Abstract
In Escherichia coli, the Min system, consisting of three proteins, MinC, MinD, and MinE, negatively regulates FtsZ assembly at the cell poles, helping to ensure that the Z ring will assemble only at midcell. Of the three Min proteins, MinC is sufficient to inhibit Z-ring assembly. By binding to MinD, which is mostly localized at the membrane near the cell poles, MinC is sequestered away from the cell midpoint, increasing the probability of Z-ring assembly there. Previously, it has been shown that the two halves of MinC have two distinct functions. The N-terminal half is sufficient for inhibition of FtsZ assembly, whereas the C-terminal half of the protein is required for binding to MinD as well as to a component of the division septum. In this study, we discovered that overproduction of the C-terminal half of MinC (MinC122-231) could also inhibit cell division and that this inhibition was at the level of Z-ring disassembly and dependent on MinD. We also found that fusing green fluorescent protein to either the N-terminal end of MinC122-231, the C terminus of full-length MinC, or the C terminus of MinC122-231 perturbed MinC function, which may explain why cell division inhibition by MinC122-231 was not detected previously. These results suggest that the C-terminal half of MinC has an additional function in the regulation of Z-ring assembly.
MinC inhibits formation of the Z ring by preventing FtsZ assembly. However, the binding interactions between FtsZ and MinC are not well understood. MinC consists of two independent domains of approximately equal size separated by a short linker (2, 6). The N-terminal half is sufficient for the inhibition of FtsZ assembly (6), and the C-terminal domain is required for binding to MinD (6, 9). When fused to MalE, full-length MinC and MinC1-115 inhibited polymerization of FtsZ, whereas MinC116-231 did not (6, 7). This suggests that the N-terminal half of MinC interacts with FtsZ to inhibit the Z ring, and that the presence of MalE does not seem to interfere with this function.
Studies of the C-terminal domain of MinC showed that fusions of green fluorescent protein (GFP) to the N termini of MinC115-231 and MinC108-231 localized to Z rings when coproduced with MinD in a ΔminCDE strain (9, 10, 16). It has been proposed that targeting of MinC to midcell is an important step in its inhibition of Z-ring assembly (9, 10, 16). However, the means by which GFP fusions to the C-terminal domain of MinC target the cell division septum is not known, because until now this domain has not shown any activity other than binding to MinD or a related protein, DicB (10, 16).
The three-dimensional crystal structure of MinC from Thermotoga maritima (2) indicates that MinC consists of two different domains and forms a dimer. Interestingly, the structure shows that the N-terminal domain of MinC has weak similarity to the N-terminal domain of FtsA, a cell division protein that interacts with FtsZ directly. This is consistent with the inhibition of Z-ring assembly by the N-terminal domain of MinC, although FtsA is thought to act oppositely to promote Z-ring integrity (8, 12). This potential contradiction led us to test whether the N-terminal domain of FtsA inhibits the Z ring.
While working with a FtsA-MinC chimeric protein consisting of the N terminus of FtsA fused to the C-terminal half of MinC, we found, unexpectedly, that the C-terminal half of MinC by itself inhibited the Z-ring formation in vivo when MinD was coproduced. Although the N-terminal half of MinC carries the major Z-ring-inhibitory activity, we show here that the C-terminal half of MinC has a new and surprising role in inhibiting Z-ring assembly as well.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Escherichia coli strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) medium containing 0.5% NaCl supplemented with the appropriate antibiotics at 30°C, unless otherwise indicated. Cells were grown to mid-exponential phase (optical density at 600 nm, ∼0.5) for all microscopic examinations. Ampicillin (Amp) or chloramphenicol (Cm) was added at 100 μg or 20 μg per ml, respectively, as needed. Strains Top10 and XL1-Blue were used for plasmid constructions.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| WM1074 | MG1655 ΔlacU169 (TX3772) | 1 |
| WM1032 | WM1074 ΔminCDE::kan | 13 |
| BL21(DE3) | Protein overproduction strain for pET vectors | Novagen |
| Top10 | Invitrogen | |
| XL1-Blue | Novagen | |
| Plasmids | ||
| pDSW208 | Ptrc-gfp pBR322 derivative | 15 |
| pDSW209 | Ptrc-gfp pBR322 derivative | 15 |
| pDSW210 | Ptrc-gfp pBR322 derivative | 15 |
| pET28a(+) | Protein overproduction plasmid | Novagen |
| pBAD33 | pACYC184 derivative containing the araBAD promoter | 5 |
| pWM2060 | pDSW209 lacking GFP | 4 |
| pWM2784 | FLAG inserted between EcoRI and HindIII sites of pDSW210 | This study |
| pWM2816 | FLAG-minCDE in pWM2784 | This study |
| pWM2818 | FLAG-minC122-231 minDE in pWM2784 | This study |
| pWM2819 | FLAG-minC122-231(R172A)minDE in pWM2784 | This study |
| pWM2801 | FLAG-minC in pWM2784 | This study |
| pWM2802 | FLAG-minC122-231 in pWM2784 | This study |
| pWM2803 | FLAG-minC122-231(R172A) in pWM2619 | This study |
| pWM2844 | FLAG-minC122-231 in pDSW208 | This study |
| pWM2733 | gfp-minC in pDSW209 | This study |
| pWM2734 | gfp-minC122-231 in pDSW209 | This study |
| pWM2735 | minC-gfp in pWM2619 | This study |
| pWM2736 | minC122-231-gfp in pWM2619 | This study |
| pWM2619 | Six-His tag inserted between EcoRI and HindIII sites of pDSW210 | This study |
| pWM1682 | his-minD in pET28a | 11 |
| pWM2691 | his-minD in pBAD33 | This study |
| pWM2769 | minDE in pWM2060 | This study |
Plasmid constructions.
Oligonucleotide primers used for plasmid construction are listed in Table 2.
TABLE 2.
Oligonucleotides used for plasmid construction
| Name | Sequence |
|---|---|
| pDSW-6His | CGGAATTCCACCATCACCATCACCATGAGCTCGGTACCCGGGGATCC |
| pDSW-FLAG | CGGAATTCGACTACAAGGACGACGATGACAAAGAGCTCGGTACCCGGGGATCC |
| pDSW-reverse | CAAATTCTGTTTTATCAGAC |
| MinC-f (SacI) | GCGAGCTCTCAAACACGCCAATCGAGC |
| MinC122-f (SacI) | GCGAGCTCGTCACAAAAACGCGTTTAATAG |
| MinC-r (BamHI) | CGGGATCCTCAATTTAACGGTTGAACGG |
| MinE-r (BamHI) | CGCGGATCCTTTCAGCTCTTCTGCTTC |
| MinE-r2 (BamHI) | CGGGATCCTTATTTCAGCTCTTCTGCTTCC |
| MinC-R172A-f | CATGATGCGCGGTGCTGCGCTGGCAGGGG |
| MinC-R172A-r | CCCCTGCCAGCGCAGCACCGCGCATCATG |
| MinC-r (SalI) | ACGCGTCGACATTTAACGG |
| MinD6 | TTGAATTCGCACGCATTATTGTTG |
To insert DNA encoding the six-His tag or the FLAG peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) into the expression plasmid pDSW210 (15), a multiple-cloning site and gfp were amplified with Hi-Fidelity polymerase (Roche Scientific) using the primer pair pDSW-6His/pDSW-reverse or pDSW-FLAG/pDSW-reverse. The resulting products were digested with EcoRI and HindIII and ligated into pDSW210 cleaved with the same enzymes, yielding plasmids pWM2619 and pWM2784.
To construct pWM2816 (FLAG-minCDE), pWM2818 (FLAG-minC122-231DE), pWM2801 (FLAG-minC), and pWM2802 (FLAG-minC122-231), primers MinC-f (SacI)/MinE-r2 (BamHI), MinC122-f (SacI)/MinE-r2 (BamHI), MinC-f (SacI)/MinC-r (BamHI), and MinC122-f (SacI)/MinC-r (BamHI) were used to amplify minCDE, minC122-231DE, minC, and minC122-231, respectively. The resulting products were digested with SacI and BamHI and ligated into pWM2784 cleaved with the same enzymes. The EcoRI-HindIII fragment of pWM2802 was cloned into same sites of pDSW208 to construct pWM2844 (FLAG-minC122-231). Plasmid pWM2844, derived from pDSW208, has a stronger promoter than pWM2802, derived from pDSW210.
Site-directed mutagenesis [FLAG-minC122-231(R172A)minDE and FLAG-minC122-231(R172A)] was performed according to the two-step PCR method using the primers pDSW-FLAG/MinC(R172A)-r and pDSW-reverse/MinC(R172A)-f, respectively. The resulting products were digested with SacI and BamHI and ligated into pWM2784 cleaved with the same enzymes, yielding pWM2819 [FLAG-minC122-231(R172A)DE] and pWM2803 [FLAG-minC122-231(R172A)], respectively. To construct pWM2769 (minDE), primers minD-f (EcoRI) and minE-r (BamHI) were used to amplify minDE. The resulting products were digested with EcoRI and BamHI and ligated into pWM2060 (4) cleaved with the same enzymes.
The EcoRI-HindIII fragments of pWM2622 (His-MinC) and pWM2624 (His-MinC122-231) (D. Shiomi and W. Margolin, unpublished data) were cloned into EcoRI-HindIII-cleaved pDSW209 to yield pWM2733 (GFP-MinC) and pWM2734 (GFP-MinC122-231), respectively. minC and minC122-231 were amplified using plasmids pWM2622 and pWM2624 as templates and primers pDSW-f and MinC-r (SalI), respectively. The products were digested with EcoRI and SalI and cloned into pWM2619 cleaved with the same enzymes to yield pWM2735 (MinC-GFP) and pWM2736 (MinC122-231-GFP), respectively. The XbaI-HindIII fragment of pWM1682 (pET-his-minD) was cloned into pBAD33 cleaved with the same enzymes to yield pWM2691 (pBAD-his-minD).
Microscopy and immunoblotting.
Immunoblotting with rabbit anti-FtsZ, rabbit anti-MinD, or mouse anti-FLAG M2 (Sigma) antibodies was performed with blots containing equal amounts of cell protein per lane, as described previously (1). Purified MinD protein from strain BL21(DE3) carrying pWM1682 (pET28a-his-minD) was used to raise rabbit anti-MinD polyclonal antiserum (Pacific Immunology, Ramona, CA). Band intensities on blots were measured by ImageJ version 1.32 (http://rsb.info.nih.gov/ij/); the lowest intensities in each blot were normalized to 1. Microscopic examination of immobilized live cells, anti-FtsZ immunofluorescence microscopy, cell fixation, and staining with DAPI (4′,6′-diamidino-2-phenylindole) were done essentially as described previously (4). Briefly, WM1074 (wild-type [WT]) cells or WM1032 (ΔminCDE) cells carrying one of the plasmids that encode FLAG-MinC derivatives were grown overnight in LB medium at 30°C. An overnight culture was diluted 1:100 in LB medium supplemented with Amp and incubated at 30°C for 4 h with various concentrations (0, 0.1, or 1 mM) of isopropyl-thiogalactoside (IPTG) to induce the Trc99 promoter on the plasmid. The cells were spun down, resuspended in LB medium, mixed 1:1 with 2% LB agarose, spotted onto a coverslip, and observed under an epifluorescence microscope (Olympus BX60) equipped with a 100× immersion oil objective and a GFP filter cube. Images were captured with a Photometrics CoolSnap fx cooled charge-coupled device (CCD) camera driven by QED image-capturing software and saved as Adobe Photoshop TIF files. Cell lengths were measured with Object Image software (Norbert Vischer).
RESULTS AND DISCUSSION
The C terminus of MinC (MinCC) has cell division-inhibitory activity.
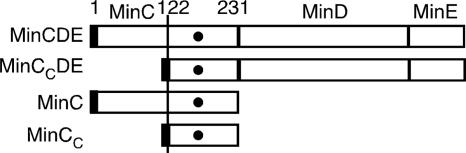
Based on weak structural similarity between an N-terminal domain of FtsA and the N-terminal half of MinC (MinCN) (2), we initially investigated whether fusing this MinCN-like domain of FtsA directly to the C terminus of MinC might yield a protein that would act like full-length MinC and inhibit cell division. As a control, we also constructed a plasmid encoding the C-terminal half of MinC (MinC122-231, herein referred to as MinCC) (Fig. 1).
FIG. 1.
Outline of minC constructs used in this study. All constructs encode a FLAG tag at the N terminus (black rectangles). Filled circles represent the position of the R172A mutation.
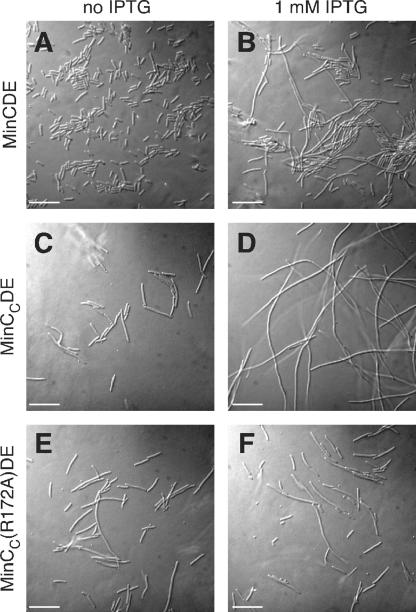
While testing this hypothesis, we found to our surprise that coproduction of MinCC, MinD, and MinE from an IPTG-inducible promoter on a plasmid caused cell filamentation. In the absence of IPTG, WM1032 (ΔminCDE) cells carrying a plasmid expressing the WT minCDE operon (encoding a FLAG tag at the N terminus of MinC) were normal and did not display a minicell phenotype, suggesting that MinC, MinD, and MinE were produced at levels sufficient to complement ΔminCDE cells and that the N-terminal FLAG tag did not interfere with MinC function (Fig. 2A). In the presence of IPTG, cells of this strain became filamentous (Fig. 2B). This was probably caused by the presence of large amounts of MinC at the membrane, preventing Z-ring assembly efficiently at all sites in the cell (3). In the absence of IPTG, WM1032 cells carrying a plasmid expressing minCCDE using the same promoter, ribosome-binding site, and FLAG tag produced minicells (Fig. 2C). This suggested either that cellular levels of MinCC, MinD, and MinE were not sufficient to complement ΔminCDE or, more likely, that MinCC did not inhibit cell division as well as full-length MinC. However, WM1032 cells carrying a plasmid expressing minCCDE became filamentous in the presence of IPTG (Fig. 2D), strongly suggesting that MinCC had cell division-inhibitory activity.
FIG. 2.
Effects of expression of minC, minCC, or minCC(R172A) in the context of the minCDE operon on cell division of a Δmin strain. Shown are micrographs of WM1032 (ΔminCDE) cells carrying a plasmid expressing the minCDE operon (pWM2816) in the absence (A) and presence (B) of 1 mM IPTG, a plasmid expressing minCCDE (pWM2818) in the absence (C) and presence (D) of 1 mM IPTG, and a plasmid expressing minCC(R172A)DE (pWM2819) in the absence (E) and presence (F) of 1 mM IPTG. All three plasmids have identical IPTG-inducible promoters and ribosome-binding sites and encode a FLAG tag at the N terminus of MinC or MinCC. Bars, 10 μm.
To exclude the possibility that MinD and MinE in the complete absence of MinC can cause cell filamentation in our expression system, we deleted minC from the minCDE plasmid in WM1032. WM1032 cells producing MinD and MinE from plasmid pWM2769 did not become filamentous even in the presence of IPTG (data not shown), indicating that filamentation of cells that produced MinCCDE specifically required the MinCC component.
To demonstrate further that overproduction of MinCC led to cell filamentation and rule out effects of the FLAG tag, we changed the minCC gene so that it encoded an alanine instead of arginine at position 172. It has been reported that the R172A mutation in MinC inhibits the interaction of MinC with the unknown septal component, although the mutant MinC still interacts with MinD (16), indicating that it has partial activity. WM1032 cells producing FLAG-MinCC(R172A) and MinDE from the same plasmid system did not become more filamentous even in the presence of IPTG (Fig. 2E and F). This supports the idea that the cell division inhibition is specific to MinCC and raised the possibility that an interaction between MinCC and FtsZ could cause the observed cell filamentation.
Because overproduction of full-length MinC alone is lethal to WT cells such as WM1074 (the min+ parent of WM1032), we reasoned that overproduction of MinCC alone might also be lethal in WM1074 if it had sufficient cell division-inhibitory activity. To test this, WM1074 cells carrying plasmids encoding various N-terminally FLAG-tagged Min constructs under IPTG control (Fig. 1) were streaked onto LB agar in the absence or presence of IPTG (Fig. 3M). Growth of cells carrying plasmids containing minC was inhibited in the presence of 0.5 or 1 mM IPTG. On the other hand, growth of cells carrying a plasmid containing minCC was inhibited only in the presence of 1 mM IPTG (Fig. 3M), suggesting that overproduction of MinCC inhibited cell division efficiently but the activity of MinCC was less than that of full-length MinC, as suggested by the data in Fig. 2. As expected, the strain overproducing MinCC(R172A) was viable even in the presence of 1 mM IPTG (Fig. 3M).
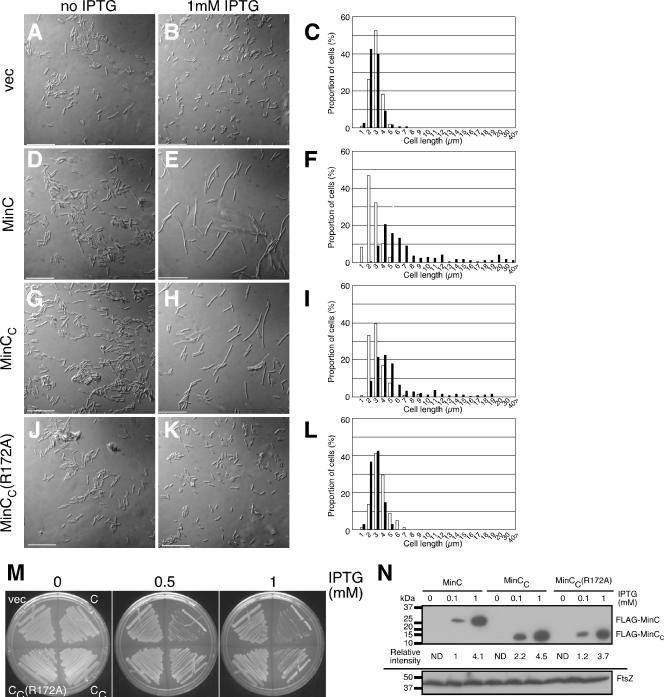
FIG. 3.
Effects of expression of minC, minCC, or minCC(R172A) on cell division of a WM1074 (WT) strain. Shown are micrographs of WM1074 cells carrying empty plasmid vector (pWM2784) (A and B) or plasmids expressing FLAG-tagged minC (pWM2801) (D and E), minCC (pWM2802) (G and H), or minCC(R172A) (pWM2803) (J and K) in the absence (A, D, G, and J) and presence (B, E, H, and K) of 1 mM IPTG. The corresponding distribution of cell lengths in the population without IPTG (open bars) or with IPTG (filled bars) is shown on the right (C, F, I, and L). (M) Plates supplemented with either 0, 0.5, or 1 mM IPTG and streaked with the cells analyzed in panels A to L to measure their viabilities. (N) Immunoblot of extracts of WM1074 cells carrying pWM2801 (FLAG-minC), pWM2802 (FLAG-minCC), or pWM2803 [FLAG-minCC (R172A)] grown in the presence of 0, 0.1, or 1.0 mM IPTG and probed with anti-FLAG antibody (top) or anti-FtsZ (bottom). Protein size markers are shown to the left.
To determine whether the lethality of overproducing MinC or MinCC in wild-type WM1074 was a result of cell division inhibition, we observed cell morphology directly. Addition of IPTG to WM1074 cells carrying the plasmid vector did not affect cell morphology (Fig. 3A and B; Table 3). Consistent with the failure to form colonies on plates, wild-type WM1074 cells overproducing MinC or MinCC in liquid culture became filamentous in 1 mM IPTG (Fig. 3E and H), in contrast to the short cells in the absence of IPTG (Fig. 3D and G). We measured the lengths of WM1074 cells producing MinC or MinCC (Fig. 3F and I; Table 3). WM1074 cells overproducing MinC in the presence of 1 mM IPTG became about threefold longer on average, whereas WM1074 cells overproducing MinCC from the same promoter and ribosome-binding site in the presence of 1 mM IPTG became about twofold longer on average. In both cases, however, there was a significant proportion of very long filaments (Fig. 3F and I), which were absent in the negative controls (Fig. 3C and L). Average lengths of WM1074 cells producing MinCC in the presence of 0.1 mM IPTG were comparable with those in the absence of IPTG (Table 3), consistent with the idea that MinCC has less activity than full-length MinC. Overproduction of the R172A derivative of MinCC had little effect on average cell length, similar to the vector control (Fig. 3C and L).
TABLE 3.
Lengths of WM1074 (WT) cells producing MinC, MinCC, or MinCC(R172A)
| Protein (plasmid) | Avg cell length (μm)a (n)
|
||
|---|---|---|---|
| No IPTG | 0.1 mM IPTG | 1 mM IPTG | |
| None (vector) | 3.42 ± 0.72 (99) | ND | 3.41 ± 1.57 (108) |
| MinC (pWM2801) | 3.01 ± 0.82 (109) | 4.43 ± 1.93 (124) | 8.86 ± 7.19 (165) |
| MinCC (pWM2802) | 3.61 ± 1.21 (148) | 3.67 ± 0.95 (102) | 6.53 ± 4.56 (248) |
| MinCC(R172A) (pWM2803) | 4.01 ± 1.05 (102) | ND | 3.29 ± 0.82 (101) |
Only cells which lacked visible septa were measured. ND, not determined.
To test whether levels of MinC and MinCC needed for the cell filamentation observed in Fig. 3 were similar, we examined levels of FLAG-tagged MinC, MinCC, and MinCC(R172A) in extracts of cells used for the microscopy in Fig. 3. Immunoblots were probed with monoclonal anti-FLAG antibody (Fig. 3N, top) and anti-FtsZ as a loading control (Fig. 3N, bottom). We were unable to detect any FLAG-MinC derivatives in the absence of IPTG, but proteins were easily detectable in the presence of IPTG (Fig. 3N, top). Measurement of band intensities indicated that the levels of FLAG-MinC derivatives were roughly equivalent, within a factor of 2, at 0.1 mM IPTG and again at 1 mM IPTG. Importantly, there was only a 10% difference between levels of FLAG-MinC and FLAG-MinCC at 1 mM IPTG. This suggests that the effects on cell division and viability we observed at 1 mM IPTG result from the intrinsic properties of the MinC derivatives and not from a disparity in protein levels.
We could not detect the FLAG-tagged proteins in the absence of IPTG and thus were not able to estimate the extent of induction with IPTG. Because 25-fold overproduction of MinC is required to inhibit cell division in a ΔminCDE strain (3), we estimate that MinC induction with 1 mM IPTG in our experiments is likely less than 25-fold, because MinC expressed from this plasmid (pWM2801) cannot inhibit cell division of strain WM1032 (ΔminCDE) (see below). With 0.1 mM IPTG in WT WM1074 derivatives, MinC levels were about twofold lower than MinCC levels (Fig. 3N), and yet cells producing MinC became slightly filamentous (∼1.5-fold longer), whereas cells producing MinCC remained normal (Table 3). In the presence of 1 mM IPTG, levels of MinC, MinCC, and MinCC(R172A) were comparable. In sum, these results lend support to the idea that the activity of MinCC is less than that of full-length MinC at equivalent levels of protein.
Overproduction of MinCC inhibits Z-ring formation in vivo.
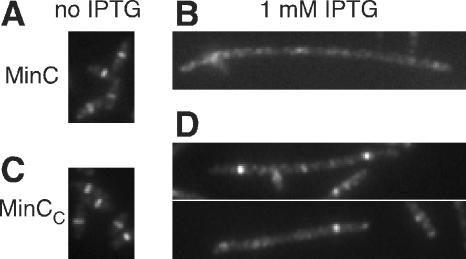
Next, we examined whether MinCC inhibits cell division by blocking Z-ring assembly. In the absence of IPTG, staining for FtsZ by immunofluorescence microscopy showed that the Z ring was formed correctly at midcell when FLAG-tagged MinC or MinCC from plasmids was produced at basal levels without IPTG induction (Fig. 4A and C). However, upon induction with IPTG, filamentous cells overproducing MinC exhibited diffuse FtsZ staining throughout the cytoplasm, as expected (Fig. 4B), indicating that Z-ring assembly was inhibited by MinC. Overproduced MinCC also resulted in diffuse FtsZ staining after IPTG induction, although many of the filamentous cells still had a Z ring at one of several potential division sites (Fig. 4D). This suggested that some Z rings were depolymerized by MinCC but others were not, consistent with the idea that MinCC is a less effective inhibitor of Z-ring assembly than full-length MinC.
FIG. 4.
Overproduced MinCC, like MinC, inhibits assembly of the Z ring. FtsZ was visualized by immunofluorescence microscopy in WM1074 cells carrying plasmids expressing FLAG-tagged minC (pWM2801) (A and B) or minCC (pWM2802) (C and D) in the absence (A and C) and presence (B and D) of 1 mM IPTG.
To exclude the possibility that the differences in observed Z-ring assembly with MinC and MinCC were a result of differences in FtsZ levels, we examined levels of FtsZ by immunoblotting with extracts from the same cells used for microscopy with anti-FtsZ. As shown in Fig. 3N, cells overproducing MinCC had levels of FtsZ comparable to those observed with cells overproducing MinC. These results support the idea that cell division inhibition by MinCC occurs via inhibition of Z-ring assembly.
Cell division inhibition by MinCC requires MinD.
Although MinC requires MinD for effective inhibition of cell division, it can inhibit cell division without MinD when it is overproduced about 25-fold (3, 6). Therefore, we tested whether higher levels of MinCC were sufficient to inhibit cell division. WM1032 (ΔminCDE) cells carrying an IPTG-inducible plasmid containing minC or minCC did not exhibit any filamentation even in the presence of IPTG (data not shown), probably because the proteins were not sufficiently overexpressed from the relatively weak Trc99 promoter (Fig. 3N). Therefore, we cloned minC and minCC under the control of a strong arabinose-inducible promoter. WM1032 cells carrying a plasmid containing minC (pBAD33-minC) became filamentous in the presence of 0.2% arabinose as expected (data not shown), while WM1032 cells carrying a plasmid containing minCC (pBAD33-minCC) remained normal in length with 0.2% arabinose (data not shown), suggesting that the activity of MinCC is low and that MinCC requires MinD for efficient activation.
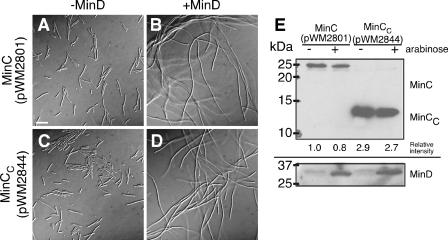
To confirm that cell division inhibition by MinC or MinCC produced from pWM2801 (MinC) or pWM2802 (MinCC) requires MinD, MinC and MinCC (induced with IPTG) were coproduced with MinD (induced with arabinose) from two separate plasmids in WM1032. As a control, overproduction of MinD with arabinose resulted in a typical minicell phenotype (data not shown), indicating that excess MinD by itself did not inhibit cell division. As expected, a minicell phenotype was observed when MinD was not induced with arabinose, but overproduction of MinC (Fig. 5A) and MinCC (data not shown) was induced with IPTG. In the presence of arabinose and IPTG, which induced overproduction of both MinD and MinC, cells became filamentous (Fig. 5B). However, when the same conditions were applied to cells carrying the MinCC plasmid pWM2802, the minicell phenotype was maintained (data not shown), suggesting that overproduced levels of MinCC were too low to inhibit cell division even in the presence of excess MinD. Therefore, we cloned FLAG-MinCC into pDSW208, which has a stronger promoter than pWM2802, to yield pWM2844. As expected, we observed a minicell phenotype upon induction with 0.1 mM IPTG in the absence of arabinose (Fig. 5C). However, in the presence of 0.2% arabinose and 0.1 mM IPTG, cells became markedly filamentous and minicells were absent (Fig. 5D), suggesting that the FtsZ-inhibitory activity of MinCC is dependent on MinD.
FIG. 5.
Inhibition of cell division by MinCC is dependent on MinD. (A to D) Micrographs of WM1032 (ΔminCDE) cells carrying plasmids expressing his-tagged minD (pWM2691) from an arabinose-inducible promoter and either FLAG-minC (pWM2801) or FLAG-minCC (pWM2844) from IPTG-inducible promoters. Cells were grown in 0.1 mM IPTG (for minCC, expressed from a stronger promoter) or 1 mM IPTG (for minC, expressed from a weaker promoter) and either without (A and C) or with (B and D) 0.2% arabinose (for expression of minD) at 30°C. Bar, 10 μm. (E) Immunoblots of extracts from the cells in panels A to D, probed with anti-FLAG to detect FLAG-MinC or FLAG-MinCC (top) or anti-MinD to detect MinD in the corresponding extracts (bottom). The relative intensities of the FLAG-MinC and FLAG-MinCC bands from extracts of the filamentous cells in 0.2% arabinose are shown below the anti-FLAG blot. Protein size markers are on the left.
To determine the levels of MinC, MinCC, and MinD protein during these experiments, we probed immunoblots of extracts of cells used for the experiments shown in Fig. 5A to D with anti-FLAG to detect FLAG-MinC or FLAG-MinCC and with anti-MinD to detect MinD. At a given IPTG level, there were no significant differences in cellular levels of MinD in the WM1032 strain producing MinC or MinCC (Fig. 5E, bottom). We then compared production levels of FLAG-MinC (pWM2801) and FLAG-MinCC (pWM2844) (Fig. 5E, top). Cellular levels of these proteins were independent of whether MinD was induced or not induced. The relative intensities of the protein bands indicated that an approximately threefold higher concentration of FLAG-MinCC relative to FLAG-MinC was sufficient to inhibit cell division efficiently in the presence of overproduced MinD.
GFP tags inhibit function of MinC and MinCC.
In previous studies with MinCC, no inhibition of FtsZ assembly or cell division had been detected. In those studies, MinCC had been fused with either MalE or GFP (6, 9, 10, 16). In contrast, our MinC and MinCC constructs carry a FLAG tag at their N termini that did not seem to perturb MinC function significantly (Fig. 2A and B). Therefore, we hypothesized that the large MalE and GFP tags fused to the N terminus of MinCC in previous studies may have inhibited full function of the smaller MinCC domain.
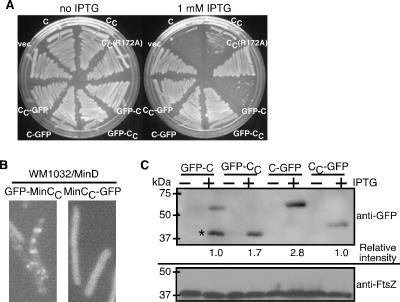
To test whether MinCC inhibited FtsZ assembly when GFP was fused to the C terminus of MinCC, we constructed MinC-GFP and MinCC-GFP in the same plasmids and under the same promoter control as the N-terminally tagged versions. Overproduction of MinC, MinCC, and GFP-MinC with IPTG inhibited colony formation of WT WM1074 derivatives (Fig. 6A). However, at the same level of IPTG, MinC-GFP did not inhibit growth of WM1074. We next overproduced GFP-MinCC or MinCC-GFP with MinD in the ΔminCDE strain WM1032. Under these conditions, GFP-MinCC localized to potential division sites, consistent with previous reports (9, 10, 16), whereas MinCC-GFP localization was diffuse and did not localize to midcell (Fig. 6B). Interestingly, although MinD was coproduced, MinCC-GFP did not detectably localize to the membrane either.
FIG. 6.
A C-terminal GFP tag inhibits function of MinC and MinCC. (A) Streak plates of WM1074 (WT) carrying a plasmid expressing FLAG-minC (pWM2801), FLAG-minCC (pWM2802), FLAG-minCC(R172A) (pWM2803), gfp-minC (pWM2733), gfp-minCC (pWM2734), minC-gfp (pWM2735), or minCC-gfp (pWM2736) supplemented with 0 or 1 mM IPTG. (B) Localization of GFP-MinCC from pWM2734 (left) or MinCC-GFP from pWM2736 (right) in cells of the ΔminCDE strain WM1032, which also produced MinD from pWM2691. Cultures were induced with 1 mM IPTG to produce the GFP fusions and 0.002% arabinose to produce MinD; live cells were examined by fluorescence microscopy. (C) Immunoblot of extracts from WM1032 (ΔminCDE) cells producing GFP-MinC, GFP-MinCC, MinC-GFP, or MinCC-GFP in either 0 (−) or 1 mM (+) IPTG, probed with anti-GFP (top), or FtsZ, probed with anti-FtsZ (bottom). Protein size markers are on the left, and the relative intensities of the GFP fusion proteins are shown below the anti-GFP blot. The asterisk highlights a prominent truncated product of GFP-MinC that was not included in the relative intensity calculation.
To test whether the failure of some of the GFP fusions to inhibit cell division resulted from lower levels or degradation of protein, we examined cellular levels of GFP-MinCC and MinCC-GFP by probing immunoblots with anti-GFP (Fig. 6C). As in Fig. 3N, GFP-tagged proteins were undetectable in the absence of IPTG. In the presence of 1 mM IPTG, GFP-MinC was approximately one-third the level of MinC-GFP. Nevertheless, at these levels, the former inhibited colony formation while the latter did not (Fig. 6A). Levels of MinCC-GFP were ∼3-fold lower than those of MinC-GFP but were comparable with that of GFP-MinC. Despite being at higher levels than GFP-MinC, MinC-GFP and GFP-MinCC were unable to inhibit cell division. We conclude that GFP fused to the N terminus of MinCC prevents Z-ring assembly inhibition by MinCC. Moreover, our results indicate that GFP fused to the C terminus of MinC (or MinCC) may inhibit full function of MinC.
Conclusions and insights from this study.
To explain how GFP-MinCC could localize to midcell but not inhibit Z-ring assembly, MinCC was previously proposed to target to an uncharacterized septal component. This targeting was dependent on MinD but independent of ZipA and FtsA (10). As localization of ZipA and FtsA is dependent on FtsZ, and we have now shown that MinCC with a FLAG tag indeed inhibits assembly of the Z ring, we propose that the target of MinCC is likely to be FtsZ, although it is still possible that an unknown protein is involved.
Which portion of MinCC is required for inhibition of Z-ring assembly? As this inhibition is prevented when GFP is fused to the N terminus of MinCC, it is likely that the N-terminal portion of MinCC is important for the inhibitory activity. Based on the crystal structure of T. maritima MinC, which is a good model for E. coli MinC, the MinCC domain forms a β-barrel-like structure (2). The Arg172 residue within MinCC, which is required for targeting MinC to the septal component, is located on the external surface of MinCC (16). Assuming that MinCC depolymerizes FtsZ, it should interact with FtsZ during this process even if the interaction is transient. Consequently, it is likely that the external surface of the MinC dimer is responsible for FtsZ depolymerization and net disassembly of the Z ring. Although there is a linker between GFP and MinCC in our construct, it is likely that GFP interferes with the interaction between MinCC and FtsZ.
How might GFP, when fused with the C terminus of full-length MinC, inhibit MinC function? One possibility is that GFP inhibits the binding of MinC to MinD. If MinCC-GFP were localized preferentially to the membrane like GFP-MinCC(R172A) (16), MinD would be the likely membrane target. However, as MinCC-GFP did not localize to midcell or preferentially to the cytoplasmic membrane (Fig. 6B), it is reasonable to propose that MinCC-GFP does not bind to MinD. The failure of MinCC-GFP to localize to midcell or the membrane was not due to protein degradation, as immunoblots probed with anti-GFP showed no major differences (less than twofold) in protein levels between MinCC-GFP and the successfully targeted GFP-MinCC (Fig. 6C).
Another possibility is that the C-terminal GFP tag inhibits dimerization of MinCC. In the MinC crystal structure, the C terminus of MinC faces another subunit of MinC in the MinC dimer, causing the C terminus to be buried in the dimer. Thus, it is possible that a C-terminal GFP tag may inhibit dimerization of full-length MinC. However, it has been speculated that dimerization of MinC is not necessary for its membrane localization, although cytoplasmic MinC may be poised at the monomer-dimer equilibrium (14). In summary, we propose that fusing GFP to the C terminus of MinC may inhibit binding of MinC to MinD and/or dimerization of MinC.
The main conclusion of the present study is that MinCC has inhibitory activity against the Z ring when overproduced. This activity is coupled to MinCC targeting to the ring, as no inhibition was observed with similar levels of the R172A mutant MinCC, which cannot target the ring. We consistently found that the inhibitory activity of MinCC was lower than that of full-length MinC, although in WT cells, similar protein levels resulted in cell filamentation. One question for the future is whether the C-terminal domain of intact MinC normally inhibits assembly of the Z ring. Although MinCN, when fused with MalE, is sufficient to prevent FtsZ polymerization in vitro (6), MinCC may enhance this activity of MinCN in vivo. Directly testing this model would be difficult, because depolymerization of FtsZ by MinCN may be dominant over any depolymerization activity by MinCC. However, it was previously reported that MinCN is about 50% as active as full-length MinC in an in vitro FtsZ depolymerization assay (6). This suggests that the MinCC domain may indeed enhance the anti-FtsZ activity of full-length MinC, which is consistent with our in vivo results.
Acknowledgments
This work was supported by grant R01-GM61074 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 3 November 2006.
REFERENCES
- 1.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordell, S. C., R. E. Anderson, and J. Löwe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geissler, B., and W. Margolin. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58:596-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, S. O., L. S. Thompson, and E. J. Harry. 2005. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J. Bacteriol. 187:6536-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, J. E., L. L. Lackner, C. A. Hale, and P. A. de Boer. 2004. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J. Bacteriol. 186:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mileykovskaya, E., I. Fishov, X. Fu, B. D. Corbin, W. Margolin, and W. Dowhan. 2003. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J. Biol. Chem. 278:22193-22198. [DOI] [PubMed] [Google Scholar]
- 12.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun, Q., and W. Margolin. 2001. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, H., and J. Lutkenhaus. 2005. MinC mutants deficient in MinD- and DicB-mediated cell division inhibition due to loss of interaction with MinD, DicB, or a septal component. J. Bacteriol. 187:2846-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]