Abstract
Pseudomonas putida encodes 20 extracytoplasmic sigma factors (ECFs). In this study, we show that one of these ECFs, known as ECF-Pp12 (PP3006), plays a role in tolerance of toluene and other organic solvents. Based on this finding, we have called the gene that encodes this new ECF rpoT. The rpoT gene forms an operon with the preceding gene and with the gene located downstream. The translated gene product of the open reading frame PP3005 is an inner membrane protein, whereas the PP3007 protein is periplasmic. A nonpolar ΔrpoT mutant was generated by homologous recombination, and survival of the mutant was tested under various stress conditions. The mutant strain was hypersensitive to toluene and other solvents but just as tolerant as the wild type of stress imposed by heat, antibiotics, NaCl, paraquat, sodium dodecyl sulfate, H2O2, and benzoate. In the ΔrpoT mutant background, expression of around 50 transcriptional units was affected: 31 cistrons were upregulated, and 23 cistrons were downregulated. This indicates that about 1% of all P. putida genes are under the direct or indirect influence of RpoT. The rpoT gene controls the expression of a number of membrane proteins, including components of the respiratory chains, porins, transporters, and multidrug efflux pumps. Hypersensitivity of the P. putida RpoT-deficient mutant to organic solvents can be attributed to the fact that in the ΔrpoT strain, expression of the toluene efflux pump ttgGHI genes is severalfold lower than in the parental strain.
In natural environments, microorganisms are exposed to changing conditions and have therefore developed a series of strategies to cope with these stressors (17, 48). With world industrialization, humans have synthesized a large number of chemicals, many of which reach the biosphere and constitute a new burden for the environment. Among the most toxic chemicals are organic solvents, such as toluene, xylenes, and styrene, which dissolve in the cell membrane, disorganize it, cause the loss of lipids and proteins, and eventually lead to cell death (8, 45, 56). Microorganisms have developed various mechanisms to resist the lethal effects of these chemicals. One of the most relevant of these mechanisms is the active reduction of their entry into the cells through the action of membrane efflux pumps, which belong to the group of multidrug resistance pumps (45). These efflux pumps extrude a broad range of structurally unrelated synthetic and natural chemicals and thus constitute an effective barrier against toxic chemicals.
Pseudomonas putida DOT-T1E exhibits the unusual property of being highly tolerant of organic solvents (39, 47). Solvent tolerance in this strain is ultimately the result of an interplay between three efflux systems known as TtgABC, TtgDEF, and TtgGHI, which have been assigned to the root nodulation cell division family (15, 45, 46, 55). Of these three efflux pumps, the TtgGHI pump seems to be the most critical for the removal of solvents from a quantitative point of view (51). The ttgGHI and the ttgDEF operons are induced in response to a variety of solvents (16; W. Terán, M. A. Felipe, M. E. Guazzaroni, T. Krell, R. Ruíz, J. L. Ramos, and M. T. Gallegos, submitted for publication), whereas the TtgABC efflux pump is expressed constitutively. The latter pump protects the cell, not only from solvents, but also from antibiotics, monocationic compounds (e.g., ethidium bromide), and toxic secondary products of plants (12, 61, 62).
Domínguez-Cuevas et al. (11) have shown that at the transcriptional level, changes in the response of P. putida KT2440 to toluene occur without significant changes in the levels of mRNAs of RNA polymerase components (core or sigma factors). This suggests that a certain amount of “roaming” RNA polymerase is involved in setting up these new programs. In eubacteria, σ70, encoded by rpoD, is responsible for the expression of most of the housekeeping genes required for normal cellular metabolism during exponential growth. To maintain cellular homeostasis in a variety of stress environments, bacteria use alternative sigma factors to redirect RNA polymerase and selectively express discrete subsets of genes. Among these are the extracytoplasmic function (ECF) subfamily of σ factors, which are divergent in sequence with respect to nonessential and secondary sigma factor groups (17, 19). In many cases, the ECF σ factor is cotranscribed with a transmembrane anti-σ factor with an extracytoplasmic sensory domain and an intracellular inhibiting domain and often control functions associated with aspects of the cell surface or transport (17, 31).
Twenty-four sigma factors have been identified in the genome of P. putida, 20 of which correspond to the ECF subfamily (35). Of these 20 sigma factors, 13 are potentially involved in iron acquisition in P. putida. Of the remaining seven, roles have been assigned to AlgU, also known as σE (RpoE), which is involved in the biosynthesis of alginate and in thermoresistance (36), and to σH (RpoH), which is engaged in heat and oxidative-stress responses. The functions of the other sigma factors, ECF-Pp1, ECF-Pp10, ECF-Pp11, ECF-Pp12, and ECF-Pp13, are unknown.
We hypothesized that the set of responses to toluene in P. putida could involve one or more of these alternative sigma factors, since this solvent induces the misfolding of periplasmic and membrane proteins. The work reported here demonstrates that the protein originally designated ECF-Pp12 (35), now called σT because of its involvement in toluene resistance, is required for P. putida to endure solvent shocks. The ttgGHI operon, which encodes the major efflux pump in terms of solvent extrusion, is a member of a σT regulon that also includes about 1% of the P. putida genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Pseudomonas putida DOT-T1E (Rifr Tolr) (47) and DOT-T1E-ΔrpoT (Rifr Kmr Tols ΔrpoT) (this study) were grown at 30°C in LB medium or in M9 minimal medium supplemented with glucose (0.5% [wt/vol]) or citrate (10 mM) as the carbon source. Escherichia coli DH5α and E. coli CC118λpir cells were grown on LB medium at 37°C. The former was used for cloning experiments and the latter to replicate plasmids based on pKNG101 (18).
Plasmid pGEMT was used for cloning PCR-amplified DNA fragments. Plasmid pRK600 (Cmr mob+ tra+; ColE1 replicon) was used as a helper for the mobilization of tra mob+ plasmids (18). A derivative of the suicide plasmid pKNG101 (Kmr mob+; ColE1 replicon) was used to generate a knockout of the rpoT gene in vivo through a double-recombination event without exerting polar effects on the downstream genes (37; see below). Plasmid pUCH218 (59) was used for the construction of the different ′phoA fusions. Subsequently, the corresponding ′phoA fusions were subcloned into pBBR1MCS-5 (25) to transfer them to P. putida. The broad-host-range tetracycline-resistant pMP220 plasmid that carries a promoterless ′lacZ gene was used as a promoter-probe vector (58).
The antibiotics used were ampicillin, 100 μg/ml; kanamycin (Km), 50 μg/ml; piperacillin, 100 μg/ml; rifampin (Rif), (20 μg/ml); streptomycin, 50 to 100 μg/ml; and tetracycline, 15 μg/ml. Gentamicin was used at a concentration of 5 μg/ml for E. coli and 20 μg/ml for P. putida.
Stress assays using double-diffusion plates.
To test the effects of a number of stressor agents, we used the double-diffusion plate assay (14) so that the concentration of the chemical ranged between zero and the highest concentration desired. Compounds and ranges were as follows: ampicillin (0 to 1,000 μg/ml), chloramphenicol (0 to 100 μg/ml), paraquat (0 to 0.3% [wt/vol]), benzoate (0 to 20 mM), NaCl (0 to 1.5 M), and SDS (0 to 1% [wt/vol]).
H2O2 and solvent shock assays in liquid culture medium.
Cells were grown overnight in 30 ml LB medium with or without toluene in the gas phase. On the following day, the cultures were diluted 1:100 in the same medium and grown under the same conditions. When the cultures reached the mid-exponential growth phase (turbidity, 0.8 ± 0.05 at 660 nm), they were divided into three aliquots. We added 0.3% (vol/vol) toluene to one of the aliquots and 1.5% (vol/vol) H2O2 to another, and the third was kept as a control. The number of viable cells was determined as CFU/ml before the stressor agent was added and 10, 30, and 60 min later. These assays were run in duplicate and repeated at least three times. The values are the averages of at least six determinations.
DNA techniques.
Plasmid DNA was isolated according to the alkaline lysis method with the QIAprep Spin Plasmid Minipreps Kit. Total DNA was isolated as described by Ramos-González et al. (49). DNA digestions with restriction enzymes, ligations, and transformations were performed with standard procedures (53). For PCRs, the standard mixture (25 μl) contained 10 ng of DNA, 200 μM of each deoxynucleoside triphosphate, 50 pmol of each primer, 2 μl of dimethyl sulfoxide, and 0.25 U of Taq polymerase. The PCR conditions were as follows: 4 min at 95°C and then 35 cycles at 60°C for 45 s, 72°C for 30 to 180 s, and 94°C for 4 s, followed by a final 5-min step at 72°C. Southern hybridization under high-stringency conditions (50% [vol/vol] formamide at 42°C) was carried out with digoxigenin-labeled probes and immunologically developed using Roche's kit.
Generation of a ΔrpoT mutant by insertional inactivation.
To generate a ΔrpoT::Km P. putida mutant strain, a 2,660-bp DNA fragment covering the rpoT gene and adjacent DNA was amplified by PCR from the P. putida DOT-T1E chromosome using primers 5′-GGAAAGCCAGTAGCGACTTC-3′ and 5′-CTTCAGCGTGGCTTCCTTGCCG-3′. The amplified product was cloned in pGEMT to yield pGEMT-rpoT. Then, DNA was digested with PmlI and Eco47III, which removed 400 bp from the rpoT gene, and the aphA3 gene encoding kanamycin resistance (37) was inserted between these sites to yield pGEMTrpoTaph3. This plasmid was digested with NotI, and the resulting 3.3-kb NotI fragment was cloned at the NotI site of pKNG101 (23) to yield plasmid pEDX-1. This plasmid was designed for marker exchange mutagenesis because it replicates in E. coli CC118λpir but not in P. putida. Transfer of pEDX-1 to P. putida DOT-T1E, selection of merodiploids, and subsequent resolution and generation of mutants after homologous double recombination were carried out as described before (50). A random mutant clone, called DOT-T1E-ΔrpoT, was retained for further studies. The absence of the wild-type allele in DOT-T1E-ΔrpoT and the presence of the rpoT::aphA3 allele were confirmed by PCR and Southern blotting (not shown). Consistency of the mutation was confirmed by sequencing DNA.
Gene fusions to ′phoA.
To construct the PP3007′::′phoA translational fusion, a KpnI-BamHI-digested PCR fragment amplified from P. putida DOT-T1E genomic DNA with primers PP3007-5 (5′-CAGGTACCTAGTGGGCGGCCTTTGAC-3′) and PP3007-3 (5′-TAGGATCCCATTGAGCACTGCCTTGG-3′) was cloned into KpnI-BamHI-cut pUCH218 and transformed into E. coli CC118λpir. Two PP3005 gene in-frame ′phoA fusions were generated in the same way as described above but using primers PP3005-5 (5′-CAGGTACCAAGAGGACGGGAGTGTCTGGAATG-3′) and PP3005-3S (5′-TAGGATCCACAGGCGGGGGCTTGGG-3′) to amplify the first 70 codons of PP3005 and primers PP3005-5 and PP3005-3L (5′-TAGGATCCGCTCGCCCTTGGTCCAGAAC-3′) to amplify the first 171 PP3005 codons. Subsequently, the ′phoA fusion constructs were excised as KpnI-HindIII fragments, cloned into pBBR1MCS-5 digested with the same enzymes, and transformed into E. coli CC118λpir to render plasmids pB3007AP (PP3007::phoA fusion), pB3005AP-S [PP3005(1-70)::phoA fusion], and pB3005AP-L [PP3005(1-171)::phoA fusion]. The KpnI-HindIII fragment from pUCH218 carrying the ′phoA gene was also cloned into pBBR1MCS-5, and the resulting plasmid, designated pBBRphoA, was used as a negative control. All four plasmids were further transferred into P. putida DOT-T1E via triparental mating using pRK600 as the helper plasmid, as described previously (49). Colonies containing active ′phoA fusions were screened for blue color on LB plates containing 5-bromo-4-chloro-3-indolyl phosphate (BCIP) at 40 μg/ml.
Cell fractionation and Western blot analysis.
P. putida cells harboring the different ′phoA fusion constructs were grown in LB medium with gentamicin to reach an optical density at 660 nm of ∼1.0. The cultures (2 ml) were then centrifuged (10,000 × g; 5 min; 4°C), and the resulting pellet fractions were solubilized in 100 μl of Laemmli sample buffer (28), treated with Benzonase (Merck, Madrid, Spain) for 10 min to degrade the DNA, and heated for 5 min at 95°C. The resulting sample was designated a whole-cell lysate. For cell fractionation experiments, cells from 1 ml of culture were harvested by centrifugation as described above and gently suspended in 1 ml of an ice-cold solution containing 10 mM Tris-HCl (pH 8), 0.5 M sucrose, 1 mM EDTA, lysozyme (100 μg/ml), and a mixture of protease inhibitors (Complete EDTA-free; Roche; catalog no. 11873580001). After a 10-min incubation on ice, spheroplasts were recovered by centrifugation (5,000 × g; 10 min; 4°C), and the periplasmic proteins in the supernatant were acid precipitated by the addition of 0.1 volume of trichloroacetic acid (TCA). After incubation on ice for 30 min, the precipitated proteins were sedimented by centrifugation at 20,000 × g for 30 min at 4°C, washed with 80% (vol/vol) acetone to remove residual TCA, air dried, resuspended in 100 μl of Laemmli buffer, and boiled for 10 min. To obtain the cytoplasmic and membrane fractions, the spheroplast pellet was suspended in 1 ml of an ice-cold solution containing 10 mM Tris-HCl (pH 8), 1 mM EDTA, 10 mM MgCl2, and protease inhibitors and subjected to three freeze-thaw cycles in liquid nitrogen and a 37°C water bath. To digest DNA, Benzonase (5 U) was added to the suspension and incubated on ice for 10 min. Following centrifugation of the sample at 5,000 × g for 10 min at 4°C to remove unbroken spheroplasts, the supernatant was centrifuged at 30,000 × g for 30 min at 4°C to separate the membrane fraction (pellet) from the soluble cytosolic fraction (supernatant). The pellet fraction was resuspended in 100 μl of Laemmli buffer and boiled for 10 min. The cytoplasmic proteins were TCA precipitated from the supernatant and processed as described above for the periplasmic proteins. Samples (15 μl) of the different fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting as described previously (30). The nitrocellulose blots were probed with an anti-PhoA monoclonal antibody (1:1,000 dilution; Caltag Laboratories; reference ME6200) and an alkaline phosphatase-conjugated rabbit anti-mouse secondary antibody (1:5,000 dilution; Calbiochem; catalog no. 401262), and developed in 10 ml of developing buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2) with 33 μl of nitroblue tetrazolium (100 mg/ml) and 33 μl of BCIP (50 mg/ml).
Pseudomonas putida microarrays.
P. putida arrays (Progenika, Spain) consisting of 5,539 gene-specific oligonucleotides (50-mer) spotted in duplicate onto γ-aminosylane-treated 25 by 75 microscope slides and linked to the slide with UV light and heat (60) were used. The oligonucleotides represented 5,350 of the 5,421 predicted open reading frames (ORFs) annotated in the P. putida KT2440 genome (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org_search=pse&org=gpp). In addition, 140 of the 148 putative ORFs predicted for the TOL plasmid pWW0 were also represented, together with the genes coding for the toluene efflux pumps of P. putida DOT-T1E (ttgABC, ttgDEF, and ttgGHI) and their corresponding regulatory genes (ttgR, ttgT, and ttgV), plus a suite of commonly used reporter genes and antibiotic resistance markers. The chips were also endowed with homogeneity controls consisting of oligonucleotides for the rpoD and rpoN genes spotted at 20 different positions, as well as duplicated negative controls at 203 predefined positions. Further details of the array characteristics were reported elsewhere by Yuste et al. (68). Assays under each set of conditions were replicated at least four times (4, 13).
RNA isolation and preparation of labeled cDNA.
P. putida DOT-T1E and DOT-T1EΔ rpoT cells were grown in LB medium to an optical density at 660 nm of 0.5. The cultures were then divided into two halves with toluene (gas phase) added to one of them and were further incubated at 30°C for another 20 min. The exposure time was long enough to allow the complete transcription of all P. putida operons but was short enough to disclose the initial response of the cells to the solvent shock. After the 20-min incubation, cells from 12-ml culture samples were harvested by centrifugation at 4°C in tubes precooled in liquid nitrogen. After centrifugation, the cell pellets were immediately immersed in liquid nitrogen, and total RNA was isolated using TRI Reagent (Ambion; reference 9738) as recommended by the manufacturer. The RNA preparations were then subjected to DNase treatment, followed by purification with RNeasy columns (QIAGEN; catalog no. 74104). The RNA concentration was determined spectrophotometrically, and its integrity was assessed by agarose gel electrophoresis.
For the preparation of fluorescently labeled cDNA, 25 μg of RNA was primed with 7.5 μg of random hexamers (Amersham; catalog no. 27-2166-01). Probe synthesis was performed at 42°C for 2 h in a 30-μl reaction volume containing 0.5 mM (each) dATP, dCTP, and dGTP; 0.25 mM (each) dTTP and aminoallyl-dUTP (Sigma; catalog no. A0410); 10 mM dithiothreitol; 40 U of RNaseOUT (Invitrogen; reference 10777-019); 400 U of SuperScript II reverse transcriptase (RT) (Invitrogen; reference 18064-014) in RT reaction buffer. The reaction was stopped by adding 10 μl of 50 mM EDTA, and the RNA template was hydrolyzed by adding 10 μl of 1 N NaOH, followed by incubation at 65°C for 15 min. Samples were then neutralized by adding 25 μl of 1 M HEPES (pH 7.5), and the hydrolyzed RNA and residual deoxynucleoside triphosphates were removed using QIAquick PCR purification columns (QIAGEN; reference 28104) according to the manufacturer's recommendations, except that the Tris buffers were replaced with phosphate wash (5 mM K2HPO4, pH 8.0, 80% [vol/vol] ethanol) and elution buffers (4 mM K2HPO4, pH 8.5) to avoid interference with the subsequent labeling. cDNA samples were dried in a Speed-Vac to completion. The dried aminoallyl-labeled cDNA was resuspended in 9 μl of 0.1 M sodium carbonate buffer (pH 9.0), mixed with either Cy3 or Cy5 fluorescent dye (mono-reactive N-hydroxysuccinimide esters; Amersham Biosciences; catalog no. PA23001 and PA25001, respectively), and allowed to couple for 2 h at room temperature in the dark. After the coupling, the reaction was quenched with 4.5 μl of 4 M hydroxylamine for 15 min. Then, 35 μl of 100 mM sodium acetate (pH 5.2) was added to the reaction mixtures, and they were again purified with QIAquick PCR purification columns, but this time using the supplied buffers. Labeling efficiency was assessed using a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies).
Microarray hybridization and data analysis.
Prior to the hybridization process, the microarray slides were blocked by immersion in 5× SSC (1× SSC is 0.15 M NaCl, 15 mM sodium citrate, pH 7), 0.1% (wt/vol) SDS, 1% (wt/vol) bovine serum albumin for 1 h at 42°C. Then, the slides were washed by two successive immersions in MilliQ water at room temperature, followed by a final wash with isopropanol. The slides were spin dried by centrifugation at 1,500 × g for 5 min and used within the next hour. Equal amounts of Cy3- and Cy5-labeled cDNAs, one of them corresponding to the control and the other to the problem to be analyzed, were mixed, dried in a Speed-Vac, and reconstituted in 35 μl of hybridization buffer (5× SSC, 25% [vol/vol] formamide, 0.5% [wt/vol] SDS, 5× Denhardt's solution, 5% [wt/vol] dextransulfate) preheated to 42°C. The labeled probe was denatured at 98°C for 3 min, applied to the microarray slide, and covered with a glass coverslip. The slide was then introduced into a humidified hybridization chamber (AHC ArrayIt Hybridization Cassette; Telechem International, Inc.) and incubated for 18 to 20 h in a water bath at 42°C, preserved from light. Following hybridization, the microarrays were washed by gentle agitation in 2× SSC, 0.1% [wt/vol] SDS at 42°C for 5 min, followed by a 5-min wash at room temperature in 1× SSC, two 5-min washes in 0.2× SSC, and a final 5-min wash in 0.1× SSC. Finally, the slides were spin dried in a centrifuge at 1,500 × g for 5 min and scanned on a GenePix 4100A scanner (Axon Instruments, Inc.). Images were acquired at 10-μm resolution, and the background-subtracted median spot intensities were determined using GenePix Pro 5.1 image analysis software (Axon Instruments, Inc.). Signal intensities were normalized by applying the lowest intensity-dependent normalization method (65) and statistically analyzed using the Almazen System software (Alma Bioinformatics S. L.). To allow appropriate statistical analysis of the results, RNA preparations from at least four independent cultures were tested for each strain (4). P values were calculated by using Student's t test. A particular ORF was considered differentially expressed if (i) the change was at least 1.8-fold and (ii) the P value was 0.05 or lower.
Computational methods.
Protein signal sequences and transmembrane domains were predicted by using the SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (2) and TMHMM v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) servers (27), respectively. Kyte-Doolittle hydropathy plots were generated with DNA Strider v1.4f5 (34) by using a window size of 9 (for PP3007) or 19 (for PP3005).
RESULTS
The gene that encodes ECF-Pp12 is clustered with genes encoding PP3005 and PP3007.
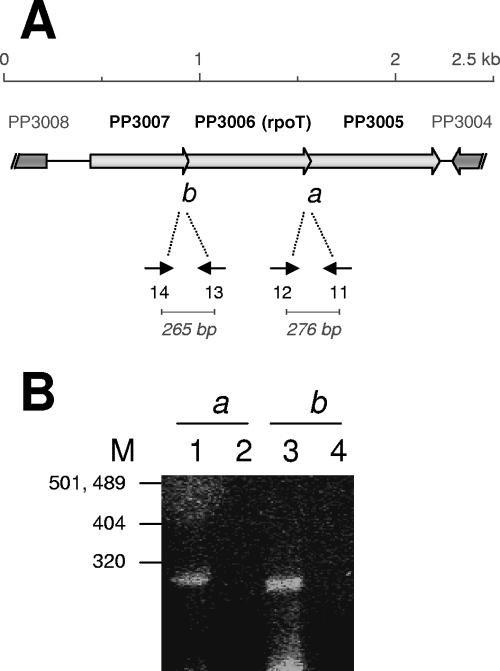
Martínez-Bueno et al. (35) suggested that ECF-Pp12 is related to alternative sigma factors involved in stress endurance (9, 19, 54, 57, 60, 63, 67); however, no clues to the role of ECF-Pp12 are available. The gene encoding ECF-Pp12 is located in a chromosomal cluster with two ORFs (PP3007 and PP3005) that are transcribed in the same direction as rpoT (Fig. 1A). Located divergently with respect to PP3005 is a cluster of two genes involved in aromatic-acid biosynthesis (aroQ and aroE) and an ORF that encodes a protein (PP3004) of unknown function. Divergent with respect to PP3007 is a cluster of four genes that encode proteins PP3008 to PP3011 of unknown function (41). PP3005 and PP3007 show no significant homology to any other known proteins, including anti-sigma factors. Sequence analysis revealed that the PP3006 and PP3005 ORFs overlap by 13 bp, and PP3006 and PP3007 overlap by 3 bp, suggesting a putative operon structure for the three genes (GenBank DQ338456). To test whether the three ORFs indeed formed a transcriptional cluster we performed RT-PCR extension using primers based on the 3′- and 5′-terminal ends of adjacent genes. In all cases, we found mRNA covering orf3007 and orf3006 on one hand and orf3006 and orf3005 on the other (Fig. 1B). Furthermore, primers based on PP3005 and PP3007 were also positive, confirming the operon structure of the cluster (not shown).
FIG. 1.
Physical map of the area around ORF3006 and RT-PCR assays. (A) Physical organization of the DNA region containing ORFs that encode PP3005, RpoT, and PP3007 of P. putida DOT-T1E. (B) Proof of operon structure of rpoT with adjacent genes. Total RNA was isolated from the wild-type strain P. putida DOT-T1E, and oligonucleotides 14 (5′-CGCAGAACAAAGGTGCCAGGC-3′) and 13 (5′-CGTGATCGTAGAGCGCCTGC-3′), as well as 12 (5′-GCAGCCACGCCGTGATAGCC-3′) and 11 (5′-TGCGGGGTAGACAGCGCGG-3′), were used to generate cDNA, which was separated on agarose gels. Lanes 2 and 4 correspond to the control, which contained the same amounts of RNA, primers, and Taq polymerase as the other samples but no reverse transcriptase. The positions of the molecular size markers (in bp) are indicated.
The P. putida DOT-T1E PP3007 protein is located in the periplasm, whereas PP3005 is located in the inner membrane.
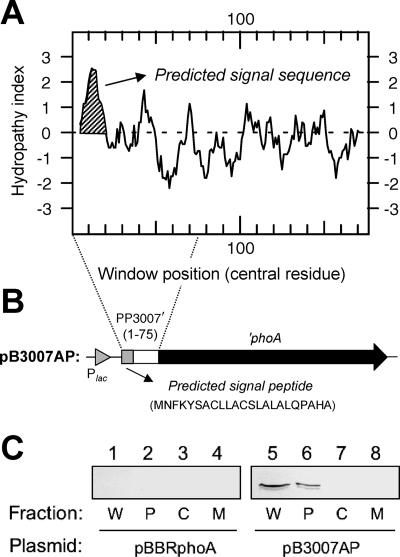
Kyte-Doolittle hydropathy analysis of the peptide sequence deduced for P. putida DOT-T1E PP3007 (18.6 kDa; 175 residues) showed a rather hydrophilic protein with a hydrophobic amino-terminal region (Fig. 2A) that resembled a signal sequence. A more detailed analysis of the PP3007 amino-terminal sequence (MNFKYSACLLACSLALALQPAHAQT) with the SignalP program revealed the presence of a putative signal peptide with a predicted cleavage site between positions 23 and 24 (AHA/QT, residues 21 through 25 [the shill indicates the cleavage site]). Based on these observations, we predicted that PP3007 was a periplasmic protein. To check this hypothesis, we constructed a translational fusion with the E. coli ′phoA gene lacking its own signal sequence. This hybrid protein should have shown alkaline phosphatase activity only if it was exported through the inner membrane to the periplasm (33). A DNA fragment encoding the first 75 amino acids of the PP3007 protein was fused to the E. coli ′phoA gene (Fig. 2B), and the construct was cloned into the broad-host-range plasmid pBBR1MCS-5 to yield pB3007AP and transferred into P. putida DOT-T1E. Then, the transconjugant strain was tested for alkaline phosphatase activity on solid LB medium containing the chromogenic substrate BCIP. Positive (blue) colonies were observed, which proved that the amino-terminal region of PP3007 was sufficient for targeting the hybrid protein to the periplasm. To further confirm that PP3007′-′PhoA was located in the P. putida periplasm, we performed subcellular fractionation on DOT-T1E cells carrying the plasmid mentioned above. Pseudomonas putida DOT-T1E(pBBRphoA) cells were used as a negative control. Western blot analysis of the different fractions using an anti-PhoA antibody (Fig. 2C) showed the presence of a band (about 54 kDa, corresponding to the mature protein without the predicted signal sequence) in the periplasmic fraction (lane 6) of P. putida DOT-T1E(pB3007AP), but not in the cytoplasmic or membrane fraction (lanes 7 and 8). In the extract of whole cells, the mature protein was also detected (lane 5). Taken together, these results strongly suggest that PP3007 is a soluble periplasmic protein.
FIG. 2.
Subcellular localization of P. putida DOT-T1E PP3007. (A) Hydrophobicity (Kyte-Doolittle) plot of the PP3007 protein showing the putative signal peptide (window size, 9). The average hydrophobicity of the amino acids included in the window is plotted in the midpoint of the window. (B) Schematic view of the PP3007′-′phoA fusion under the control of the Plac promoter in plasmid pB3007AP (positive for alkaline phosphatase activity). (C) Immunoblot detection of PP3007′-′PhoA in P. putida DOT-T1E(pP3007AP) cell fractions (lanes 5 to 8). P. putida DOT-T1E(pBBRphoA) was used as a negative control (lanes 1 to 4). Protein samples were subjected to electrophoresis (12.5% [wt/vol] SDS-PAGE), followed by Western blotting with an anti-PhoA antibody. Lanes 1 and 5, whole-cell lysate (W); lanes 2 and 6, periplasmic fraction (P); lanes 3 and 7, cytoplasmic fraction (C); and lanes 4 and 8, membrane fraction (M). The positions of molecular mass markers are indicated (in kilodaltons) on the left.
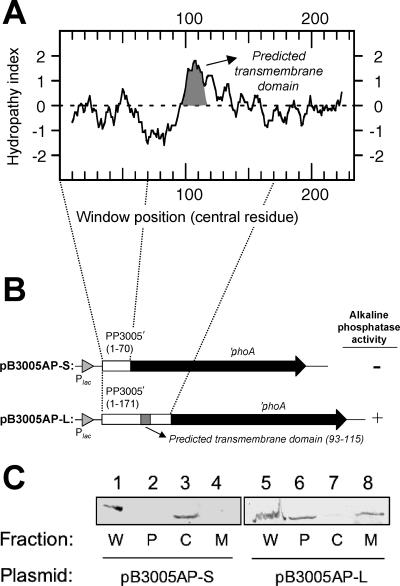
The hydropathy plot of the deduced amino acid sequence of P. putida DOT-T1E PP3005 (25.3 kDa; 233 residues) revealed the presence of a central hydrophobic region (Fig. 3A). The transmembrane prediction program TMHMM predicted a putative transmembrane domain within this region (extending from residues 93 to 115) with the N terminus facing the cytoplasm. To validate this hypothesis, two translational fusions to the ′phoA gene were constructed, one upstream of the putative transmembrane segment (at residue 70) and another downstream (at residue 171). Only the longest fusion gave rise to blue colonies on solid LB medium with BCIP, both in E. coli (not shown) and in P. putida (Fig. 3B), which implied that the C-terminal region of PP3005 was exposed to the periplasm. Immunoblot analysis of P. putida DOT-T1E(pB3005AP-S) and P. putida DOT-T1E(p3005AP-L) cell fractions with an anti-PhoA antibody verified the expression of two fusion proteins of about 54.0 and 66 kDa (Fig. 3C, lanes 1 and 5). The results also showed that whereas PP3005(1-70)-PhoA was recovered from the soluble cytoplasmic fraction (Fig. 3C, lane 3), PP3005(1-171)-PhoA was found predominantly in the membrane fraction (Fig. 3C, lane 8).
FIG. 3.
Subcellular localization of P. putida DOT-T1E(PP3005). (A) Hydrophobicity (Kyte-Doolittle) plot of the PP3005 protein showing a potential transmembrane segment (window size, 19; peaks with scores greater than 1.8 indicate putative transmembrane α-helices). (B) Schematic view of two different PP3005′-′phoA fusions under the control of the Plac promoter in plasmids pB3005AP-S and pB3005AP-L, indicating the expression of alkaline phosphatase activity. The numbers indicate the positions of the first and last amino acid residues of the proposed transmembrane domain. (C) Immunoblot detection of the PP3005′-′PhoA fusions in P. putida DOT-T1E cell fractions. Protein samples were subjected to electrophoresis (12.5% [wt/vol] SDS-PAGE), followed by Western blotting with an anti-PhoA antibody. Lanes 1 through 4, P. putida DOT-T1E(pB3005AP-S); lanes 5 through 8, P. putida DOT-T1E(pP3005AP-L). Lanes 1 and 5, whole-cell lysate (W); lanes 2 and 6, periplasmic fraction (P); lanes 3 and 7, cytoplasmic fraction (C); and lanes 4 and 8, membrane protein fraction (M).
Deletion of rpoT leads to a decrease in solvent resistance in P. putida.
To study the role of the P. putida Pp-ECF-12 protein (PP3006), we generated DOT-T1E-ΔrpoT so that an internal portion of the gene encoding PP3006 was deleted and replaced by a promoterless Kmr cassette that exerted no polar effect on the expression of downstream genes (37) (see Materials and Methods for construction details). No significant differences in the growth of the DOT-T1E-ΔrpoT mutant and its wild-type strain were observed when the cells were grown on LB or M9 minimal medium with citrate or glucose as the sole carbon source at 30°C (data not shown).
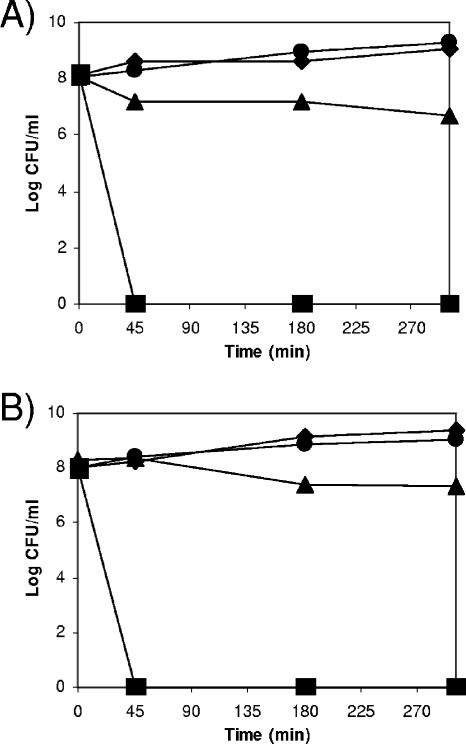
ECF-Pp12 belongs to the ECF sigma factor group (17). Members of this subfamily are involved in responses to different stresses. For instance, the ECF rpoE sigma factor product is involved in heat, metal, and desiccation stresses in E. coli (13, 20, 64). Mycobacterium tuberculosis rpoE mutants are sensitive to envelope perturbations caused by polymyxin and SDS and are also more sensitive to high temperatures than the wild-type strain (22, 32, 67). A Bacillus subtilis sigM null mutant lacking the ECF σM protein was sensitive to paraquat, a superoxide-generating reagent (38). In Listeria monocytogenes, σB contributes to cell survival under energy stress conditions imposed by carbonyl cyanide m-chlorophenylhydrazone (5). Yersinia enterocolitica rpoE mutant strains exhibited susceptibility to increased osmolarity (16, 19, 26). To identify the role of ECF-Pp12, we subjected the wild-type DOT-T1E and its isogenic DOT-T1E-ΔrpoT mutant to a series of tests in which perturbations in the cell envelope were produced. We carried out a series of double-diffusion assays in which antibiotics (ampicillin and chloramphenicol), SDS, NaCl, paraquat, and benzoate were the disturbing agents. To test tolerance to toluene, cells were spread on LB plates with solutions with different toluene concentrations (0, 0.1, and 1% [vol/vol]). We found that the DOT-T1E-ΔrpoT mutant was as tolerant as the wild-type strain of all these chemicals except toluene, since the wild-type strain formed a lawn when solid plates were overlaid with 1% (vol/vol) toluene whereas the mutant did not (see below). To test the putative effect of heat on the survival of the DOT-T1E-ΔrpoT mutant, wild-type and mutant cells in the mid-exponential phase growing on LB at 30°C were split into four aliquots and incubated at 30°C, 37°C, 42°C, and 50°C, and CFU/ml were counted over time. We found that the mutant and the wild type behaved similarly. Incubation of DOT-T1E and DOT-T1E-ΔrpoT at 30°C and 37°C had no effect on the survival of the two strains (Fig. 4), but survival was affected at 42°C (the number of CFU decreased by about 1 order of magnitude after 5 h of incubation) and a rapid decline in the number of CFU/ml was observed at 50°C, so that neither the wild-type nor the DOT-T1E-ΔrpoT cells survived after 45 min of incubation at that temperature (Fig. 4).
FIG. 4.
Survival of P. putida and its ΔrpoT mutant upon sudden temperature shock. (A) Wild-type DOT-T1E and (B) DOT-T1E-ΔrpoT. Cells were grown in 40 ml LB medium at 30°C, and when indicated, four aliquots were made and incubated at 30°C (⧫), 37°C (•), 42°C (▴), and 50°C (▪). The viable cells were counted at the indicated times. The data are the averages of six determinations, with standard deviations below 5% of the given values.
In our laboratory, we have optimized freeze-drying conditions for Pseudomonas strains and have shown that survival of the desiccation process is highly influenced by the composition of C17-cyclopropane and the lyoprotectant used (40). We tested the survival of DOT-T1E and DOT-T1E-ΔrpoT in the lyophilization process and found no significant differences in the rates of survival with cells in the exponential or stationary phase and with myo-inositol or trehalose as the lyoprotectant (not shown).
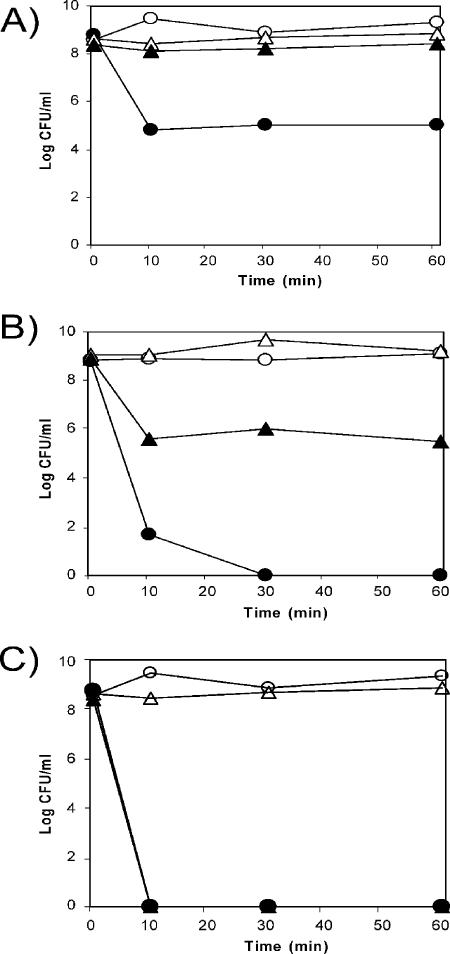
The above-mentioned series of results indicate that DOT-T1E-ΔrpoT cells were specifically sensitive to toluene. To quantify the effect of toluene, we carried out tests with DOT-T1E and DOT-T1E-ΔrpoT cells in the mid-exponential phase. We reported before that only about 1 out of 104 CFU/ml of DOT-T1E tolerated a sudden 0.3% (vol/vol) toluene shock, whereas 50 to 100% of the cells tolerated the toluene shock if preinduced with sublethal toluene concentrations. This behavior was confirmed here (Fig. 5A). In contrast with this behavior, we found that DOT-T1E-ΔrpoT mutant cells were very sensitive to toluene, and survival of noninduced cells was below 1 out of 109 CFU/ml. About 1 out of 103 to 104 cells survived the sudden toluene shock if the cultures were preinduced with sublethal toluene concentrations (Fig. 5B). This suggested that ECF-Pp12 sigma factor was required for full cellular defense against toluene. We therefore called the gene rpoT to indicate its role in sensitivity to toluene.
FIG. 5.
Survival of P. putida DOT-T1E and its ΔrpoT mutant upon solvent shock. The strains used were P. putida DOT-T1E (A), DOT-T1E-ΔrpoT (B), and DOT-T1E-PS28 (C). Cells were grown in 30 ml LB medium (circles) or LB medium with toluene in the gas phase (triangles) until the culture reached a turbidity of about 0.8 at 660 nm. The culture was divided into two halves, and to one we added 0.3% (vol/vol) toluene (closed symbols) while the other was kept as a control (open symbols). The numbers of viable cells were determined at the indicated times. The data are the averages of 6 to 10 determinations, with standard deviations in the range of 3 to 8% of the given values.
Global gene expression analysis and further phenotypic analysis of the ΔrpoT mutant.
To determine the full impact of the ΔrpoT deletion on solvent homeostasis in P. putida, we analyzed global cellular expression with microarrays. Cells of the P. putida DOT-T1E wild-type strain and its ΔrpoT isogenic deletion mutant strain were exposed or not to 1 mM toluene for 20 min prior to RNA isolation. The concentration of toluene used allowed 100% survival of wild-type and mutant cells, and enough time was allowed for the cells to induce solvent tolerance mechanisms as before (52). In each microarray, 5,350 gene-specific spots were analyzed. Based on the up- and downregulated genes, a series of physiological analyses were performed to further characterize the ΔrpoT mutant. For microarrays, each condition was tested at least four times, including two independent cultures of the P. putida strains and a dye swap experiment. The mean values for the four normalized expression ratios were then evaluated to compare the ΔrpoT mutant and the wild-type cells with and without toluene addition. A change in expression of more than 1.8-fold in either direction, when the difference between the two signals was larger than the sum of the two signal deviations, was considered a significant change.
Comparison of the ΔrpoT mutant and wild-type cells grown in the absence and in the presence of the solvent revealed that a relatively low number of genes were influenced by ΔrpoT (Tables 1 to 4). In fact, we found that in the ΔrpoT mutant cells growing without toluene, the expression of 53 genes was upregulated (Table 1) and the expression of 26 genes was downregulated (Table 2). This indicated that about 1% of the genes were under the influence of rpoT. When the physical organization of the genes was analyzed, we found that in many cases the genes formed operons, which brought the number of transcriptional units directly or indirectly regulated by rpoT to 31 upregulated units and 23 downregulated units (Tables 1 and 2). Therefore, the altered mRNA concentrations of these genes could be considered an effect of the ΔrpoT deletion.
TABLE 1.
Genes upregulated in the DOT-T1E rpoT mutant (versus the wild-type strain) before toluene additiona
| TIGR identifier | Gene product | Gene name | Change (fold) |
|---|---|---|---|
| PP0257 | Formate dehydrogenase accessory protein FdhD | fdhD | 2.1 |
| PP0504 | Outer membrane protein OprG | oprG | 2.0 |
| PP0792 | Fructose transport system repressor FruR | fruR | 1.8 |
| PP0812 | Cytochromeoubiquinol oxidase, subunit II | cyoA | 2.2 |
| PP0813 | Cytochromeoubiquinol oxidase, subunit I | cyoB | 2.3 |
| PP0814 | Cytochromeoubiquinol oxidase, subunit III | cyoC | 2.1 |
| PP0816 | Protoheme IX farnesyltransferase | cyoE-2 | 2.3 |
| PP0887 | Sensor histidine kinase | 2.1 | |
| PP0888 | Response regulator | 1.9 | |
| PP0951 | Sigma-54 modulation protein | 1.8 | |
| PP1002 | Lysine permease | lysP | 2.0 |
| PP1110 | Serine O-acetyltransferase; putative | 4.5 | |
| PP1111 | Synthetase; putative | 4.7 | |
| PP1112 | Conserved hypothetical protein | 2.3 | |
| PP1113 | Pyridoxal-phosphate-dependent enzyme family protein | 2.3 | |
| PP1149 | Hypothetical protein | 2.1 | |
| PP1157 | Acetolactate synthase; catabolic; putative | 1.8 | |
| PP1188 | C4-dicarboxylate transport protein | dctA | 2.7 |
| PP2246 | Conserved hypothetical protein | 2.2 | |
| PP2379 | Sco1/SenC family protein | 2.8 | |
| PP2471 | Integration host factor, alpha subunit | ihfA | 1.9 |
| PP2648 | Universal stress protein family | 2.0 | |
| PP2656 | Phosphate ABC transporter; periplasmic phosphate-binding protein | 2.1 | |
| PP2916 | Riboflavin synthase, alpha subunit | ribE-2 | 2.1 |
| PP3005 | Hypothetical protein | 4.2 | |
| PP3007 | Hypothetical protein | 5.1 | |
| PP3079 | Peptidyl-prolyl cis-trans isomerase C | ppiC-2 | 1.9 |
| PP3122 | CoA-transferase, subunit A; putative | 3.0 | |
| PP3255 | Conserved hypothetical protein | 4.4 | |
| PP3365 | Acetolactate synthase; catabolic; putative | 1.9 | |
| PP3547 | Oxidoreductase; short-chain dehydrogenase/reductase family | 1.9 | |
| PP3754 | Beta-ketothiolase | 2.5 | |
| PP3839 | Alcohol dehydrogenase; zinc containing | 1.9 | |
| PP4064 | Isovaleryl-CoA dehydrogenase | ivd | 2.8 |
| PP4066 | Enoyl-CoA hydratase; putative | 1.8 | |
| PP4067 | Acetyl-CoA carboxylase; biotin carboxylase; putative | 1.8 | |
| PP4227 | Conserved domain protein | 2.2 | |
| PP4228 | Hypothetical protein | 2.4 | |
| PP4250 | Cytochromecoxidase, cbb3-type, subunit I | ccoN-1 | 4.6 |
| PP4251 | Cytochromecoxidase, cbb3-type, subunit II | ccoO-1 | 2.4 |
| PP4252 | Cytochromecoxidase, cbb3-type, CcoQ subunit | ccoQ-1 | 3.5 |
| PP4253 | Cytochromecoxidase, cbb3-type, subunit III | ccoP-1 | 3.2 |
| PP4403 | 2-Oxoisovalerate dehydrogenase; lipoamide acyltransferase component | bkdB | 1.9 |
| PP4561 | Conserved hypothetical protein | 1.8 | |
| PP4739 | Hypothetical protein | 2.2 | |
| PP4870 | Azurin | 2.0 | |
| PP4933 | Conserved hypothetical protein | 2.8 | |
| PP5029 | Formiminoglutamase | hutG | 2.0 |
| PP5030 | Imidazolonepropionase | hutI | 3.7 |
| PP5031 | Proline permease | proY | 4.6 |
| PP5032 | Histidine ammonia-lyase | hutH | 6.4 |
| PP5033 | Urocanate hydratase | hutU | 3.7 |
| PP5035 | Histidine utilization repressor | hutC | 2.0 |
Genes putatively located in the same transcriptional unit are in boldface.
TABLE 4.
Genes downregulated in the DOT-T1E rpoT mutant (versus the wild-type strain) after toluene shocka
| TIGR identifier | Gene product | Gene name | Change (fold) |
|---|---|---|---|
| PP0136 | Conserved hypothetical protein | −2.2 | |
| PP0266 | Conserved hypothetical protein | −2.3 | |
| PP0267 | Outer membrane ferric siderophore receptor; putative | −2.8 | |
| PP0290 | Glutamine amidotransferase | hisH | −1.9 |
| PP0291 | Conserved hypothetical protein | −1.8 | |
| PP0379 | Coenzyme PQQ synthesis protein B | pqqB | −2.9 |
| PP0663 | Transcriptional regulator AsnC family | −1.8 | |
| PP0700 | Transmembrane sensor; putative | −1.9 | |
| PP0794 | 1-Phosphofructokinase | fruK | −2.6 |
| PP0861 | Outer membrane ferric siderophore receptor | −2.9 | |
| PP1009 | Glyceraldehyde 3-phosphate dehydrogenase | gap-1 | −3.1 |
| PP1023 | 6-Phosphogluconolactonase | pgl | −4.2 |
| PP1024 | 2-Dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2- | eda | −3.5 |
| PP1399 | Hypothetical protein | −2.2 | |
| PP1400 | Dicarboxylate MFS transporter | −1.8 | |
| PP1545 | Hypothetical protein | −1.9 | |
| PP1673 | Cobyrinic acid a c-diamide synthase | cobB | −1.8 |
| PP1827 | Modification methylase, HemK family | −2.3 | |
| PP1891 | Type 1 pilus subunit FimI | fimI | −1.8 |
| PP2307 | Conserved hypothetical protein | −2.0 | |
| PP2310 | Methyl-accepting chemotaxis transducer | −2.3 | |
| PP2794 | Oxidoreductase, short-chain dehydrogenase/reductase family | −2.2 | |
| PP2885 | Transporter; putative | −2.0 | |
| PP2886 | Cytochrome b561; putative | −4.0 | |
| PP2887 | Catalase; putative | −3.9 | |
| PP3397 | Hypothetical protein | −1.9 | |
| PP3642 | Hypothetical protein | −2.7 | |
| PP3963 | Conserved domain protein | −2.5 | |
| PP4186 | Succinyl-CoA synthetase, beta subunit | sucC | −1.9 |
| PP4675 | Membrane protein; putative | −2.0 | |
| PP4676 | Conserved hypothetical protein | −2.4 |
Genes putatively located in the same transcriptional unit are in boldface.
TABLE 2.
Genes downregulated in the DOT-T1E rpoT mutant (versus the wild-type strain) before toluene additiona
| TIGR or GenBank identifier | Gene product | Gene name | Change (fold) |
|---|---|---|---|
| PP0267 | Outer membrane ferric siderophore receptor; putative | −2.0 | |
| PP0291 | Conserved hypothetical protein | −2.1 | |
| PP0315 | Rieske 2Fe2S family protein | −1.9 | |
| PP0794 | 1-Phosphofructokinase | fruK | −2.3 |
| PP1166 | Membrane protein; putative | −1.8 | |
| PP1399 | Hypothetical protein | −2.4 | |
| PP1868 | ATP-dependent RNA helicase, DEAD box family | −2.0 | |
| PP2420 | Outer membrane ferric siderophore receptor | −1.8 | |
| PP2885 | Transporter; putative | −3.6 | |
| PP2886 | Cytochrome b561; putative | −4.2 | |
| PP2887 | Catalase; putative | −3.0 | |
| PP3151 | Aldehyde dehydrogenase family protein | −1.9 | |
| PP3219 | Alkanesulfonate monooxygenase; putative | −1.8 | |
| PP3245 | Hypothetical protein | −1.8 | |
| PP3319 | Sensory box protein/GGDEF domain protein | −2.1 | |
| PP3397 | Hypothetical protein | −1.8 | |
| PP3480 | Hypothetical protein | −2.1 | |
| PP3612 | TonB-dependent receptor; putative | −2.0 | |
| PP3642 | Hypothetical protein | −1.8 | |
| PP3731 | Transcriptional regulator, TetR family | −1.8 | |
| PP3963 | Conserved domain protein | −2.1 | |
| PP4047 | Hypothetical protein | −2.2 | |
| PP4465 | Porin; putative | −2.1 | |
| PP4675 | Membrane protein; putative | −3.2 | |
| PP4676 | Conserved hypothetical protein | −4.2 | |
| AF299253b | Inner membrane component of the TtgGHI efflux pump | ttgH | −2.5 |
| Membrane fusion protein of the TtgGHI efflux pump | ttgG | −2.5 | |
| Outer membrane component of the TtgGHI pump | ttgI | −2.5 |
Genes putatively located in the same transcriptional unit are in boldface.
GenBank identifier.
A significant number of genes encoded membrane proteins, such as PP1166, PP2420, and PP4675. These genes, together with the TtgGHI efflux pump, were among the downregulated units. The downregulation of the ttgGHI efflux genes could be responsible for the observed sensitivity to toluene. To confirm this, we compared the killing rate by toluene (0.3% [vol/vol]) between the ΔrpoT strain and P. putida DOT-T1-PS28, which carries a mini-Tn5 insertion in ttgH. Figure 5C shows that killing of ttgH was more acute than in ΔrpoT, which implies that the low level of expression of ttgGHI still conferred some, albeit limited, survival of toluene. Other downregulated genes included those for 1-phosphofructokinase and catalase (Table 2). To test whether the decrease in catalase activity influenced the survival of P. putida when exposed to H2O2, we cultured P. putida DOT-T1E and its isogenic mutant ΔrpoT in LB until the turbidity reached 0.8 at 660 nm. We then added H2O2 to reach 1.5% (vol/vol) and determined the number of CFU/ml. We found that ΔrpoT was as sensitive to H2O2 as the parental strain, since almost 10−2 wild-type and mutant cells survived after 60 min of incubation. Since 1-fructokinase was found to be repressed, we tested whether ΔrpoT was able to grow with fructose (0.5% [vol/vol]). The mutant ΔrpoT and the parental strain grew at similar rates (tg = 112 ± 5 min), perhaps because in the presence of fructose, the FruR repressor becomes inactive and the levels of 1-phosphofructokinase are consequently similar in the wild-type and the mutant cells.
Among the upregulated genes, it is worth noting that the operon including rpoT was expressed at a higher level in the ΔrpoT mutant, which suggested that the RpoT sigma factor is somehow involved in the control of its own expression. We corroborated this finding by fusing the promoter region of the PP3007 ORF to ′lacZ in pMP220 and determining the β-galactosidase activities in RpoT-deficient and RpoT-proficient backgrounds. Our β-galactosidase results corroborated the findings obtained with microarrays: in the RpoT-deficient background β-galactosidase activity was fourfold higher than in the parental strain. To further corroborate these data, we performed quantitative RT-PCR analysis, and we confirmed that PP3007 expression was threefold higher in the ΔrpoT background than in the parental background. We consequently achieved good correlation between microarray data and quantitative-RT-PCR and gene fusion analyses.
Other sets of upregulated genes include the hut operon, which encodes proteins involved in histidine catabolism; oprG, which encodes a putative lipoprotein; a number of transporters for nutrients (amino acids and C4-dicarboxylate); a number of proteins with potential folding properties, such as PP3079 (peptidyl cis-trans isomerase C); and the o-ubiquinol oxidase and cytochrome c oxidase cbb-3-type genes of the respiratory chains (Table 1). The increased expression of hut genes did not result in higher growth rates (90 ± 4 min for wild-type versus 92 ± 3 min for mutant cells) or in higher yields (about 0.75 ± 0.05 units at 600 nm) with 5 mM histidine as the sole C and N source for both strains.
When toluene-treated ΔrpoT mutant cells were compared to toluene-treated wild-type cells, most of the genes that were expressed at a higher level than in the wild type in the absence of toluene were also expressed at similar levels in the presence of toluene. However, for five genes, the level of induction was at least doubled: PP3005 (the gene adjacent to rpoT), PP5029 and PP5030 (involved in histidine metabolism), a coenzyme A (CoA)-transferase (PP3122), and a protein of unknown function (PP4228). Furthermore, 23 other new genes (most with unknown specific functions) were induced. Of these, two were induced more than 7-fold: PP1740 (7.3-fold) and PP5390 (17-fold). It is of interest that an efflux transporter (PP0178) belonging to the MFP subfamily and the universal stress protein PP2648 (Table 3) were induced about threefold. The induction of these proteins indicates that toluene stress responses involve several different sets of genes and that the set of 23 genes induced in the ΔrpoT background in response to toluene is not under the direct control of RpoT.
TABLE 3.
Genes upregulated in the DOT-T1E rpoT mutant (versus the wild-type strain) after toluene shocka
| TIGR identifier | Gene product | Gene name | Change (fold) |
|---|---|---|---|
| PP0153 | Conserved hypothetical protein | 2.7 | |
| PP0155 | Pyridine nucleotide transhydrogenase, beta subunit | pntB | 2.2 |
| PP0178 | Efflux transporter; membrane fusion protein; putative | 2.4 | |
| PP0504 | Outer membrane protein OprG | oprG | 3.7 |
| PP0529 | Exodeoxyribonuclease VII small subunit | xseB | 2.1 |
| PP0530 | 3,4-Dihydroxy-2-butanone 4-phosphate synthase | ribB | |
| PP0788 | Conserved hypothetical protein | 2.1 | |
| PP0810 | Cyoups1 protein | cyoups1 | 1.9 |
| PP0812 | Cytochromeoubiquinol oxidase, subunit II | cyoA | 3.0 |
| PP0813 | Cytochromeoubiquinol oxidase, subunit I | cyoB | 1.9 |
| PP0814 | Cytochromeoubiquinol oxidase, subunit III | cyoC | 2.1 |
| PP0816 | Protoheme IX farnesyltransferase | cyoE-2 | 2.2 |
| PP0887 | Sensor histidine kinase | 2.2 | |
| PP0888 | Response regulator | 2.5 | |
| PP0889 | Conserved hypothetical protein | 2.2 | |
| PP1110 | Serine O-acetyltransferase; putative | 5.5 | |
| PP1111 | Synthetase; putative | 7.3 | |
| PP1112 | Conserved hypothetical protein | 2.0 | |
| PP1113 | Pyridoxal-phosphate-dependent enzyme family protein | 2.0 | |
| PP1157 | Acetolactate synthase; catabolic; putative | 1.9 | |
| PP1188 | C4-dicarboxylate transport protein | dctA | 3.4 |
| PP1196 | Hypothetical protein | 1.9 | |
| PP1747 | Conserved hypothetical protein | 7.3 | |
| PP2257 | Aerotaxis receptor Aer-1 | aer-1 | 1.8 |
| PP2379 | Sco1/SenC family protein | 3.2 | |
| PP2554 | 4-Hydroxyphenylpyruvate dioxygenase putative | 2.5 | |
| PP2648 | Universal stress protein family | 3.3 | |
| PP2656 | Phosphate ABC transporter; periplasmic phosphate-binding protein | 1.8 | |
| PP2737 | Oxidoreductase; short-chain dehydrogenase/reductase family | 2.0 | |
| PP2855 | Conserved hypothetical protein | 3.0 | |
| PP2916 | Riboflavin synthase alpha subunit | ribE-2 | 1.9 |
| PP3005 | Hypothetical protein | 13.1 | |
| PP3007 | Hypothetical protein | 5.5 | |
| PP3122 | CoA-transferase subunit A; putative | 11.3 | |
| PP3255 | Conserved hypothetical protein | 2.9 | |
| PP3365 | Acetolactate synthase; catabolic; putative | 1.9 | |
| PP3632 | Transcriptional regulator LysR family | 2.2 | |
| PP3633 | N-Acetyl-gamma-glutamyl-phosphate reductase | argC | 2.6 |
| PP3656 | Aromatic compound-specific porin; putative | 2.1 | |
| PP3657 | Nitrobenzoate reductase; putative | 2.8 | |
| PP3754 | Beta-ketothiolase | 4.0 | |
| PP4066 | Enoyl-CoA hydratase; putative | 2.3 | |
| PP4067 | Acetyl-CoA carboxylase; biotin carboxylase; putative | 2.9 | |
| PP4225 | DNA-binding response regulator | 3.0 | |
| PP4227 | Conserved domain protein | 1.8 | |
| PP4228 | Hypothetical protein | 7.0 | |
| PP4240 | Microcin b17-processing protein mcbd; putative | 1.8 | |
| PP4250 | Cytochromecoxidase, cbb3 type, subunit I | ccoN-1 | 6.6 |
| PP4251 | Cytochromecoxidase, cbb3 type, subunit II | ccoO-1 | 1.8 |
| PP4252 | Cytochromecoxidase, cbb3 type, CcoQ subunit | ccoQ-1 | 1.8 |
| PP4253 | Cytochromecoxidase, cbb3 type, subunit III | ccoP-1 | 2.3 |
| PP4261 | Cation-transporting P-type ATPase | 2.1 | |
| PP4403 | Oxoisovalerate dehydrogenase lipoamide acyltransferase component | bkdB-2 | 2.0 |
| PP4523 | Agmatinase; putative | 2.0 | |
| PP4548 | Oxidoreductase; putative | 2.0 | |
| PP4624 | Hydrolase, alpha/beta fold family | 2.7 | |
| PP4870 | Azurin | azu | 2.0 |
| PP5028 | Proline iminopeptidase | pip | 2.0 |
| PP5029 | Formiminoglutamase | hutG | 2.3 |
| PP5030 | Imidazolonepropionase | hutI | 10.0 |
| PP5031 | Proline permease | proY | 13.0 |
| PP5032 | Histidine ammonia-lyase | hutH | 6.7 |
| PP5033 | Urocanate hydratase | hutU | 7.5 |
| PP5036 | Chlorohydrolase family protein | 2.1 | |
| PP5390 | Hypothetical protein | 17.0 | |
| PP5392 | Conserved hypothetical protein | 2.0 |
Genes putatively located in the same transcriptional unit are in boldface.
In response to toluene, 31 genes were downregulated in ΔrpoT compared to the wild-type strain. Of these, 12 genes were already found to be downregulated in the absence of toluene (Table 4). These genes included the PP2885-PP2887 cluster and a number of hypothetical proteins of unknown function. Among the genes that were newly downregulated in the ΔrpoT background were two regulators, one a sensor histidine kinase (PP0700) and the other a member of the AsnC family of transcriptional regulators (PP0663). Some members of the new set of downregulated genes encoded oxidoreductases (PP0379, PP1009, and PP2794).
DISCUSSION
The Pseudomonas putida alternative ECF sigma factor originally annotated as ECF-Pp12 (PP3006 in The Institute for Genomic Research [TIGR] annotation) was identified in this study as involved in the response to toluene; for this reason, we named it RpoT. The number of genes under the direct or indirect influence of this sigma factor is modest: microarray analyses indicated that about 1% of all P. putida genes were involved. The rpoT gene is the second in an operon that includes PP3005 downstream and PP3007 upstream, and the expression of this operon seems to be influenced by the lack of RpoT and solvents. In the ΔrpoT mutant background, expression from the operon promoter was 4- to 5-fold higher than in the wild-type background, and when sublethal concentrations of toluene were added, the difference between backgrounds reached 13-fold. In the wild-type background, expression from the PP3007 promoter in response to toluene also increased almost threefold, as determined with a fusion of the promoter region of orf3007 to ′lacZ (our own observations). Therefore, the increased expression of the operon that encodes RpoT in response to this solvent clearly indicates that this ECF is involved in endurance of toluene (organic-solvent)-induced stress. Analysis of the microarray assays for the rest of the genes encoding ECFs revealed no significant variations in the levels of mRNA in response to toluene.
Blast analysis of the ECF-Pp12 (PP3006) sigma factor against all entries in GenBank, excluding P. putida annotations, revealed that it exhibits high identity/similarity to ECF sigma factors of soil microorganisms, such as Pseudomonas syringae (57%/70%), Agrobacterium tumefaciens (45%/76%), Xanthomonas axonopodis (36%/52%), Mesorhizobium etli (32%/51%), and Ralstonia solanacearum (37%/54%). The role this sigma factor may play in these microorganisms remains unexplored. Adjacent to the sigma factor in all of these microorganisms was a gene which also exhibited high similarity to PP3005. At the protein level, the identity of PP3005 with the corresponding protein in P. syringae was 52%, whereas in A. tumefacines, X. axonopodis, M. etli, and R. solanacearum, identity was in the range of 25% to 28%. Although a protein highly similar to PP3007 is also present in P. syringae, no such similar protein was found in the other microorganisms mentioned above. We therefore propose that in all these microorganisms, the ECF sigma factor, like PP3006, may work in conjunction with a protein similar to PP3005.
In silico analysis of PP3005 suggests that it has a transmembrane α-helix that extends from residues 93 to 115. The N-terminal end was thought to be located in the cytoplasmic space, whereas the C-terminal end was probably located in the periplasmic space. This was proved experimentally using ′phoA fusions. Indeed, fusions of the N-terminal end to ′phoA yielded colorless colonies on plates with BCIP, whereas those fused to the C-terminal end turned blue on plates with BCIP. The in silico analysis of PP3007 suggests that it exhibits a signal peptide that directs it to the periplasmic space. ′phoA fusions to the ORF encoding PP3007 confirmed this assumption (Fig. 2).
The activities of alternative ECF sigma factors are often controlled posttranscriptionally through their interaction with anti-sigma factors (3, 29). As a general trend, most ECF sigma factors are cotranscribed with one or more negative regulators (44). Often, these include a transmembrane protein functioning as an anti-sigma factor that binds and inhibits the cognate sigma factor. Upon receiving a stimulus from the environment, the sigma factor is released and binds to RNA polymerase to stimulate transcription. This mechanism coordinates a cytoplasmic transcriptional response to signals perceived by the proteins at the membrane level (17, 21). For instance, RseA and RseB, encoded together with RpoE in the rpoE rseABC operon, form a signal pathway that allows E. coli to respond to protein unfolding upon periplasmic or envelope stress, especially under heat shock conditions that lead to the controlled proteolysis of RseA (1, 6, 24). Interestingly, localization studies demonstrated that PP3005 has a single transmembrane domain located around its central region, which would place a significant portion (half of the protein) on each side of the inner membrane. Although no sequence similarity to previously characterized proteins was found, the membrane topology of PP3005 resembles that of ECF anti-sigma factors. Although the hypothesis is thus far untested, PP3005 may interact with RpoT to regulate its function, and we cannot rule out the possibility that the periplasmic protein PP3007 might also play a role in this regulation.
As mentioned before, the P. putida RpoT protein exhibits a certain similarity to P. aeruginosa AlgU and E. coli σE protein; however, RpoT is clearly a distinct sigma factor at the sequence level, and particularly at the functional level, because in both E. coli and P. aeruginosa, the σE pathway is critical for protection against oxidative stress and temperature shocks (43), whereas an RpoT mutant of P. putida was as tolerant as the wild type of oxidative stress mediated by paraquat, H2O2, and heat shock treatments (see Fig. 4).
The σE regulon of E. coli was defined with a genetic strategy that allowed the identification of around 20 promoters that were upregulated in response to σE overexpression. The genes include rpoH, htrA, oprE itself, ostA, nlpB, and genes that encode proteins involved in LPS biogenesis and protein folding and inner and outer membrane proteins (1, 7, 20, 42, 66). In nonstressed P. putida cells, σT seemed to directly or indirectly control 54 promoters, 31 of which were upregulated and 23 downregulated. The regulated set of genes included those for cell envelope proteins and membrane proteins involved in electron transfer and substrate transport, chaperone-related proteins, regulators, and proteins of unknown function, some of which are predicted to be membrane proteins. Therefore, like E. coli RpoE, the RpoT protein of P. putida is involved in the control of envelope-related functions.
A relevant feature in the ΔrpoT mutant background in the absence of toluene was the fact that the expression of the main toluene extrusion pump (ttgGHI) was downregulated. Rojas et al. (51) showed that TtgGHI is the main toluene (organic solvents) extrusion pump from a quantitative point of view. The reduced expression of the main efflux pump explains in part the higher sensitivity of the ΔrpoT mutant to a sudden toluene shock. Indeed, in noninduced cells, a sudden exposure to ΔrpoT led to a survival rate below 10−8 (Fig. 5B), which is similar to that observed for the ΔttgGHI mutant. However, it should be noted that the killing rate of the mutant deficient in RpoT is less acute than that of the TtgGHI-deficient mutant (See Fig. 5B versus Fig. 5C). Therefore, the observed 2.5-fold decrease in the ttgGHI level in the ΔrpoT background is of physiological significance. Dinamarca et al. (10) found that inactivation of the cytochrome ubiquinol oxidase system (Cyo system) reduced catabolite repression at the PalkB and PalkS2 promoters when cells were grown on LB medium. These authors proposed that the Cyo system participates in a transfer cascade signal so that high levels of Cyo components lead to the repression of certain pathways. They suggested that the genes potentially under the control of this cascade were those encoding a catalase involved in oxygen stress (PP2887) and integral membrane electron transport proteins, such as PP2886 and PP0315. In DOT-T1E-ΔrpoT, increased expression of the Cyo system (by about twofold) appeared concomitantly with the repression (two- to fourfold) of the genes encoding catalase and the electron transport proteins, both in the absence and in the presence of toluene. Although in P. putida expression of the catalase encoded by PP2887 was downregulated, the strain was catalase positive and as tolerant as the parental strain of up to 1.5% (vol/vol) H2O2, indicating that expression of other catalases is RpoT independent.
Up- or downregulation of genes in the ΔrpoT mutant does not necessarily imply a direct role for this ECF in their expression, since regulation can also be the consequence of indirect effects resulting from cascades in the regulatory networks. In this regard, we found that in the ΔrpoT background, a number of genes were downregulated. A clear example was 1-phosphofructokinase (fruA), which was downregulated in ΔrpoT cells exposed or not to toluene, concomitant with the increased expression of the repressor FruR (PP0792). However, this effect on transcription was not translated into a significant physiological difference between wild-type and ΔrpoT cells, probably because the FruR repressor becomes inactive in the presence of fructose and the fructose catabolic genes are consequently expressed at a level high enough to allow maximum growth rates.
A surprising finding was that the genes that were downregulated in ΔrpoT were very different with and without toluene, with only 12 genes in common under both growth conditions (Tables 2 and 4). The lower expression of these 12 genes was probably the result of the direct control of RpoT, whereas the variation in the expression of the other genes may have been due to indirect regulatory events. In contrast, in the ΔrpoT mutant, almost all the genes that were upregulated in the absence of toluene remained induced at similar levels in the presence of toluene, indicating that they are part of the rpoT regulon. In the presence of toluene, about 40 other genes were upregulated in response to the solvent, so the expression of these genes is most probably RpoT independent.
In short, RpoT seems to control a limited number of genes in P. putida, some of which are involved in stress endurance against toluene. We suggest that the increased toluene sensitivity of the ΔrpoT mutant can be explained by a reduction in the expression level of one of the key solvent efflux pumps.
Acknowledgments
This study was supported by grants from the CICYT (BIO-2003-00515, GEN2001-4698-CO5-03, and BIO-2006-05668) and a grant from the European Commission (QLK3-CT-2002-01923).
We thank Carmen Lorente for secretarial assistance and Karen Shashok for improving the use of English in the manuscript.
Footnotes
Published ahead of print on 27 October 2006.
REFERENCES
- 1.Ades, S. E., I. L. Grigorova, and C. A. Gross. 2003. Regulation of the alternative σE during initiation, adaptation, and shut off of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, M., P. Dunman, J. Kormanec, C. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—towards standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 5.Chaturongakul, S., and K. J. Boor. 2006. σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 72:5197-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collinet, B., H. Yuzawa, T. Chen, C. Herrera, and D. Missiakas. 2000. RseB binding to the periplasmic domain of RseA modulates the RseA: σE interaction in the cytoplasm and the availability of σE RNA polymerase. J. Biol. Chem. 275:33898-33904. [DOI] [PubMed] [Google Scholar]
- 7.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 8.De Smet, M. J., J. Kingman, and B. Witholt. 1978. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli. Biochim. Biophys. Acta 506:64-80. [DOI] [PubMed] [Google Scholar]
- 9.DeVries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinamarca, M. A., A. Ruíz-Manzano, and F. Rojo. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domínguez-Cuevas, P., J. E. González-Pastor, S. Marqués, J. L. Ramos, and V. de Lorenzo. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 281:11981-11991. [DOI] [PubMed] [Google Scholar]
- 12.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100-1106. [DOI] [PubMed] [Google Scholar]
- 13.Egler, M., C. Grosse, G. Grass, and D. H. Nies. 2005. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godoy, P., M. I. Ramos-González, and J. L. Ramos. 2004. Pseudomonas putida mutants in the exbBexbDtonB gene cluster are hypersensitive to environmental and chemical stressors. Environ. Microbiol. 6:605-610. [DOI] [PubMed] [Google Scholar]
- 15.Grosse, C., G. Grass, A. Anton, S. Franke, A. Navarrete Santos, B. Lawley, N. L. Brown, and D. H. Nies. 1999. Transcriptional organization of the czc heavy metal homoeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 181:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guazzaroni, M. E., T. Krell, A. Felipe, R. Ruíz, C. Meng, X. Zhang, M. T. Gallegos, and J. L. Ramos. 2005. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA. J. Biol. Chem. 280:20887-20893. [DOI] [PubMed] [Google Scholar]
- 17.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 18.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heusipp, G., M. A. Schmidt, and V. L. Miller. 2003. Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol. Lett. 226:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Hiratsu, N., M. Amemura, H. Nashimoto, H. Shinawaga, and K. Makino. 1995. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J. Bacteriol. 177:2918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 22.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaniga, K., I. Delar, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 24.Klein, G., C. Dartigalongue, and S. Raina. 2003. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol. Microbiol. 48:269-285. [DOI] [PubMed] [Google Scholar]
- 25.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 26.Kovacikova, G., and K. Skorupski. 2002. The alternative sigma factor σE plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, E. J., Y.-H. Cho, H.-S. Kim, B.-E. Ahn, and J.-H. Roe. 2004. Regulation of σB by an anti- and an anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. 186:8490-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llamas, M. A., J. L. Ramos, and J. J. Rodríguez-Herva. 2000. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J. Bacteriol. 182:4764-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 33.Manoil, C., J. J. Mekalanos, and J. Beckwith. 1990. Alkaline phosphatase fusions: sensors of subcellular location. J. Bacteriol. 172:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marck, C. 1988. ‘DNA Strider': a ′C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez-Bueno, M., R. Tobes, M. Rey, and J. L. Ramos. 2002. Detection of multiple extractoplasmatic function (ECF) sigma factors in the genome of Pseudomonas putida KT2440 and their counterparts in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 4:842-855. [DOI] [PubMed] [Google Scholar]
- 36.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ménard, R., P. J. Sansonetti, and C Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min Cao, M., C. M. Moore, and J. D. Helmann. 2005. Bacillus subtilis paraquat resistance is directed by σM, an extracytoplasmic function sigma factor, and is conferred by YqjL and BcrC. J. Bacteriol. 187:2948-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosqueda, G., and J. L. Ramos. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J. Bacteriol. 182:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz-Rojas, J., P. Bernal, E. Duque, P. Godoy, A. Segura, and J. L. Ramos. 2006. Involvement of cyclopropane fatty acids in the response of Pseudomonas putida KT2440 to freeze-drying. Appl. Environ. Microbiol. 72:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, et al. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 42.Raina, S., D. Missiakas, and C. Georgopolus. 1995. The rpoE gene encoding the σE/σ24 heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raivio, T. L. 2005. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 56:1119-1128. [DOI] [PubMed] [Google Scholar]
- 44.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 45.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-González, A. Rojas, W. Terán, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 46.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramos, J. L., E. Duque, M. J. Huertas, and A. Haïdour. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos, J. L., M. T. Gallegos, S. Marqués, M. I. Ramos-González, M. Espinosa-Urgel, and A. Segura. 2001. Responses of gram-negative bacteria to certain environmental stressors. Curr. Opinin. Microbiol. 4:166-171. [DOI] [PubMed] [Google Scholar]
- 49.Ramos-González, M. I., E. Duque, and J. L. Ramos. 1991. Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids. Appl. Environ. Microbiol. 57:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Herva, J. J., and J. L. Ramos. 1996. Characterization of an OprL null mutant of Pseudomonas putida. J. Bacteriol. 178:5836-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojas, A., A. Segura, M. E. Guazzaroni, W. Teran, A. Hurtado, M. T. Gallegos, and J. L. Ramos. 2003. In vivo and in vitro evidence that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J. Bacteriol. 185:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 54.Schurr, M. J., H. Yu, J. M. Martínez-Salazar, J. C. Boucher, and V. Deretic. 1995. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segura, A., A. Hurtado, E. Duque, and, J. L Ramos. 2004. Transcriptional phase variation at the flhB gene of Pseudomonas putida DOT-T1E is involved in response to environmental changes and suggests the participation of the flagellar export system in solvent tolerance. J. Bacteriol. 186:1905-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269:8022-8028. [PubMed] [Google Scholar]
- 57.Silo-Suh, L., S.-J. Suh, P. V. Phibbs, and D. E. Ohman. 2005. Adaptations of Pseudomonas aeruginosa to the cystic fibrosis lung environment can include deregulation of zwf, encoding glucose-6-phosphate dehydrogenase. J. Bacteriol. 187:7561-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spaink, H. P., R. J. H. Oker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRJ151. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 59.Strom, M. S., and S. Lory. 1987. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J. Bacteriol. 169:3181-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tart, A. H., C. Matthew, C. Wolfgang, and Daniel J. Wozniak. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187:7955-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terán, W., A. Felipe, A. Segura, A. Rojas, J. L. Ramos, and M. T. Gallegos. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terán, W., T. Krell, J. L. Ramos, and M. T. Gallegos. 2006. Effector-repressor interactions. Binding of a single effector molecule to the operator bound TtgR homodimer mediates derepression. J. Biol. Chem. 281:7102-7109. [DOI] [PubMed] [Google Scholar]
- 63.Wu, W., H. Badrane, S. Arora, H. V. Baker, and S. Jin. 2004. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto, K., and, A. Ishihama. 2005. Transcriptional response of Escherichia coli to external zinc. J. Bacteriol. 187:6333-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young, J. C., and F. V. Hartl. 2003. A stress sensor for the bacterial periplasm. Cell 113:1-2. [DOI] [PubMed] [Google Scholar]
- 67.Yu, H., J. C. Boucher, N. S. Hibler, and V. Deretic. 1996. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (σE). Infect. Immun. 64:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuste, L., A. B. Hervás, I. Canosa, R. Tobes, J. I. Jiménez, J. Nogales, M. M. Pérez-Pérez, E. Santero, E. Díaz, J. L. Ramos, V. de Lorenzo, and F. Rojo. 2006. Growth phase-dependent expression of the Pseudomonas putida KT2440 transcriptional machinery analysed with a genome-wide DNA microarray. Environ. Microbiol. 8:166-177. [DOI] [PubMed] [Google Scholar]