomal peptide synthetase genes, are encoded on a large (>200-kb) plasmid with similarity to pXO1 based on sequence data limited to the region flanking the cereulide biosynthetic cluster (12, 22).
B. cereus isolates that are associated with periodontal disease constitute an intriguing group of isolates because they are poorly characterized at the molecular level. A collection of human isolates was previously characterized by using multilocus enzyme electrophoresis (18) and multilocus sequence typing (20). The majority of the periodontal isolates grouped with other clinical B. cereus isolates and were distinct from the environmental B. cereus and B. thuringiensis (18). In addition, Helgason et al. demonstrated that 51 of 62 clinical isolates carried extrachromosomal elements ranging in size from 15 to 600 kb. Hybridization studies showed that many of the periodontal isolates contained similar plasmid profiles (18). These data suggest that there are distinct plasmid-containing subgroups of B. cereus that are associated with human disease.
Over the past 2 years, numerous B. cereus genome and plasmid sequencing projects have significantly increased our understanding of the biological role of plasmids in B. cereus and questioned the classifications within the B. cereus group. Our group described a 208-kb pXO1-like plasmid, pBC10987, from B. cereus ATCC 10987 that was originally isolated from a dairy source (33). Ivanova et al. (23) reported on the sequence of B. cereus ATCC 14579 and that of an ∼15-kb linear molecule which was shown to be similar to the Bam35 group linear phage (38). A recent study by Hoffmaster et al. described B. cereus G9241, an isolate with unique biochemical and phenotypic traits, from a patient with an anthrax-like disease (21). This isolate harbored pBCXO1, a plasmid that is 99.6% identical to B. anthracis plasmid pXO1. B. cereus G9241 also contained two additional plasmids: a 52-kb linear element with similarity to phages of the B. cereus group, including that of B. cereus ATCC 14579, and pBC210, a large circular plasmid that was not similar to B. anthracis pXO1, pXO2, or any other known B. cereus group plasmid. Although pBC210 did not encode the anthrax poly-γ-d-glutamic acid capsule biosynthetic genes, it did contain a putative polysaccharide biosynthetic gene cluster thought to be responsible for the production of observed encapsulation of B. cereus G9241 (21). The genome sequences of B. thuringiensis serovar Konkukian strain 97-27 and B. cereus E33L (previously referred to as B. cereus ZK) revealed that each contained unique plasmid profiles (16). The B. thuringiensis isolate contained a 77-kb plasmid named pBT9727, whereas B. cereus E33L contained five plasmids ranging in size from ∼5 to 466 kb (named pE33L5, pE33L8, pE33L9, pE33L54, and pE33L466). It is unclear whether the plasmids in B. cereus E33L or B. thuringiensis serovar Konkukian strain 97-27 provide a competitive advantage in the environment or whether they have any link to pathogenicity. These plasmid sequences encompass the known diversity found among the plasmids of the B. cereus group.
To the best of our knowledge, the present study represents the first systematic sequencing and comparative analysis of plasmids from clinically relevant B. cereus isolates. We explore the evolutionary relationship of the large plasmids of the B. cereus group through comparative sequence analysis. A highly conserved core region was found in all pXO1-like plasmids in addition to a less-conserved region. The pathogenicity island contained within the B. anthracis pXO1 plasmid (pXO1-PI) was absent in all of the plasmids from clinical B. cereus isolates; however, in each case it was replaced with a novel sequence, suggesting niche-specific adaptation. Screening of B. cereus group isolates for the presence of these plasmids demonstrates that the pXO1-like plasmids are more often associated with clinical isolates compared to their environmental counterparts. These plasmids could be used to define and characterize these subgroups of clinical B. cereus isolates.
MATERIALS AND METHODS
Bacterial strains.
The large plasmids from B. cereus AH818 and AH820, two periodontal isolates distinct both geographically and temporally, are included in the present study. Briefly, B. cereus AH818 was isolated in September 1995 from a case of apical periodontitis in a 38-year-old female in Curitiba, Brazil, following a root canal procedure. B. cereus AH820 was isolated in October 1995 in Akershus, Norway, from the periodontal pocket of a 76-year-old female with marginal periodontitis. These two isolates were first described by Helgason et al. (18) and shown by hybridization studies to contain a large plasmid of approximately 300 kb. Sequencing revealed that these plasmids, now named pPER272, are ∼272 kb in size (see below and Table 1) .
TABLE 1.
Plasmids examined in this study
| Plasmid | Species and strain | Associated pathogenesis | Size (kb) | Predicted copy no. | pXO1-likee | Origin coordinates | Conserved core coordinates | GenBank accession no. | Source or reference |
|---|---|---|---|---|---|---|---|---|---|
| pXO1 | B. anthracis Ames Ancestor | Anthrax | 181,677 | 1-3a | Y | 55777-55934 | 52066-102495 | AE017336 | 29 |
| pXO2 | B. anthracis Ames Ancestor | Anthrax | 94,830 | 2-5a | N | NAf | NA | AE017335 | 28 |
| pBC210 | B. cereus G9241 | Anthrax-like pneumonia | 209,385b | ∼1 | N | NA | NA | DQ889679 | 21 |
| pBCXO1 | B. cereus G9241 | Anthrax-like pneumonia | 190,861b | ∼1 | Y | 113884-114041 | 113884-114041 | DQ889680 | 21 |
| pBC10987 | B. cereus ATCC 10987 | None; environmental/dairy source | 208,369 | ∼2 | Y | 69201-69108 | 14101-73090 | AE017195 | 33 |
| pE33L466 | B. cereus E33L | Unknown | 466,370 | NDc | N | NA | NA | CP000040 | 31 |
| pE33L54 | B. cereus E33L | Unknown | 53,501 | NDc | N | NA | NA | CP000042 | 31 |
| pBCAH818 (pPER272) | B. cereus AH818 | Periodontal disease | 272,145 | NDd | Y | 56262-56419 | 52377-119432 | DQ889678 | This study |
| pBCAH820 (pPER272) | B. cereus AH820 | Periodontal disease | 272,145 | NDd | Y | 56262-56419 | 52377-119432 | DQ889677 | This study |
| pCER270 | B. cereus AH187 | Emetic disease | 204,529 | NDd | Y | 93686-93843 | 89801-166332 | DQ889676 | This study |
| pBT9727 | B. thuringiensis 97-27 | Unknown | 77,112 | NDc | N | NA | NA | CP000047 | 31 |
| pAW63 | B. thuringiensis HD73 | None | 71,777 | NDd | N | NA | NA | DQ025752 | 44 |
The predicted copy number is based on read coverage from whole-genome shotgun sequencing projects of multiple B. anthracis genomes. This range represents seven diverse B. anthracis genomes sequenced at The Institute for Genomic Research.
These are revised sizes based on the completed sequencing and closure of the plasmids in this isolate. Therefore, they do not match the sizes cited in Hoffmaster et al. (21), which represented incomplete plasmid molecules.
The copy number of these plasmids cannot be determined (ND) with the read coverage method since the primary sequence reads are not available.
The copy number of these plasmids cannot be determined (ND) since the plasmids were sequenced independently of the chromosome (45).
Y, yes; N, no.
NA, not applicable.
B. cereus AH187 (also known as B. cereus F4810/72) was isolated by the Public Health Laboratory Service, London, United Kingdom, in 1972 during an outbreak and is the type strain for emetic-disease-causing B. cereus isolates (12). B. cereus AH187 contains pCER270, a large plasmid (∼270 kb) that encodes the cereulide biosynthetic gene cluster (renamed from pBCE4810 for consistent nomenclature [12]). The strain is positive for cereulide (emetic toxin) formation and negative for the hemolysin BL hemolytic enterotoxin and cytotoxin K but does encode the nonhemolytic Nhe enterotoxin. The strain was recently characterized by Ehling-Schulz et al. (14) using a multilocus sequence typing (MLSTDB scheme; http://pubmlst.org/bcereus/ [5]) and is sequence type 26. This is a sequence type commonly found among emetic B. cereus strains (13).
Sequencing strategies.
Plasmids pPER_AH820 and pCER270 were assembled from a whole-genome shotgun project of B. cereus AH820 and B. cereus AH187, respectively. Sequencing for these plasmids was performed from three libraries: (i) a 3- to 4-kb insert size (3 to 4 kb) library, (ii) an 8- to 10-kb insert size (8 to 10 kb) library, and (iii) a 30- to 40-kb insert size fosmid library. Plasmids from B. cereus AH818 were separated by pulsed-field gel electrophoresis, and DNA was isolated from the gels. In addition, AH818 plasmids were prepared by using a QIAGEN large construct kit (QIAGEN, Valencia, CA). Small and large insert libraries (3 to 4 kb and 8 to 10 kb) were then constructed as previously reported (33, 34) and sequenced by using an 3730xl DNA analyzer (Applied Biosystems) at the Institute for Genomic Research's Joint Technology Center. After the initial assembly, using methods previously described (16, 23, 25, 33, 34), scaffolds and contigs were identified that belong to the large plasmids compared to previously sequenced plasmids (including B. anthracis pXO1 and pBC10987 [Table 1]) using BLAST (3) and MUMmer (10). Closure was performed as previously described (33), followed by manual annotation (see Tables S1 and S2 in the supplemental material). These efforts resulted in closed circular molecules of 272,145 bp for pPER272 from B. cereus AH820 and AH818 (see below for naming of these plasmids as pPER272), as well as a molecule of 270,082 bp for pCER270 (Table 1).
BLAST score ratio analysis.
The plasmid coding sequences (CDS) were compared by using the BLAST score ratio analysis (32). For each predicted CDS in a reference plasmid, the BLASTP raw score was collected for the alignment against itself (REF_SCORE) and the most similar CDS (QUE_SCORE) in each of the query plasmids analyzed. These scores were normalized by dividing the QUE_SCORE obtained for each query plasmid CDS by the REF_SCORE. Synteny plots (see Fig. 4 and 5) were generated in which each point indicates a single CDS plotted at the 5′ end of the coding region in the reference plasmid genome and the best match in the query plasmid. The color of each point indicates the level of similarity between the reference and query CDS and reflects the BLAST score ratio (see legends for Fig. 4 and 5). CDS with a normalized ratio of <0.4 were considered to be nonhomologous.
FIG. 4.
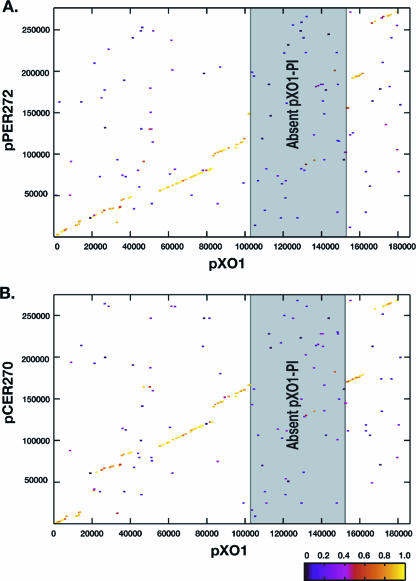
BLAST score ratio-based synteny plots of the novel plasmids with B. anthracis pXO1 demonstrate a lack of conservation of the pXO1 pathogenicity island. (A) Comparison of pXO1 and pPER272. (B) Comparison of pXO1 and pCER270. The gray box in each panel indicates the location of the pXO1-pathogenicity island, which is absent in each of the plasmids analyzed. Each point on the figure represents an individual peptide in pXO1 compared to the proteome of the novel plasmid. The associated color of each point indicates the level of similarity between the compared peptides.
FIG. 5.
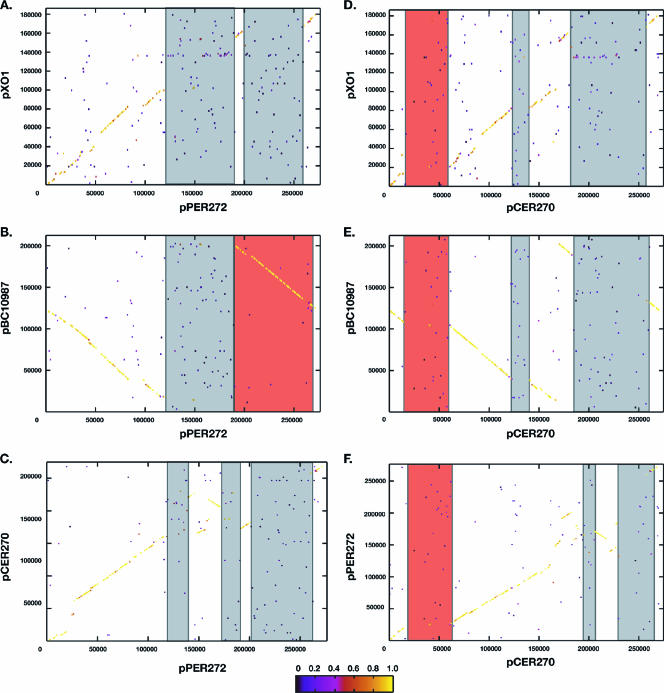
BSR-based synteny plots of the novel plasmids with other pXO1-like plasmids identifying regions of similarity and dissimilarity. (A to C) pPER272. Gray boxes indicate regions unique to pPER272. The red box in panel B identifies a region in the pPER272 plasmids that is shared with pBC10987, both in gene content and synteny. (D to F) All comparisons are to pCER270. The red box in all three panels illustrates the location of the cereulide biosynthetic gene cluster. The gray boxes indicate unique regions to pCER270.
Evolutionary studies.
The coding sequences of a highly conserved region from the pXO1-like plasmids were concatenated into a single ∼40-kb fragment (locations in Table 1). Only coding sequences present in all five plasmids were included in this analysis. These sequences were compared by using a Tamura-Nei genetic distance model (39), and a neighbor-joining tree was constructed.
PCR screening of B. cereus and B. thuringiensis isolates.
Regions that were specific to each plasmid or group of plasmids were amplified by PCR with genomic DNA from a panel of 33 isolates from the B. cereus group using the primers listed in Table S3 in the supplemental material. Targeted regions were amplified from purified genomic DNA by using DyNAzyme I DNA polymerase (5 U; Finnzymes), a 0.4 μM concentration of each primer, and a 0.2 mM concentration of each deoxynucleoside triphosphate in 1× DyNAzyme reaction buffer. DNA polymerase and reaction buffer was added after heating each reaction to 94°C. Thirty cycles of amplification were performed, each consisting of 1 min of denaturation (94°C), 1 min of annealing (58°C), and 2 min of polymerization (72°C). A final 7-min polymerization step (72°C) was added after the cycling procedure. Amplified products were examined by gel electrophoresis using 0.8% agarose gels stained with ethidium bromide (0.5 μg/ml). The presence of an amplified product of the appropriate size was scored as positive. The lack of any amplified product was scored as negative (Table S4 in the supplemental material).
The five primer sets were designed to distinguish each of the unique pXO1-like plasmids. The B. anthracis pXO1 primer set targets the edema factor gene (GBAA_pXO1_142/pXO1-122) and identifies anthrax toxin subunit containing B. anthracis or B. cereus, such as B. cereus G9241, which contains the toxin subunit on pBCXO1 (21). The pCER270 primer set was designed to amplify a region of the cereulide peptide synthetase gene, cesA, identified only among emetic-toxin-producing B. cereus group isolates (BcAH187_pCER270_0023) (12). The primer set for the B. cereus ATCC 10987 pBC10987 plasmid was designed to amplify BCEA_0211, which encodes a tyrosinase. This gene was previously identified in the unique region of pBC10987 compared to pXO1 (33). This pBC10987 region that is shared with pPER272, described in the present study, represents a common region of the pXO1-like plasmids that is not represented on pXO1. The unique region targeted by the pPER272 primer set encodes pPER272_0144, a conserved hypothetical protein conserved between the two periodontal plasmids sequenced in the present study but not shared by pBC10987. The conserved pXO1-like plasmid primer set amplifies pXO1-58, a gene encoding a hypothetical protein. We selected this region since it was highly conserved among all of the pXO1-like plasmids but did not appear to be part of the replication region, allowing for the possibility of different types of replication machinery in unknown plasmids.
RESULTS
General plasmid features.
The general features of the B. cereus group plasmids sequenced in the present study and those previously sequenced are outlined in Table 1. The plasmids can be separated into two groups based on their nucleotide sequence similarity to the B. anthracis plasmid pXO1. The first group includes plasmids that are similar to pXO1 and is comprised of pBCXO1 (B. cereus G9241), pBC10987 (B. cereus ATCC 10987), pPER_AH818 (B. cereus AH818), pPER_AH820 (B. cereus AH820), and pCER270 (B. cereus AH187). This pXO1-like group of plasmids ranges in size from ∼181 to 272 kb and, interestingly, the B. anthracis pXO1 is the smallest of the group (Table 1). The second group of plasmids does not show any similarity to pXO1 and includes pXO2 (B. anthracis), pBC210 (B. cereus G9241), pE33L466 and pE33L54 (B. cereus E33L), as well as pAW63 (B. thuringiensis AW63), and pBT9727 (B. thuringiensis serovar Konkukian strain 97-27), (Table 1). These non-pXO1-like plasmids range in size from ∼54 to 466 kb. A series of smaller cryptic plasmids, less than 20 kb, have been identified in plasmid DNA preparations of several B. cereus and B. thuringiensis isolates; however, these plasmids are not included in the present study.
For plasmids derived from whole-genome shotgun sequencing projects we were able to estimate the plasmid copy number. Plasmid copy numbers were obtained by comparing the average plasmid sequence coverage to that of the chromosome. The plasmid sequence coverage was normalized to the chromosome sequence coverage (i.e., if the plasmid coverage was 20X and the chromosomal coverage was 10X, the plasmid copy number was 2). In all cases, pXO1-like plasmids have a copy number that is between 1 and 3, whereas plasmid pXO2 has a copy number ranging from two to five copies per chromosome (estimated from seven B. anthracis genome projects [J. Ravel et al., unpublished]). The difference in copy number may be attributed to differences in the plasmid size (181 kb versus 95 kb for B. anthracis pXO1 and pXO2, respectively) or replication and maintenance mechanisms as described by Tinsley et al. (42, 43).
Sequence comparisons of the periodontal plasmids.
The two periodontal isolates, B. cereus AH818 and B. cereus AH820, were originally selected for sequencing for different but complementary reasons. B. cereus AH818 was selected since it was shown to contain a large plasmid (pPER_AH818 [18]). B. cereus AH820 was also known to contain a large plasmid (pPER_AH820) and represented the closest phylogenetically known B. cereus isolate to B. anthracis as measured by a 13-locus multilocus sequence typing schema (20). Comparative sequence analysis revealed that the two plasmids were identical at every base but one. In plasmid pPER_AH818 position 195,841 is variable with an equal number of sequencing reads containing an “A” or “G,” whereas pPER_AH820 contains reads with only an “A” at this location. Figure S1 in the supplemental material shows the result of the comparison using the whole-genome alignment tool MUMmer demonstrating that these sequences are syntenic and identical. These plasmids are collectively referred to as pPER272 (the annotation for this plasmid is presented in Table S1 in the supplemental material since the single base variation does not affect the annotation).
Comparison of B. cereus plasmids to B. anthracis pXO1.
Whole-genome shotgun sequencing of B. cereus group isolates and targeted B. cereus group plasmid sequencing projects have provided several plasmid sequences for comparative studies (Table 1). Comparison of the pXO1-like plasmids to B. anthracis plasmid pXO1 using the BLAST score ratio analysis revealed a highly conserved core region (Fig. 1). This highly conserved region of pXO1 (∼50 kb, Table 1) contains genes that are thought to be involved in plasmid replication and maintenance (labeled as Rep in Fig. 1). In a recent report, Tinsley and Khan subcloned ∼5 kb of this conserved region from B. anthracis Sterne pXO1 and demonstrated that one particular coding sequence, renamed repX (also known as pXO1-45), was required for the initiation of replication (42). Alignment of the RepX homolog from each of the pXO1-like plasmids demonstrates that this peptide is highly conserved (>98% identity among the examined isolates; Fig. 2A). The RepX homologs from the pXO1-like plasmids differ at only eight amino acid positions. All B. anthracis pXO1 plasmids have identical RepX sequence. One amino acid change is conserved among the B. cereus pXO1-like plasmid RepX proteins compared to the B. anthracis RepX protein sequence (Fig. 2A). Of the remaining seven amino acid changes, six are conserved in B. cereus pXO1-like plasmids, excluding pBCXO1. The pPER272 RepX contains a single amino acid change that is not conserved in any of the other pXO1-like plasmids.
FIG. 1.
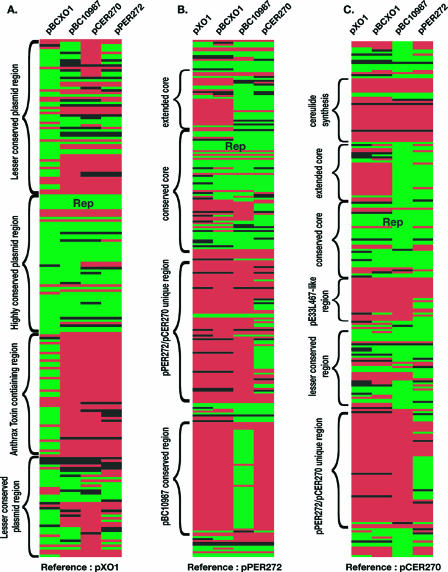
Similarity of newly identified plasmids to B. anthracis pXO1 and other pXO1-like plasmids. For panel A, a BLAST score ratio analysis was performed using the proteome of B. anthracis pXO1 as reference. Panels B and C use pPER272 and pCER270, respectively, as the reference. Each block represents an individual reference plasmid CDS compared to the most similar CDS in the query plasmid. The names of the query plasmids are indicated at the top of each panel. A green block indicates a highly conserved CDS (BSR > 0.8) on the reference and query plasmids. A red box indicates a CDS that is not conserved between the reference and query plasmids (BSR < 0.4). Black boxes indicate that the two CDS share some similarity but are divergent (0.4 < BSR < 0.8). “Rep” in panels A to C indicates the location of the putative replication machinery. Regions with similar or distinct features are highlighted to the left of each panel.
FIG. 2.
Conserved replication protein and initiation of replication region. (A) Peptide alignment of RepX (pXO1-45) from pXO1-like plasmids. Black boxes and asterisks indicate the locations of amino acid substitutions. None of the differences are within the active sites predicted by Tinsley and Khan (42). Panel B, The putative origin of replication of the pXO1-like plasmids as identified by Tinsley et al. (42). The alignment was constructed by using CLUSTAL W (40) and displayed with Boxshade (http://bioweb.pasteur.fr/docs/softgen.html#BOXSHADE). The gray boxes and arrows represent the palindromic sequences putatively thought to facilitate the initiation of replication. The nucleotides in the black boxes indicate differences among the compared putative plasmid origins. Notably, the palindrome is exact in only the B. anthracis Sterne pXO1 plasmid, whereas all other sequenced plasmids have differences in the palindromic region.
Tinsley and Khan also identified a 158-bp region (nucleotide [nt] 55726 to 55883 of B. anthracis Sterne pXO1 [GenBank accession no. AF065404]) that appeared to be required for the initiation of replication of the B. anthracis Sterne pXO1 plasmid (42). The putative origin is contained within the highly conserved core replication region in all pXO1-like plasmids (Fig. 2B). In this region, B. anthracis Sterne pXO1 contains a perfect 24-bp palindrome (42). However, in the other pXO1 plasmid sequences, as well as the B. cereus pXO1-like plasmids, the palindrome is imperfect with a 2-bp mismatch (Fig. 2B, nucleotide locations are in Table 1). The effect, if any, of these differences on plasmid replication is unclear.
The majority of the pXO1-like core plasmid region (Table 1) is annotated as containing hypothetical and conserved hypothetical proteins (46 of 55 genes [83.6%]); however, the remaining nine annotated genes for which a function could be assigned appear to be involved in surface expression of plasmid-encoded factors. Three of these conserved genes encode putative membrane proteins; the remaining genes encode a putative lipoprotein, two peptidases or proteases, two homologs to type IV secretion proteins, and a surface layer protein (see Table S5 in the supplemental material).
In addition to the highly conserved region in pXO1, there is one region of lesser conservation (two regions are shown in Fig. 2A since the molecule is circular, and this region spans the boundaries of the molecule). The gene conservation in this region varies among these plasmids (Fig. 2), however, based on gene presence; pBC10987 is more similar to pXO1 than is pPER272, with pCER270 being the least similar. The pXO1 region of lesser conservation contains mostly hypothetical and conserved hypothetical proteins, which does not provide any functional clues for the conservation, or lack thereof, of this region.
Evolution of the pXO1-like plasmids.
The identification of the highly conserved core region of the pXO1-like plasmids (Fig. 1) allowed for the direct evolutionary comparison of the plasmids. The conserved coding sequences were aligned, and a neighbor-joining tree was constructed by using the Tamura-Nei genetic distance model (39). From this analysis, it is clear that plasmids pXO1 and pBCXO1 are closely related, and the other plasmids are significantly divergent (Fig. 3). Interestingly, pBC10987 is more closely related to the pXO1/pBCXO1 cluster than the pathogenic pXO1-like plasmids, pPER272 and pCER270, suggesting that it has diverged from the pXO1/pBCXO1 group more recently than the plasmids from pathogenic B. cereus isolates. This evolutionary comparison mirrors the results obtained by the BLAST score ratio analysis.
FIG. 3.
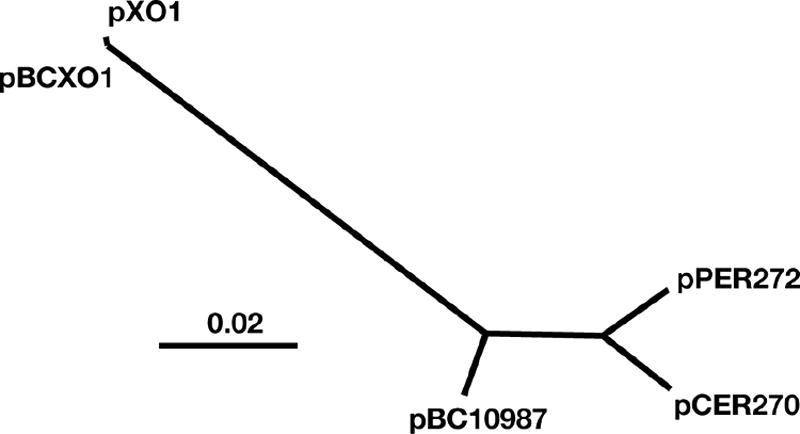
Phylogenetic analysis based on the conserved core region of the pXO1-like plasmids. A neighbor-joining tree was constructed by using the conserved core nucleotide sequences of 40 conserved genes. The tree shows that, based on this region, pBC10987 is more similar to pXO1 than either of the pathogenesis related plasmids, pCER270 or pPER272.
Comparison of plasmids pPER272 and pCER270 to the pXO1-like plasmids.
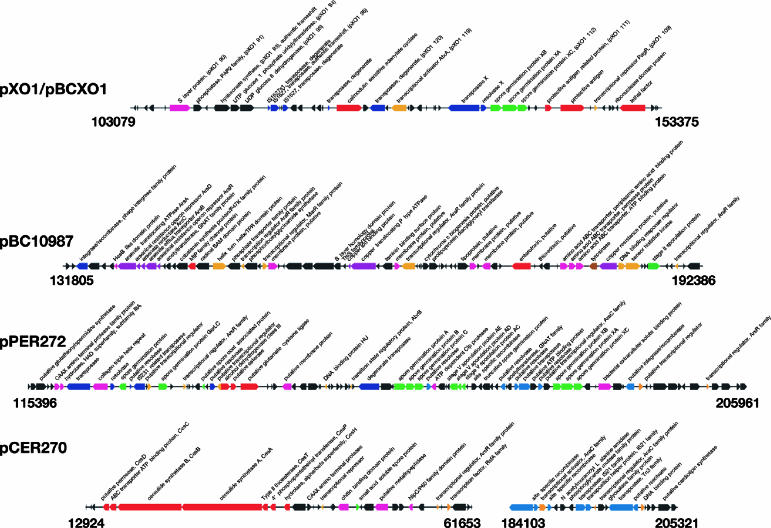
Comparative sequence analysis of the pXO1-like plasmids using BLAST score ratio analysis (32) reveals interesting trends in terms of gene acquisition and loss. It is of note that the pXO1-PI previously discovered on pBCXO1 (21) has not been found in any of the novel pXO1-like plasmids analyzed in the present study (Fig. 4). In each case the pXO1-PI is replaced by a novel sets of genes. The pXO1-PI is flanked by transposable elements which have been implicated in its inversion (35). The unique regions of pBC10987, pPER272, and pCER270 are not flanked by transposable elements; however, it must be noted that most of these unique regions do contain mobile genetic elements such as transposase, recombinase, and resolvase genes (colored blue in Fig. 6). The functional capabilities of these putative mobile elements are unclear.
FIG. 6.
Unique regions from each of the B. cereus group plasmids analyzed. Genes are colored according to function: red, toxin or virulence related; green, sporulation and germination; blue, transposon or mobile element related; orange, regulation; pink, membrane associated; brown, proteases; black with annotation, genes for which no functional class could be assigned; and black with no annotation, hypothetical and conserved hypothetical proteins.
Conserved regions between the pPER272 and PCER270 plasmids.
The BLAST score ratio analysis and the nucleotide comparison of the conserved region of the pXO1-like plasmids indicated that pBC10987, pPER272, and pCER270 were more closely related to each other than to pXO1 (Fig. 1 and 3). An additional ∼20-kb region of conservation adjacent to the highly conserved core region was identified on pBC10987, pPER272, and pCER270 (>90% nucleotide identity; pPER272, nt 25914 to 45091; pCER270, nt 61700 to 81660 [Fig. 1]). This region may represent an extended core region among this subgroup of plasmids. This region, much like the core, consists of genes for which no function can be currently assigned.
A region of conservation (∼23 kb) was identified between pPER272 and pCER270 (>90% nucleotide identity, pPER272, nt 150722 to 170610; pCER270, nt 206131 to 229202 [Fig. 5]) that is also present on the chromosomes of the B. cereus group (Fig. 1) (16, 23, 33). This region contains stage V sporulation genes (AC-AE), germination genes and a formaldehyde detoxification gene cluster. Stage V sporulation genes are required for the utilization and incorporation of dipicolinic acid during spore formation in Bacillus subtilis (44). Mutagenesis of these genes results in spores that are unstable and sensitive to wet heat (30), and this gene cluster had only been found on the chromosomes of Bacillus species (16, 23, 25, 33, 34). In association with the sporulation and germination genes is a conserved three-gene cluster involved in detoxification of formaldehyde (17). The role of these conserved gene clusters are unclear; however, their presence on two divergent plasmids from two distinct clinical presentations is intriguing.
pPER272 unique regions.
The pPER272 plasmid contains ∼140 kb of DNA that is not present in pXO1 (nt 115396 to 205961and 206537 to 260044 [Fig. 1 and 5]), of which ∼52 kb is shared (>99% nucleotide identity (nt 206537 to 258652 [Fig. 5B, red box]) with a unique region that was previously identified in pBC10987 (33). This shared region encodes a number of hypothetical and conserved hypothetical proteins that seem to be limited to the pXO1-like plasmids. This region also encodes an amino acid ABC transporter, an arsenic resistance gene cluster, a major intrinsic protein (MIP) family channel protein that may function as a porin, and a number of regulatory elements. pPER272 also contains an additional unique ∼90-kb region that is not present in pBC10987 or pXO1, except for a few isolated genes (nt 115396 to 205961 [Fig. 1 and 6]). These adjacent regions replace pXO1-PI in pPER272. The unique regions of pPER272 do not encode genes with an obvious link to virulence; however, as mentioned above, the stage V sporulation genes and germination genes may provide an improved survival mechanism. There are at least nine germination-associated homologs and ten regulatory genes (DNA-binding motif-containing proteins) in pPER272 that may increase the ability of these isolates to respond to a variety of environmental signals, potentially providing growth or metabolic advantages over plasmidless spores. IS231 and IS605 elements are both represented on this plasmid and have been commonly found in B. cereus group isolates (8, 33). The presence of 12 transposon-related genes, resolvases, and transposases suggests that some of the unique DNA found within this plasmid may have been acquired from other species via lateral gene transfer.
pCER270 unique genes.
Sequence comparison of the pCER270 plasmid and other pXO1-like plasmids reveals additional genetic variation in pCER270 (Fig. 5D to E). There is an ∼77-kb insertion that replaces the pXO1-PI (nt 182380 to 258941). This region contains the pPER272 shared sporulation and germination region described above. This similarity is not syntenic with the pCER270 plasmid, as indicated by the change in slope of the line on the BSR synteny plot (Fig. 5F). This pattern is often seen as a result of secondary insertions or deletions. There are a number of transposable elements at the 3′ end of this region, including transposases (BcAH187_pCER270_0188) and site-specific resolvases (BcAH187_pCER270_0182 and BcAH187_pCER270_0184) that may have been responsible for the insertion of this unique DNA at this location.
The most distinguishing feature of pCER270 is an ∼46-kb region containing the cereulide biosynthesis gene cluster (nt 15094 to 61653). The ∼24-kb (nt 15094 to 38668) cereulide synthesis (ces) gene cluster encodes seven proteins involved in the synthesis of the cereulide toxin (Fig. 6). This gene cluster and limited flanking regions have been previously described (12). Upstream of the ces gene cluster is a region that is similar to many of the pXO1-like plasmids (85 to 95% nucleotide conservation) and is part of the lesser similarity regions (Fig. 1). A 5-kb region downstream of the ces gene cluster has similarity to other pXO1-like plasmids and is followed by an ∼18-kb region that is unique to pCER270 (nt 43839 to 61653). The region encodes mostly proteins of unknown function; however, a chitin-binding protein (BcAH187_pCER270_0034), small acid-soluble spore protein (BcAH187_pCER270_0036), and a putative metallopeptidase (BcAH187_pCER270_0038) are present in this region. Downstream of the conserved core region is a pCER270 region encoding hypothetical proteins also found on pE33L466 (∼82% nucleotide identity, nt 122833 to 127778), which until now had only shared similarity with pBC210, a non-pXO1-like plasmid from B. cereus G9241 (21).
Screening of B. cereus isolates for plasmid genes.
We designed five sets of primers to unique regions in each of the pXO1-like plasmids (see Table S3 in the supplemental material) to determine the prevalence in environmental and clinical populations of B. cereus (see Table S4 in the supplemental material). Each of the unique regions was selected to provide a discriminative assay for the screening of isolates with an unknown plasmid content. We did not find any B. cereus isolates, clinical or environmental, other than the controls that contained the pXO1 edema factor gene or the cereulide biosynthetic gene cluster (Table 2). These results were not surprising, since we did not have any B. cereus clinical isolates known to be associated with respiratory or emetic diseases in our panel of 33 isolates. The target sequence of the highly conserved core region of the pXO1-like plasmids was found in 72.7% (16/22) clinical B. cereus isolates, whereas it was only found in 18.2% (2/11) environmental isolates. The pBC10987 plasmid target was found in 86% of clinical isolates (15/22), and none of environmental isolates except the control isolate, B. cereus ATCC 10987 (1/11). The majority of isolates containing the conserved plasmid target sequence also contained the target sequence from pBC10987. Three clinical isolates were positive for the conserved target sequence but not the pBC10987 target: the emetic toxin strain B. cereus AH187(F4810/72); B. cereus AH817, a periodontal isolate; and B. cereus AH1134, an endophthalmitis isolate. Two environmental isolates, B. cereus AH1135 and B. cereus AH546, soil isolates from France and Norway, respectively, showed a similar profile. Since the panel contained 16 clinical isolates from periodontal sources, it was surprising that the pPER272 target sequence was only found in the control isolates, B. cereus AH818 and B. cereus AH820. This suggests that the selected region was exclusive to these periodontal isolates and may represent a subgroup within the B. cereus periodontal plasmid isolates. Overall, these results indicate that the clinical isolates are more likely to harbor pXO1-like plasmids that are similar to pBC10987 than are environmental isolates.
TABLE 2.
Identification of conserved plasmid regions in B. cereus and B. thuringiensis isolates
| PCR primer set | No. of:
|
|
|---|---|---|
| Clinical isolates (n = 22) | Environmental isolates (n = 11) | |
| Anthrax toxin containinga | 0 | 0 |
| Conserved pXO1-like backboneb | 16 | 2 |
| pBC10987/periodontalc | 15 | 1 |
| Periodontal plasmidd | 2 | 0 |
| Cereulide-encoding plasmide | 1 | 0 |
The primer set will amplify a region of the edema factor of the B. anthracis pXO1 plasmid (GBAA_pXO1_0142) and other anthrax toxin encoding plasmids.
The nucleotide sequence of pXO1-58 is highly conserved in all plasmids previously sequenced, and the amplified product will be indicative of the presence of a pXO1-like plasmid.
The primer set target for this group of plasmids is shared between the pBC10987 and pPER272 due to the duplication of the non-pXO1 region and is designed to BCE_A0211 in pBC10987.
The primer set is specific to pPER272. It is designed to a conserved hypothetical gene, pPER272_0144 in pPER272.
The primer set is designed to amplify a region of the cereulide peptide synthetase BcAH187_pCER270_0023 from pCER270.
DISCUSSION
Prior to the present study, detailed sequence analysis of large B. cereus group plasmids was mostly limited to those of B. anthracis and of a few environmental isolates of B. cereus or B. thuringiensis (16, 23, 33). The present study identifies and compares the primary sequence of pXO1-like plasmids harbored by two periodontal and an emetic B. cereus with the goal of determining the role of plasmid-encoded genes in virulence.
The pXO1-like plasmids range in size from ca. 181 to 272 kb and, interestingly, pXO1, is the smallest. This suggests that pXO1 has undergone or is undergoing gene loss, retaining factors that mainly are required for pathogenesis and plasmid maintenance. The pPER272 and pCER270 plasmids are the largest pXO1-like plasmids, ∼272 and ∼270 kb, respectively, potentially reflecting adaptation to multiple ecological niches in addition to encoding potential virulence factors. We sequenced two plasmids from periodontal isolates and demonstrated that these ∼272-kb plasmids were identical except for a single nucleotide (see Fig. S1 in the supplemental material). These two isolates were from obtained from distinct geographic and temporal locations, and yet they harbor essentially the same plasmid. The size of the pCER270 was previously predicted to be ∼208 kb by using pulsed-field gel electrophoresis and hybridization methods (12), whereas our sequencing effort revealed a larger plasmid of 270 kb. Both of these novel plasmids exhibit a mosaic structure and contain sets of genes previously found on the chromosomes of B. cereus group members (16, 23, 33). This observation suggests that these large plasmids of the B. cereus group, like other bacterial plasmids, can act as the vehicle for the exchange of genetic information.
Analysis of the copy number of the pXO1-like plasmids indicates that they are maintained at one to three copies per chromosome (Table 1). This value is significantly different from a report by Coker et al., who estimated that the copy number of B. anthracis pXO1 ranged from 13 to 40 copies per chromosome and that a higher copy number leads to increased virulence (9). It must be noted that different methodologies were used to calculate the plasmid copy number; we used the depth of clone coverage from random shotgun sequencing, whereas Coker et al. used a single-locus quantitative PCR method. A large number of factors could have resulted in such disparate results, including growth time, media, and growth conditions, all of which were variable between the present study and the study by Coker et al. (9). In each case, the plasmid copy number calculation, no matter how it is measured, is a narrow snapshot of the dynamic process of plasmid replication and maintenance and as such can vary from isolate to isolate.
It has been demonstrated that the chromosomes of members of B. cereus group are very similar, both in sequence similarity and synteny (19, 31), and that plasmids can play a significant adaptive role in the pathogen host range, virulence, and ecology of the isolate (16, 33, 34). The complete sequence of multiple pXO1-like plasmids enabled comparative studies to identify regions of similarity and divergence potentially responsible for the observed phenotypes (virulence and nutritional adaptation). Analysis of the pXO1-like plasmids resulted in the identification of unique virulence-associated regions in pCER270. Based on the nucleotide similarity of the conserved core region and the BLAST score ratio analysis, pCER270 is the most distantly related plasmid to pXO1 and contains the greatest proportion of unique DNA (see Table S6 in the supplemental material). The unique regions of pCER270 contain the cereulide synthesis gene cluster previously described (12), in addition to a region shared with pPER272 and the chromosomes of spore-forming bacilli. These conserved genes include sporulation and germination genes, as well as a formaldehyde detoxification locus (17, 30, 44). The presence of these genes on pCER270 and pPER272 suggests that there is genetic exchange between the chromosomes and plasmids of the B. cereus group. When the complete chromosome sequences of the strains containing pCER270 or pPER272 become available, it will be interesting to look for the presence or absence of chromosomal homologs for these genes. One can speculate that if these genes are only present on the plasmids, they might represent a mechanism to guarantee plasmid maintenance. However, it is still unclear whether these plasmid-encoded chromosomal genes provide a competitive advantage in virulence, metabolic, or environmental adaptation for these isolates.
Examination of pPER272 did not result in the conclusive identification of classical virulence factors. It is still debatable as to what role the plasmids play in periodontal disease, but a large pXO1-like plasmid is more often associated with periodontal B. cereus isolates than environmental isolates, suggesting a possible connection (Table 2) (18). Plasmid-encoded genes were identified that may be associated with the periodontal phenotype by comparing pPER272 with pXO1-like plasmids from clinical and environmental B. cereus isolates. For example, pPER272 contains a region previously described in pBC10987 that replaced pXO1-PI (33). This region encodes an MIP channel homolog and other membrane proteins that may allow interaction with epithelial cells in the oral cavity. Adjacent to this shared region, pPER272 contained a unique additional 90 kb of sequence not found in pXO1 (Fig. 1 and 6). It would be difficult to determine whether pBC10987 has lost the pPER272 unique region over time in the environment or whether pPER272 acquired the additional region to adapt to other ecological niches. Alternatively, the shared pPER272/pBC10987 region may encode factors associated with periodontal virulence, and the categorization of pBC10987 as an environmental plasmid is misleading. Screening B. cereus isolates for pXO1-like plasmid regions supports the latter hypothesis (Table 2), since no periodontal B. cereus isolates outside the ones carrying pPER272 contained the pPER272-specific sequence. In contrast, 15 of the 22 clinical isolates contain the pBC10987 region, a region shared with pPER272 (Table 2). Further molecular analysis is required to determine whether any virulence determinants are encoded on pPER272 and/or pBC10987.
The pXO1-like plasmids have a highly conserved region that contains the putative origin and a replication initiation protein (42) (Fig. 2). RepX, a pXO1 protein required for initiation of plasmid replication is highly conserved among the pXO1-like plasmids (Fig. 2A). This observation suggests that these plasmids might have evolved from a common ancestral plasmid, forming a unique plasmid family and a distinct incompatibility group. Comparison of the RepX sequences revealed a high level of identity with only eight variable amino acid positions, further suggesting that the replication mechanism is highly conserved among the pXO1-like plasmids. Based on the conserved core coding sequences in this region, pBC10987 is more closely related to pXO1 (and pBCXO1) than the pathogenic B. cereus plasmids pPER272 and pCER270 (Fig. 3). Additional analysis using BLAST score ratio revealed that pBC10987, pPER272, and pCER270 share an extended core region that is not shared with pXO1 (Fig. 2B and C), suggesting that pXO1 may have either evolved further or may represent a more ancestral form of the plasmid. In either case, pXO1 is more distantly related to the other pXO1-like plasmids.
It has been previously demonstrated that B. cereus isolates from clinical presentations form clonal groups (1, 13, 18). The plasmids may play a major role in this observed clonality, even though chromosomal markers have exclusively been used to determine the level of clonality. Clonal radiation can be initiated by events such as genomic insertion, deletion, or acquisition of plasmids and represents an increased level of fitness over the rest of the population. One can envision that a B. cereus isolate having acquired a pXO1-like plasmid may then contain a unique combination of plasmid and chromosomal factors, allowing successful exploitation of an environmental niche. Examples of highly successful B. cereus group clones are (i) B. anthracis that infects mammals (27) and (ii) B. thuringiensis that specifically infects lepidopteran worms (36). It is tempting to speculate by analogy that pCER270 in emetic-toxin-producing isolates or pPER272 in periodontal isolates are solely responsible for the observed virulence and success of these clones. Emetic B. cereus isolates, almost all of which have been shown to contain pCER270 and which belong to the same sequence type 26 group, most likely represent an example of a successful clone (1, 13). However, there must also be additional unidentified mechanisms of virulence since not all periodontal or emetic strains have been shown to contain pXO1-like plasmids.
The present study reveals that pXO1-like plasmids vary in size and copy number and are widespread throughout the B. cereus group. The pXO1-like plasmids in combination with the chromosome appear to form identifiable subgroups of B. cereus that can be associated with certain disease presentations. Although a conserved pXO1-like plasmid core has been identified, each plasmid studied contains an additional set of unique genes. Some genes have a readily identifiable role in virulence, such as the emetic toxin biosynthetic genes, whereas others cannot be linked to pathogenicity solely through sequence analysis, as is the case with pPER272. In addition, we suggest that the pXO1-like plasmids have coevolved with the chromosome to further improve pathogenesis and/or niche adaptation. Much like B. anthracis is a specialized clonal pathogen, it appears that pathogenic B. cereus may harbor specialized plasmids associated with its clinical and metabolic phenotypes.
Supplementary Material
Acknowledgments
This study was supported in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. N01-AI-30071 and by NSF grant 0242162. O.A.O. and A.B.K. were supported by grants from the Norwegian Research Council through a Strategic University Programme (SUP, project number 146534/420) and the FUGE Consortium for Advanced Microbial Sciences and Technology (FUGE-CAMST, project number 152020/310).
Footnotes
Published ahead of print on 13 October 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agata, N., M. Ohta, and M. Mori. 1996. Production of an emetic toxin, cereulide, is associated with a specific class of Bacillus cereus. Curr. Microbiol. 33:67-69. [DOI] [PubMed] [Google Scholar]
- 2.Agata, N., M. Ohta, M. Mori, and M. Isobe. 1995. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol. Lett. 129:17-19. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apetroaie, C., M. A. Andersson, C. Sproer, I. Tsitko, R. Shaheen, E. L. Jaaskelainen, L. M. Wijnands, R. Heikkila, and M. S. Salkinoja-Salonen. 2005. Cereulide-producing strains of Bacillus cereus show diversity. Arch. Microbiol. 184:141-151. [DOI] [PubMed] [Google Scholar]
- 5.Barker, M., B. Thakker, and F. G. Priest. 2005. Multilocus sequence typing reveals that Bacillus cereus strains isolated from clinical infections have distinct phylogenetic origins. FEMS Microbiol. Lett. 245:179-184. [DOI] [PubMed] [Google Scholar]
- 6.Beecher, D. J., J. L. Schoeni, and A. C. L. Wong. 1995. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63:4423-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlin, F., M. Fricker, A. Pielaat, S. Heisterkamp, R. Shaheen, M. Salkinoja Salonen, B. Svensson, C. Nguyen-The, and M. Ehling-Schulz. 2006. Emetic toxin-producing strains of Bacillus cereus show distinct characteristics within the Bacillus cereus group. Int. J. Food Microbiol. 109:132-138. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., P. Braathen, C. Leonard, and J. Mahillon. 1999. MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. Mol. Microbiol. 32:657-668. [DOI] [PubMed] [Google Scholar]
- 9.Coker, P. R., K. L. Smith, P. F. Fellows, G. Rybachuck, K. G. Kousoulas, and M. E. Hugh-Jones. 2003. Bacillus anthracis virulence in guinea pigs vaccinated with anthrax vaccine adsorbed is linked to plasmid quantities and clonality. J. Clin. Microbiol. 41:1212-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delcher, A. L., S. Kasif, R. D. Fleischmann, J. Peterson, O. White, and S. L. Salzberg. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehling-Schulz, M., M. Fricker, H. Grallert, P. Riek, M. Wagner, and S. Scherer. 2006. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehling-Schulz, M., B. Svensson, M. H. Guinebretiere, T. Lindback, M. Andersson, A. Schulz, M. Fricker, A. Christiansson, P. E. Granum, E. Martlbauer, C. Nguyen-The, M. Salkinoja-Salonen, and S. Scherer. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183-197. [DOI] [PubMed] [Google Scholar]
- 14.Ehling-Schulz, M., N. Vukov, A. Schulz, R. Shaheen, M. Andersson, E. Martlbauer, and S. Scherer. 2005. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71:105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granum, P. E., A. Andersson, C. Gayther, M. T. Giffel, H. Larsen, T. Lund, and K. OSullivan. 1996. Evidence for a further enterotoxin complex produced by Bacillus cereus. FEMS Microbiol. Lett. 141:145-149. [DOI] [PubMed] [Google Scholar]
- 16.Han, C. S., G. Xie, J. F. Challacombe, M. R. Altherr, S. S. Bhotika, D. Bruce, C. S. Campbell, M. L. Campbell, J. Chen, O. Chertkov, C. Cleland, M. Dimitrijevic, N. A. Doggett, J. J. Fawcett, T. Glavina, L. A. Goodwin, K. K. Hill, P. Hitchcock, P. J. Jackson, P. Keim, A. R. Kewalramani, J. Longmire, S. Lucas, S. Malfatti, K. McMurry, L. J. Meincke, M. Misra, B. L. Moseman, M. Mundt, A. C. Munk, R. T. Okinaka, B. Parson-Quintana, L. P. Reilly, P. Richardson, D. L. Robinson, E. Rubin, E. Saunders, R. Tapia, J. G. Tesmer, N. Thayer, L. S. Thompson, H. Tice, L. O. Ticknor, P. L. Wills, T. S. Brettin, and P. Gilna. 2006. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis. J. Bacteriol. 188:3382-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms, N., J. Ras, W. N. Reijnders, R. J. van Spanning, and A. H. Stouthamer. 1996. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178:6296-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgason, E., D. A. Caugant, I. Olsen, and A. B. Kolstø. 2000. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 38:1615-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A. B. Kolstø. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. J. Maiden, F. G. Priest, M. Barker, L. X. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoton, F. M., L. Andrup, I. Swiecicka, and J. Mahillon. 2005. The cereulide genetic determinants of emetic Bacillus cereus are plasmid-borne. Microbiology 151:2121-2124. [DOI] [PubMed] [Google Scholar]
- 23.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Lund, T., and P. E. Granum. 1996. Characterization of a non-hemolytic enterotoxin complex from Bacillus cereus isolated after a food-borne outbreak. FEMS Microbiol. Lett. 141:151-156. [DOI] [PubMed] [Google Scholar]
- 27.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 28.Okinaka, R., K. Cloud, O. Hampton, A. Hoffmaster, K. Hill, P. Keim, T. Koehler, G. Lamke, S. Kumano, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. Jackson. 1999. Sequence, assembly, and analysis of pX01 and pX02. J. Appl. Microbiol. 87:261-262. [DOI] [PubMed] [Google Scholar]
- 29.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 32.Rasko, D. A., G. S. A. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioninformatics 6: Article 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasko, D. A., J. Ravel, O. A. Økstad, E. Helgason, R. Z. Cer, L. X. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolstø, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 35.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. X. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 36.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoeni, J. L., and A. C. L. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68:636-648. [DOI] [PubMed] [Google Scholar]
- 38.Stromsten, N. J., S. D. Benson, R. M. Burnett, D. H. Bamford, and J. K. H. Bamford. 2003. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J. Bacteriol. 185:6985-6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorsen, L., B. M. Hansen, K. F. Nielsen, N. B. Hendriksen, R. K. Phipps, and B. B. Budde. 2006. Characterization of emetic Bacillus weihenstephanensis, a new cereulide-producing bacterium. Appl. Environ. Microbiol. 72:5118-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinsley, E., and S. A. Khan. 2006. A novel FtsZ-like protein is involved in replication of the Anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 188:2829-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tinsley, E., A. Naqvi, A. Bourgogne, T. M. Koehler, and S. A. Khan. 2004. Isolation of a minireplicon of the virulence plasmid pXO2 of Bacillus anthracis and characterization of the plasmid-encoded RepS replication protein. J. Bacteriol. 186:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tovar-Rojo, F., M. Chander, B. Setlow, and P. Setlow. 2002. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van der Auwera, G. A., L. Andrup, and J. Mahillon. 2005. Conjugative plasmid pAW63 brings new insights into the genesis of the Bacillus anthracis virulence plasmid pXO2 and of the Bacillus thuringiensis plasmid pBT9727. BMC Genomics 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.