Abstract
The NAD+-dependent malic enzyme (DME) and the NADP+-dependent malic enzyme (TME) of Sinorhizobium meliloti are representatives of a distinct class of malic enzymes that contain a 440-amino-acid N-terminal region homologous to other malic enzymes and a 330-amino-acid C-terminal region with similarity to phosphotransacetylase enzymes (PTA). We have shown previously that dme mutants of S. meliloti fail to fix N2 (Fix−) in alfalfa root nodules, whereas tme mutants are unimpaired in their N2-fixing ability (Fix+). Here we report that the amount of DME protein in bacteroids is 10 times greater than that of TME. We therefore investigated whether increased TME activity in nodules would allow TME to function in place of DME. The tme gene was placed under the control of the dme promoter, and despite elevated levels of TME within bacteroids, no symbiotic nitrogen fixation occurred in dme mutant strains. Conversely, expression of dme from the tme promoter resulted in a large reduction in DME activity and symbiotic N2 fixation. Hence, TME cannot replace the symbiotic requirement for DME. In further experiments we investigated the DME PTA-like domain and showed that it is not required for N2 fixation. Thus, expression of a DME C-terminal deletion derivative or the Escherichia coli NAD+-dependent malic enzyme (sfcA), both of which lack the PTA-like region, restored wild-type N2 fixation to a dme mutant. Our results have defined the symbiotic requirements for malic enzyme and raise the possibility that a constant high ratio of NADPH + H+ to NADP in nitrogen-fixing bacteroids prevents TME from functioning in N2-fixing bacteroids.
Members of the Rhizobiaceae enter into a symbiotic association with a plant host, during which time the free-living bacteria differentiate into nitrogen-fixing cells known as bacteroids. This transition is accompanied by a number of important physiological adaptations, among which are alterations to the metabolic pathways of the microbe (14). There is much evidence to suggest that bacteroids within nodules utilize C4-dicarboxylic acids as their primary carbon and energy sources and that these acids are metabolized via the tricarboxylic acid (TCA) cycle (13, 30, 31, 43). A number of reports have suggested that cycling of amino acids between the plant and bacteroid also plays a key role in carbon and nitrogen flow in N2-fixing root nodules (1, 24, 40). These reports have demonstrated that much remains to be resolved concerning C and N flow between the bacteroid and plant during symbiotic N2 fixation.
In N2-fixing bacteroids metabolizing C4-dicarboxylic acids via the TCA cycle, a pathway for the generation of acetyl coenzyme A (acetyl-CoA) is essential. One route for the formation of acetyl-CoA is via malic enzyme and pyruvate dehydrogenase (7, 8). Malic enzymes are responsible for the conversion of malate to pyruvate with the concomitant reduction of a nicotinamide cofactor. Sinorhizobium meliloti contains two malic enzymes, DME (diphosphopyridine nucleotide-dependent malic enzyme), which is dependent on NAD(P)+ (EC 1.1.1.39), and TME (triphosphopyridine nucleotide dependent malic enzyme), which utilizes NADP+ exclusively (EC 1.1.1.40) (39). S. meliloti dme mutants were shown to form root nodules which fail to fix nitrogen (7), while tme mutants formed wild-type nitrogen-fixing root nodules (7, 8). Biochemical characterization of DME and TME revealed that the enzymes have similar kinetic parameters and have almost identical Kms for substrate and cofactor, indicating that the enzymes should have similar rates of activity (39). The enzymes do differ in their responses to TCA cycle intermediates, with DME activity being enhanced by fumarate, succinate, and malate but inhibited by acetyl-CoA (9, 39).
Here we employed specific antibodies to directly quantify the levels of malic enzymes in free-living cells and bacteroids, and we found that TME protein levels in bacteroids are 20% of those in free-living cells. These data suggested that the reduced levels of TME in bacteroids could be the reason for its failure to functionally replace DME in dme mutant strains. We investigated this question by determining the symbiotic phenotypes of dme mutant strains carrying chromosomally integrated constructs in which (i) tme is expressed from the dme promoter and (ii) dme is expressed from the tme promoter.
The analysis of the symbiotic requirement for malic enzymes is complicated by the unusual bipartite structure of the DME and TME enzymes of S. meliloti. DME and TME are 770 and 761 amino acids in length, respectively, and are members of a large family of malic enzymes that contain an approximately 300-amino-acid domain at their C termini that is not present in other prokaryotic and eukaryotic malic enzymes, such as the 478- and 389-amino-acid enzymes from Bacillus stearothermophilus (19) and Streptococcus bovis (18), respectively (26). The C-terminal extension shows similarity to phosphotransacetylase enzymes (PTA) (EC 2.3.1.8) that are responsible for the interconversion of acetyl-CoA and acetyl phosphate. The widespread prevalence of the PTA-like domain suggests functional relevance; however, deletion of the C-terminal region of the S. meliloti malic enzymes did not abolish malic enzyme activity (26). We wished to determine if the C-terminal extension is required during symbiosis. To accomplish this task, two experimental procedures were carried out. First, the NAD+-dependent malic enzyme of Escherichia coli, SfcA (36), a protein that lacks the PTA-like extension, was expressed from the dme promoter in dme-deficient S. meliloti strains. Second, a truncated form of DME that lacks the C-terminal domain but still retains malic enzyme activity (26) was expressed in dme-deficient S. meliloti strains. Both constructs were analyzed in plant and enzyme assays in order to determine if either construct was able to support symbiotic nitrogen fixation.
MATERIALS AND METHODS
Bacterial strains and genetic methods.
E. coli cells were grown at 37°C in Luria-Bertani broth (LB), while S. meliloti cells were grown at 30°C in LBmc (LB supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2) and minimal medium (M9) with carbon sources at 15 mM as previously described (8). E. coli strain EJ1321 (pck dme tme) (16) and S. meliloti strain RmH194 (pckA1::Tn3HoHo pod-1 dme-1::Tn5 tme-4::ΩSp) (8) fail to grow on minimal medium with succinate as a carbon source. Complementation of this growth phenotype was used to determine whether the malic enzyme constructs created in this work were functional.
Derivatives of the gentamicin-resistant suicide vector pUCP30T (34) were mobilized into S. meliloti by triparental matings as previously described (11). Recombinants were selected on LBmc plates containing 20 μg/ml gentamicin (Gm) plus neomycin (Nm; 200 μg/ml), spectinomycin (Sp;100 μg/ml), or streptomycin (Sm; 200 μg/ml) as required. Because the pUCP30T chimeric malic enzyme constructs carried S. meliloti DNA from both the tme and dme gene regions, these constructs could recombine at either the tme or dme locus. To identify the cointegrate-recombination class, phage ΦM12 was used to transduce the Gm-resistant (Gmr) marker of pUCP30T and either the dme or tme mutation antibiotic resistance marker from individual recombinants into the wild-type strain Rm1021 (10). Approximately 30 clones from each transduction were replica plated onto LBmc plates containing neomycin (for the dme Tn5 mutations) or LBmc plates containing spectinomycin (for the tme ΩSp mutation) and gentamicin (the antibiotic marker for pUCP30T). Cotransduction of both the Gmr and the dme or tme antibiotic resistance marker demonstrated linkage between pUCP30T and the demonstrated recombination at the particular locus of interest. In all cases, recombination at the targeted structural gene was achieved with the pUCP30T vector construct containing the same structural gene, and these strains were utilized for further experimentation. Crude cell extracts and bacteroid extracts were produced according to previous protocols (7, 12).
Molecular biology techniques and introduction of SphI sites into the dme and tme genes.
General molecular biology methodologies were performed according to standard techniques. To facilitate manipulation of the tme and dme genes of S. meliloti, an SphI site was inserted at the previously identified translational start sites with the primers 5′-GATACCCGGCATGCTTCCTCAACCTC-3′ and 5′-CGCCCGTGTGCATGCCTTTGC-3′, respectively, following the procedure of Kunkel et al. (22). The templates used for site-directed mutagenesis were produced from truncated regions of tme and dme. For dme, pTH139 containing the entire dme open reading frame was digested with SalI and religated to yield plasmid pTH398, containing 868 bp of the dme open reading frame. For tme, pTH251 (26) was digested with BspHI, end filled with Klenow polymerase, and digested with EcoRI. The resulting 480-bp fragment was gel purified and ligated into pUC119 digested with SphI, treated with Klenow polymerase, and redigested with EcoRI to produce pTH399. These plasmids were then transformed into E. coli strain CJ236 (ung dut) to generate single-stranded DNA (22). The resulting plasmids were transformed into DH5α cells and then screened for the presence of an SphI site. On average, 50% of the transformants contained the desired mutation. Following DNA sequencing to verify sequence integrity, the dme and tme constructs pTH400 and pTH401B, respectively, were used in further experiments. The full-length genes were reconstituted such that both genes would retain the SphI at the translational start site. dme was reconstructed by inserting the SalI fragment from pTH139 into pTH400 to produce pTH408. tme was reconstructed by inserting a 2.5-kb EcoRI fragment from pTH392 into pTH401B, resulting in pTH407B.
Production of the pdme-tme construct.
To introduce tme downstream of the dme promoter, pTH408 was digested with SphI and KpnI, and the resulting 3.8-kb vector-dme promoter fragment was gel purified. Digesting pTH407B with SphI and KpnI and gel purifying the resulting 2.7-kb fragment isolated the tme structural gene. The two fragments were ligated to generate the plasmid pTH409. Transformants of E. coli EJ1321 carrying pTH409 grew on M9 minimal medium with succinate as the sole carbon source, verifying that pTH409 produced a functional TME product. To introduce the hybrid construct into S. meliloti, the 3.5-kb pdme-tme fragment was isolated with HindIII and KpnI and inserted into the similarly digested vector pUCP30T, to give pTH433. This cloning strategy resulted in the pdme-tme construct being inserted in the orientation opposite that of the lacZ promoter present on this plasmid, thus removing the possibility that a promoter other than dme could be responsible for expressing tme. Cointegration of pTH433 at the tme gene loci was verified by transduction.
Production of the ptme-dme construct.
pTH407B was digested with HindIII and EcoRI, producing a 487-bp tme fragment that was inserted into pBluescript KS. A 2.98-kb dme fragment from pTH408 was isolated following digestion with HindIII and inserted into a similarly digested pBluescript KS vector. This resulted in the dme structural gene being inserted in the vector such that the BamHI site was in the 3′ region of the gene. These two constructs allowed the use of the SphI site located in both fragments that were previously inserted into the translational start site of both genes. The final construct was produced by removing the 2.8-kb dme fragment via a SphI/BamHI digest followed by insertion downstream of the tme promoter in pBluescript KS, resulting in pTH596. This construct resulted in 471 bp of the tme promoter being positioned upstream of the dme open reading frame, with the translational start site of dme being fused to the translational start site of tme via the SphI site. The 3.3-kb hybrid construct was then digested with BamHI and KpnI and inserted into pUCP30T to create plasmid pTH597. A triparental mating was carried out to introduce the construct into various dme::Tn5 mutant S. meliloti strains, and transductions were carried out to verify recombination at the dme gene loci as described above.
Production of pdme-sfcA.
The sfcA gene was amplified from the plasmid pMEE1 (36) using the primers 5′-GAA ACA GAG CAT GCA ACC AAA AAC AAA AAA AC-3′ and 5′-CTC TCA TCC GCC AAA ACA GCC-3′ (bold type indicates the SphI restriction site introduced in the PCR fragment at the translational start site of sfcA). The resulting 1.8-kb fragment was digested with SphI and PstI and ligated to the plasmid pTH408, which was similarly digested. This ligation replaced the dme structural gene of pTH408 with the sfcA gene in such a way that sfcA was positioned downstream of the dme promoter. The resulting plasmid, pTH512, was digested with HindIII and PstI, and the 2.1-kb fragment (pdme-sfcA) was inserted into pUCP30T, also digested with HindIII and PstI, to generate pTH513. Plasmid pTH513 complemented RmH194 for growth on succinate verifying that the pdme-sfcA construct was functional. pTH513 was introduced into several S. meliloti malic enzyme deficient mutants through triparental matings, and transductions were carried out to verify recombination at the dme gene locus.
Production of dmeΔc construct.
pTH139 (9) carrying the entire dme open reading frame was digested with PstI and religated to remove the C-terminal region of dme and leave only the first 449 amino acids of DME. The truncated dme gene (dmeΔc) was removed from the resulting plasmid (pTH452) via digestion with HindIII and XbaI and inserted into pUCP30T digested with the same enzymes to produce pTH458. A triparental mating was carried out to introduce the construct into dme::Tn5 mutant S. meliloti strains, and transductions were carried out to verify recombination at the dme gene loci as described above.
Enzyme assays and plant tests.
For both free-living cells and bacteroid samples, pyruvate formation assays were carried out in the presence of either 1.5 mM NAD+ or NADP+ to determine DME or TME activity, respectively, as previously described (9). Malate dehydrogenase assays were utilized to verify cell and bacteroid extract integrity (9), with the change in absorption at 340 nm recorded with a Varian Cary 1E UV-visible spectrophotometer for approximately 3 to 5 min. Protein concentrations were determined using the Bio-Rad protein assay.
Plant assays with Medicago sativa var. Iroquois were carried out using Leonard assemblies following the method described by Yarosh et al. (42). Triplicate pots of 10 to 12 seedlings were grown, and 28 days postinoculation the plants were harvested. Acetylene reduction assays were carried out with three root systems per pot according to the method of Yarosh et al. (42), and the plants themselves were dried for 3 weeks, after which time the dry weights were recorded.
Native polyacrylamide gel electrophoresis (PAGE) and Western blots.
Native polyacrylamide gels (7%) were produced as previously described (26). Portions of 30 μg per sample of crude cell or bacteroid extract were electrophoresed at 4°C with a constant current of 4 mA for 1 h and 7 mA thereafter until the dye front was 1 cm from the end of the gel. Following electrophoresis, the gels were rinsed and stained for NAD+-dependent malic enzyme activity as previously described (26).
Polyclonal antibodies were obtained by repeated injection of female specific-pathogen-free rabbits with 200 μg of purified DME or TME protein (39). Polyclonal antibodies produced in this way were specific for the respective antigen and reacted with denatured protein. Proteins were transferred from 7% sodium dodecyl sulfate (SDS)-polyacrylamide gels to Immobilon P membranes (Millipore) and treated with anti-TME or anti-DME antibody (a 1/10,000 dilution) for 60 min. The membranes were then exposed to the secondary antibody (anti-rabbit antibody-peroxidase; Sigma) for 30 min and developed using enhanced chemiluminescence reagent (Amersham).
Quantification of the malic enzymes.
In order to quantify the DME and TME present in free-living cells and bacteroids, two Western blots for each protein were produced. S. meliloti cell extracts were obtained from an overnight LBmc-grown culture, while bacteroids were obtained from alfalfa plants inoculated with wild-type S. meliloti. Each SDS-PAGE gel had a constant amount of total free-living protein, with one gel containing various amounts of purified protein while the second had various amounts of total bacteroid protein. Two exposures (30 s and 1 min) were used for each Western blot, and a laser densitometer was used to measure the relative intensities of DME and TME. Two separate standard curves for the purified proteins were produced, and these were used to estimate the amount of DME or TME per 2 μg of free-living-cell extract. The values obtained for TME and DME were then used as a standard to estimate the amount of each protein in bacteroid extracts by comparing the intensity of the 2 μg of total protein from free-living cells to the standard curves of total bacteroid protein.
RESULTS
Quantification of DME and TME levels in bacteroids from alfalfa nodules.
Our previous studies suggested that tme expression and TME activity were reduced relative to DME in N2-fixing bacteroids (7, 8). To precisely determine the levels of DME and TME in free-living cells and N2-fixing bacteroids, we chose to employ antibodies raised against the S. meliloti DME and TME proteins which had been overproduced in E. coli and purified as previously described (39). These antibodies could be used to directly quantify the two enzymes in crude cell extracts, since the anti-DME antibodies failed to cross-react with TME and the anti-TME antibodies failed to cross-react with DME in Western blots of total cell extracts. Standard curves were produced using purified DME and TME, and these curves were used to estimate the amounts of the two proteins in both free-living cells and bacteroids. The results obtained from the Western blots (see Fig. S1 in the supplemental material) indicate that there is 6.8 ± 0.6 ng of DME/μg of protein from free-living cells and 4.7 ± 0.6 ng of TME/μg of protein from free-living cells. Bacteroids contained 14.2 ± 0.6 ng of DME/μg of total protein and 0.9 ± 0.003 ng of TME/μg of total protein. Hence, TME protein levels in bacteroids are reduced to 20% of the TME levels in free-living cells and are also reduced to 6% relative to the level of DME protein in bacteroids. The reduction in TME protein levels detected in bacteroids versus free-living cells verified the previous activity studies (8), and a reexamination of the activity data for free-living cells and bacteroids (e.g., see Table 3 in reference 7 and Table 4 in reference 8) revealed that in all cases the DME activity levels in bacteroids were approximately 40% higher than those in free-living cells.
TME cannot replace the need for DME in symbiotic N2 fixation.
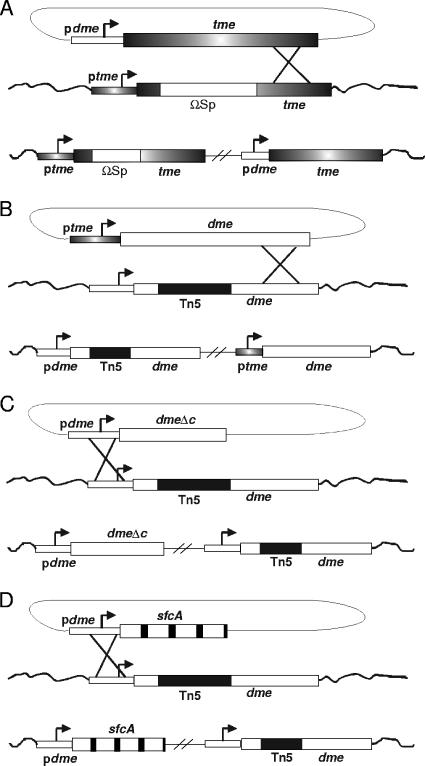
In a previous study, it was shown that increasing the TME activity in free-living cells overcame the dme-dependent succinate growth-negative phenotype of S. meliloti strain RmG443 (pckA pod1 dme-2::Tn5) (8). We therefore wished to determine whether increasing the level of TME activity in bacteroids would allow dme mutant strains to fix N2 in nodules. To accomplish this, we placed the tme structural gene under the control of the dme promoter, as this should ensure appropriate tme expression and appropriate TME activity in N2-fixing bacteroids. Since the transcriptional start site for both the tme and dme genes had been determined and since the translational start sites for both genes have been confirmed through the N-terminal sequence of purified DME and TME proteins (26, 39), we were able to interchange the malic enzyme promoters and the structural genes with minimal ambiguity. Oligonucleotide-directed mutagenesis was employed to introduce an SphI site at the ATG start codon for each gene, and the dme promoter region was then cloned upstream of the tme structural gene ATG codon. The dme promoter with the tme structural gene is referred to as pdme-tme, and this construct, in the plasmid pTH433, was inserted into the S. meliloti dme mutant genome via a single crossover recombination as outlined in Fig. 1A (see Materials and Methods).
FIG. 1.
Schematic outlining the integration of the various tme and dme constructs into the S. meliloti genome. A. Integration of the ptme-dme plasmid pTH597 into the tme::ΩSp locus. B. Integration of the pdme-tme plasmid pTH433 at the dme::Tn5 locus. C. Integration of the dmeΔc plasmid pTH458 at the dme::Tn5 locus. D. Integration of the pdme-sfcA plasmid pTH513 at the dme::Tn5 locus.
All strains in which the tme gene was expressed from the dme promoter (RmH897 and RmH898) showed elevated levels of NADP+-dependent malic enzyme in extracts from free-living cells relative to the parental wild-type and RmG455 dme mutant S. meliloti strains (Table 1). The symbiotic phenotypes of the various strains were determined by measuring the dry weights and acetylene-reducing (nitrogenase) activities of plants some 30 days after inoculation of alfalfa seedlings with these strains. Plants inoculated with dme strains expressing tme from the dme promoter (RmH897 and RmH899) had only residual acetylene reduction activity comparable to that of the plants inoculated with their dme-deficient parent S. meliloti strains (RmG455 and RmG994) (Table 1). Strain RmH898 that expressed the pdme-tme construct in a dme+ background had reduced acetylene reduction activity; however, this is likely due to experimental variation, since these plants had only marginally reduced average plant dry weights relative to plants inoculated with Rm1021. For other strains, the average plant dry weight determinations mirrored the acetylene reduction data.
TABLE 1.
S. meliloti strains expressing tme from the dme promoter
| Strain | Genotype | Activity in free-living cells
|
ARAc | Plant dry wt (mg)d | Activity in bacteroids
|
||||
|---|---|---|---|---|---|---|---|---|---|
| NAD+ dependenta | NADP+ dependenta | MDHb | NAD+ dependenta | NADP+ dependenta | MDHb | ||||
| Rm1021 | Wild type | 58 ± 4 | 74 ± 6 | 739 ± 15 | 1,221 ± 320 | 50.5 ± 0.8 | 45 ± 1 | 19 ± 1 | 2,608 ± 59 |
| RmG455 | Rm1021 dme-3::Tn5 | 21 ± 2 | 61 ± 2 | 611 ± 9 | 7 ± 3 | 10 ± 1 | ND | ND | 2,512 ± 190 |
| RmG994 | Rm1021 dme-3::Tn5 tme-4::ΩSp | 21 ± 1 | NDe | 864 ± 25 | ND | 10.6 ± 1 | ND | ND | 3,769 ± 136 |
| RmG995 | Rm1021 tme-4::ΩSp | 72 ± 5 | ND | 782 ± 19 | 1,156 ± 258 | 53.1 ± 2.8 | 50 ± 2 | 8 ± 1 | 3,269 ± 85 |
| RmH897 | RmG994::pdme-tme | 24 ± 1 | 104 ± 8 | 684 ± 12 | 24 ± 6 | 11.3 ± 0.4 | NDg | 89 ± 2g | 2,864 ± 63 |
| RmH898 | RmG995::pdme-tme | 70 ± 3 | 134 ± 5 | 704 ± 11 | 563 ± 176 | 44.8 ± 2.6 | 52 ± 3 | 60 ± 1 | 3,299 ± 72 |
| RmH899 | RmG994::pdme-tme | 14 ± 1 | 67 ± 5 | 695 ± 16 | 15 ± 5 | 12.3 ± 0.9 | —f | — | — |
| RmH900 | RmG995::pdme-tme | 47 ± 1 | 86 ± 5 | 1,062 ± 11 | — | — | — | — | — |
| None (uninoculated) | — | — | — | — | — | 8.5 ± 0.4 | — | — | — |
Specific activity, expressed as nanomoles of pyruvate formed/minute/mg protein. Values are means ± standard errors for triplicate samples.
Malate dehydrogenase (MDH) specific activity, expressed as nanomoles NADH formed/minute/mg protein. Values are means ± standard errors for triplicate samples.
ARA, acetylene reduction activity, expressed as nanomoles of ethylene produced per minute per plant and presented as means for triplicate samples containing three root systems each ± standard errors.
Means for triplicate pots containing 7 to 10 plants each ± standard errors.
ND, none detected.
—, not applicable or not tested.
Samples were incubated for 30 min at 30oC.
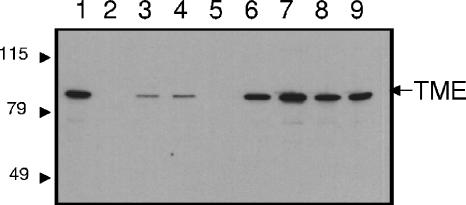
Enzyme assays carried out on extracts of bacteroids isolated from the nodules revealed elevated levels of NADP+-dependent activity in strains expressing tme from the dme promoter (RmH897 and RmH898) compared to Rm1021 bacteroids (Table 1). Similarly, analysis using TME-specific antibodies revealed elevated TME levels in all strains expressing tme from the dme promoter (RmH897, RmH898, RmH899, and RmH900) (Fig. 2, lanes 6 to 9), whereas no TME was detected in the tme mutant RmG994 (Fig. 2, lanes 2 and 5). As described above, the levels of TME in the Rm1021 and RmG455 (dme3::Tn5) bacteroids were very low (Fig. 2, lanes 3 and 4). The duplication of the dme promoter region that occurred in pdme-tme cointegrate strains did not appear to effect dme expression, as levels of DME protein, as detected with antibodies (data not shown), and DME enzyme activity levels in these strains (RmH897 to RmH900) were comparable to those in the wild type (Table 1). In summary, together these data demonstrated that TME could not function in place of DME for N2 fixation in bacteroids.
FIG. 2.
Western blot to detect TME protein present in S. meliloti extracts. Total bacteroid protein was loaded at 2 μg per sample, while 2 μg of free-living-cell extract from S. meliloti wild-type strain Rm1021 was run as a control. Lane 1, Rm1021 (wild-type free-living-cell extract); lane 2, RmG994 (Rm1021 dme-3::Tn5 tme-4::ΩSp); lane 3, Rm1021 (wild-type bacteroid extract); lane 4, RmG455 (Rm1021 dme-3::Tn5); lane 5, RmG994 (Rm1021, dme-3::Tn5 tme-4::ΩSp); lane 6, RmH897 (RmG994::pdme-tme); lane 7, RmH898 (RmG995::pdme-tme); lane 8, RmH899 (RmG994::pdme-tme); lane 9, RmH900 (RmG995::pdme-tme). Numbers on the left indicate molecular weight markers.
Reduced DME synthesis reduces symbiotic N2 fixation in S. meliloti.
The results from experiments in which tme expression was increased in bacteroids raised the question of how much DME activity is required for N2 fixation. Accordingly, we constructed strains in which the dme gene was under- and overexpressed. To reduce dme expression, the dme promoter region upstream from the dme ATG start codon was replaced by the symbiotically weak tme promoter. The resulting ptme-dme construct in the plasmid pTH597 was recombined via a single crossover at the dme locus in dme, tme, and dme tme mutant strains as outlined in Fig. 1B (also see Materials and Methods).
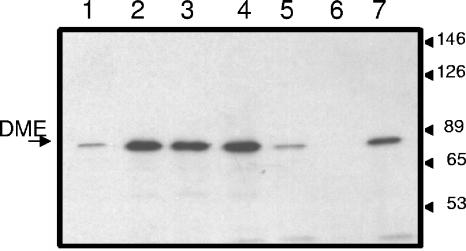
The symbiotic N2-fixing abilities and the enzyme activities in both free-living and bacteroid forms of the resulting strains are shown in Table 2. The amounts of DME protein in Western blots of bacteroid extracts as detected with anti-DME antibodies were also determined (Fig. 3). As expected, plants inoculated with dme mutant strains (RmG454 and RmG994) failed to fix N2, as demonstrated by their negligible acetylene reduction (nitrogenase) activity and dry weight values, which were similar to those of uninoculated plants. In contrast, plants inoculated with dme mutant strains that also expressed dme from the tme promoter (RmH979 and RmH980) had 20 to 30% of the acetylene reduction (nitrogenase) activity and 35% of the dry weight of plants inoculated with wild-type Rm1021 (Table 2). These plants were shorter than those inoculated with the wild-type strain but much larger and greener than the uninoculated controls or plants inoculated with the dme mutant strains. Thus, expression of dme from the tme promoter resulted in a sharp reduction in N2 fixation activity in root nodules.
TABLE 2.
Phenotypes of S. meliloti strains expressing dme from the tme promoter
| Strain | Genotype | Activity in free-living cells
|
ARAc | Plant dry wt (mg)d | Activity in bacteroids
|
||||
|---|---|---|---|---|---|---|---|---|---|
| NAD+ dependenta | NADP+ dependenta | MDHb | NAD+ dependenta | NADP+ dependenta | MDHb | ||||
| Rm1021 | Wild type | 107 ± 4 | 47 ± 2 | 313.3 ± 3.9 | 1,004 ± 2 | 46.8 ± 4.8 | 115 ± 2 | 34 ± 3 | 1,135 ± 13 |
| RmG454 | Rm1021, dme-2::Tn5 | 52.1 ± 2 | 60 ± 1 | 543.1 ± 25.5 | 13 ± 2 | 10.1 ± 0.8 | 14 ± 1 | 0.9 ± 0.1 | 62 ± 1 |
| RmG994 | Rm1021, dme-3::Tn5 tme-4::ΩSp | 54 ± 1 | 5 ± 1 | 439.3 ± 40.6 | NDe | 9.5 ± 0.5 | ND | ND | 461 ± 1 |
| RmG995 | Rm1021, tme-4::ΩSp | 114 ± 1 | 25 ± 2 | 381 ± 21.5 | 1,039 ± 69 | 54.6 ± 8.3 | 141 ± 2 | 61 ± 1 | 672 ± 23 |
| RmH979 | RmG454::ptme-dme | 77 ± 2 | 65 ± 3 | 375 ± 26.2 | 341 ± 73 | 16.6 ± 1.0 | 31 ± 1 | 7 ± 1 | 317 ± 2 |
| RmH980 | RmG994::ptme-dme | 82 ± 3 | 4 ± 1 | 352.5 ± 15 | 214 ± 34 | 16.5 ± 1.6 | 40 ± 1 | 0.2 ± 0.1 | 235 ± 7 |
| RmH981 | RmG995::ptme-dme | 124 ± 1 | 29 ± 5 | 387.9 ± 24.4 | 400 ± 106 | 46.4 ± 6.8 | 116 ± 2 | 42 ± 1 | 155 ± 3 |
| None (uninoculated) | —f | — | — | — | — | 8.5 ± 1.5 | — | — | — |
Specific activity, expressed as nanomoles of pyruvate formed/minute/mg protein. Values are means ± standard errors for triplicate samples.
Malate dehydrogenase (MDH) specific activity, expressed as nanomoles NADH formed/minute/mg protein. Values are means ± standard errors for triplicate samples.
ARA, acetylene reduction activity, expressed as nmoles of ethylene produced per minute per plant and presented as means for triplicate samples containing three root systems each ± standard errors.
Means for triplicate pots containing 7 to 10 plants each ± standard error.
ND, none detected.
—, not applicable or not tested.
FIG. 3.
Western blot to detect DME in bacteroid extracts. Total bacteroid protein was loaded at 2 μg per sample, while 2 μg of free-living-cell extract from S. meliloti wild-type strain Rm1021 was run as a control. Numbers on the right indicate molecular weight markers. Lane 1, RmH979 (RmG454::ptme-dme); lane 2, RmH981 (RmG995::ptme-dme); lane 3, RmG995 (Rm1021 tme-4::ΩSp); lane 4, Rm1021 bacteroid extract (wild type); lane 5, RmH980 (RmG994::ptme-dme); lane 6, RmG994 (Rm1021 dme-3::Tn5 tme-4::ΩSp); lane 7, Rm1021 free-living-cell extract (wild type).
The enzyme activity measurements demonstrated that while the tme promoter was as strong as the dme promoter in free-living cells, dme expression in bacteroids from this promoter was reduced, as the NAD+-dependent malic enzyme activity in RmH979 and RmH980 bacteroid extracts was less than 30% of wild-type levels (Table 2). Similarly, the amount of DME protein in the RmH979 and RmH980 bacteroid extracts was clearly smaller than in strains carrying the wild-type dme locus (Fig. 3, lanes 1 and 5). We note that despite the absence of DME protein in extracts from dme mutant strains (Fig. 3, lane 6), a high residual NAD+-dependent malic enzyme activity was detected in the crude extracts from free-living cells and bacteroids of dme strains (Table 2). This background was also observed previously and is due to cross-reacting activities, including that from malate dehydrogenase, which is present at high levels in these extracts.
To increase the level of DME in bacteroids, the dme locus was cloned into the high-copy-number plasmid pBBR5 (20). NAD+-dependent malic enzyme activities in bacteroids from Rm1021 carrying this plasmid were 657 nmol/min/mg protein. However, the dry weights of plants inoculated with this strain were not statistically different from those of plants inoculated with wild-type Rm1021, and we therefore conclude that the level of DME in the wild-type bacteroids is not limiting for N2 fixation. On the other hand, the data from strains in which dme was expressed from the tme promoter show clearly that reducing the level of DME in bacteroids renders the conversion of malate to pyruvate a rate-limiting reaction for N2 fixation. Limiting the rate of pyruvate synthesis is likely to limit the flux of malate through the TCA cycle and hence the generation of ATP and perhaps reductant for nitrogenase. It is also possible that reduced levels of pyruvate could limit alanine dehydrogenase-catalyzed alanine synthesis, and that in turn may reduce N2 fixation (1, 40).
Expression of the 3′-truncated dme gene (dmeΔc) in S. meliloti dme mutant strains.
The interpretation of the experimental results obtained from manipulations of the TME and DME proteins were complicated by the unusual structure of these proteins. This prompted us to investigate whether the C-terminal PTA-like domain of DME is required for N2 fixation, since DME deletions that lack this region maintain malic enzyme activity (26). Accordingly, we constructed a dme 3′-deletion derivative, referred to as dmeΔc, which expressed a truncated DME protein lacking the C-terminal 321 amino acids (DMEΔC). The pTH458 plasmid carrying the dmeΔc gene and dme promoter was recombined via a single crossover into dme mutant strains as illustrated in Fig. 1C and described in Materials and Methods.
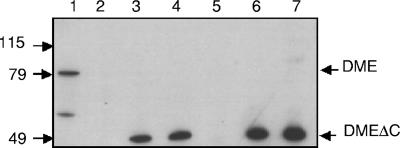
The symbiotic phenotypes of these strains were assessed as described above (Tables 1 and 2). Plants inoculated with dme-1 and dme-2 strains carrying dmeΔc were Fix+ and had dry weights of 36.4 ± 2.6 mg and 48.3 ± 0.1 mg, respectively, compared to 40.6 ± 3.7 mg for plants inoculated with the wild type strain Rm1021 and 7.1 ± 1.0 mg and 7.4 ± 0.6 mg for plants inoculated with the dme mutant strains. Enzyme assays of bacteroid extracts from dme strains carrying dmeΔc had NAD+-dependent malic enzyme activity that was comparable to wild-type levels of 71 nmol/min/mg protein. Western blots of these bacteroid extracts showed that strains expressing dmeΔc produced only the truncated malic enzyme migrating at 49 kDa, compared to the 82 kDa full-length DME protein (Fig. 4). These data verified that the symbiotic phenotype observed was due to the expression of the truncated gene, dmeΔc, and not to regeneration of an intact dme gene. These results established that the PTA-like C-terminal domain of DME is not required for N2 fixation, and this prompted us to investigate whether a heterologous NAD+ malic enzyme such as that from Escherichia coli could replace the function of DME in N2-fixing bacteroids.
FIG. 4.
Western blot to detect DME and DMEΔC proteins in bacteroid extracts. Total bacteroid protein was loaded at 2 μg per sample. Lane 1, Rm1021 (wild type); lane 2, RmG454 (Rm1021 dme-2::Tn5); lane 3, RmH996 (RmG454::dmeΔc); lane 4, RmH998 (RmG454::dmeΔc); lane 5, RmG456 (Rm1021 dme-1::Tn5); lane 6, RmH999 (RmG456::dmeΔc); lane 7, RmH1000 (RmG456::dmeΔc). Numbers on the left indicate molecular weight markers.
The Escherichia coli NAD+-dependent malic enzyme ScfA can replace DME in N2-fixing bacteroids.
The E. coli NAD(P)+malic enzyme encoded by sfcA has only limited homology to other prokaryotic malic enzymes and instead appears to be more closely related to eukaryotic malic enzymes and the Bacillus subtilis malic enzyme Mao2.bs (2, 26). The sfcA gene encodes a polypeptide of 574 amino acids and lacks the PTA-like C-terminal region present in DME and TME. An alignment of the scfA-encoded protein with the N-terminal malic enzyme region of DME revealed that while the two proteins share only 25% identity, the conserved regions common to all malic enzymes are identifiable (see Fig. S2 in the supplemental material) (26). To express the scfA gene in S. meliloti, scfA was cloned downstream of the dme promoter to give a pdme-scfA construct in the plasmid pTH513. The E. coli NAD+-dependent malic enzyme could clearly function in S. meliloti free-living cells, since cointegration of the pdme-sfcA plasmid, pTH513, in RmH194 (pckA1::Tn3HoHo pod-1 dme-1::Tn5 tme-4::ΩSp) restored the ability of this strain to grow on succinate. To investigate whether sfcA could function symbiotically in root nodules, the pTH513 plasmid was recombined at the dme locus in various dme, dme tme, and tme mutant strains. The symbiotic phenotype of these strains revealed that the E. coli NAD+-dependent malic enzyme restored near-wild-type activity to the dme mutant strains, with plant dry weights varying from 65 to 84% of that of the wild-type-inoculated plants (RmK215, RmK216, RmK217, and Rm1021) (Table 3). Student's t test showed no statistical significant difference between dry weight values for the wild-type-inoculated and the RmK215-, RmK216-, and RmK217-inoculated plants. Enzyme assays conducted with dme mutant bacteroids expressing sfcA revealed approximately half (44 to 59%) of the NAD+-dependent malic enzyme activity present in wild-type bacteroids (Table 3). Native PAGE gels stained for NAD+-dependent malic enzyme activity revealed that the scfA-encoded protein was present in the RmK215, RmK216, and RmK217 bacteroid extracts and that, as expected, no DME was present (Fig. 5. ) These data demonstrate that the E. coli NAD+-dependent malic enzyme is capable of restoring wild-type N2-fixing activity to S. meliloti dme mutant strains and confirm that the DME C-terminal PTA-like domain is not required for N2 fixation.
TABLE 3.
Phenotypes of S. meliloti strains expressing the sfcA gene under the control of the dme promoter
| Strain | Genotype | Activity in free-living cells
|
Plant dry wt (mg)c | Activity in bacteroids
|
||
|---|---|---|---|---|---|---|
| NAD+ dependenta | MDHb | NAD+ dependenta | MDHb | |||
| Rm1021 | Wild type | 64 ± 1 | 543 ± 11 | 40.6 ± 3.7 | 103 ± 2 | 3,172 ± 68 |
| RmG455 | Rm1021 dme-3::Tn5 | 26 ± 15 | 713 ± 20 | 7.4 ± 0.6 | 8 ± 1 | 3,401 ± 94 |
| RmG456 | Rm1021 dme-1::Tn5 | 29 ± 9 | 717 ± 16 | 7.1 ± 1.0 | 8 ± 2 | 3,103 ± 60 |
| RmG994 | Rm1021 dme-3::Tn5 tme-4::ΩSp | 30 ± 4 | 725 ± 37 | 8.8 ± 0.6 | NDd | 2,251 ± 72 |
| RmG995 | Rm1021 tme-4::ΩSp | 67 ± 3 | 583 ± 14 | 45.9 ± 4.1 | 94 ± 3 | 3,198 ± 139 |
| RmK215 | RmG455::pdme-sfcA | 122 ± 20 | 713 ± 44 | 34.3 ± 3.2 | 50 ± 1 | 2,909 ± 47 |
| RmK216 | RmG456::pdme-sfcA | 99 ± 10 | 598 ± 4 | 33.9 ± 6.6 | 46 ± 1 | 4,073 ± 214 |
| RmK217 | RmG994::pdme-sfcA | 85 ± 13 | 589 ± 22 | 26.3 ± 1.4 | 61 ± 1 | 2,739 ± 300 |
| RmK218 | RmG995::pdme-sfcA | 135 ± 12 | 722 ± 37 | 50.6 ± 3.5 | 77 ± 11 | 2,925 ± 55 |
| RmK219 | Rm1021::pdme-sfcA | 114 ± 11 | 612 ± 17 | 59.4 ± 11.2 | 74 ± 1 | 2,894 ± 28 |
| None (uninoculated) | —e | — | — | 6.3 ± 0.4 | — | — |
Specific activity, expressed as nanomoles of pyruvate formed/minute/mg protein. Values are means for triplicate samples ± standard errors.
Malate dehydrogenase (MDH) specific activity, expressed as nanomoles NADH formed/minute/mg protein. Values are means for triplicate samples ± standard errors.
Expressed as means for triplicate pots containing 7 to 10 plants each ± standard errors.
ND, none detected.
—, not applicable or not tested.
FIG. 5.
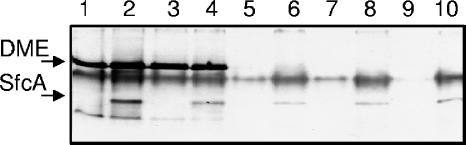
Nondenaturing PAGE to show NAD+-dependent malic enzyme in bacteroid extracts of S. meliloti expressing sfcA. Protein was loaded at 30 μg per sample, and the gel was stained to detect NAD+-dependent malic enzyme activity. Lane 1, Rm1021 (wild type); lane 2, RmK219 (Rm1021::pdme-sfcA); lane 3, RmG995 (Rm1021 tme-4::ΩSp); lane 4, RmK218 (RmG995::pdme-sfcA); lane 5, RmG455 (Rm1021 dme-3::Tn5); lane 6, RmK215 (RmG455::pdme-sfcA); lane 7, RmG456 (Rm1021 dme-1::Tn5); lane 8, RmK216 (RmG456::pdme-sfcA); lane 9, RmG994 (Rm1021 dme-3::Tn5 tme-4::ΩSp); lane 10, RmK217 (RmG994::pdme-sfcA).
DISCUSSION
Why NAD+-dependent malic enzyme (DME) is required for symbiotic N2 in alfalfa root nodules remains to be unambiguously established. We have previously suggested that the DME-NAD+ malic enzyme together with pyruvate dehydrogenase convert malate to acetyl-CoA and that this is an essential anaplerotic pathway for cells metabolizing C4-dicarboxylic acids via the TCA cycle (7, 26). However, it is also possible that DME is required to generate pyruvate as a precursor for some other important metabolic pathways in bacteroids. Pyruvate could be a precursor in the yet-to-be-defined pathway leading to the generation of reductant for nitrogenase (35). Another possible pathway could be alanine synthesis in nodules. However, this is unlikely, given that alanine dehydrogenase mutants of R. leguminosarum and M. loti form Fix+ nodules (21, 24). The finding that reduced NAD+ malic enzyme synthesis reduces N2 fixation suggests that a minimal malate-to-pyruvate flux is required in alfalfa bacteroids to maintain efficient N2 fixation. Our biochemical characterization of the DME and TME proteins and all of the physiological evidence suggest that these enzymes function solely in the synthesis of pyruvate from malate (9, 39).
The observation that low levels of NAD+ malic enzyme in bacteroids supported significant symbiotic N2 fixation is in contrast to the finding that increasing the levels of NADP+ malic enzyme in bacteroids failed to support detectable N2-fixing activity in dme mutants. This result is more striking given that the NADP+ malic enzyme can function in place of DME in allowing a S. meliloti pckA1 pod-1 dme-1::Tn5 tme-4::ΩSp mutant to grow on succinate as a carbon source. The ability of the C-terminal-deletion derivative of DME together with the ability of the E. coli NAD+ malic enzyme to restore wild-type N2-fixing activity to S. meliloti dme mutants clearly demonstrates that the PTA-like domain of DME is not required for N2 fixation. Moreover, this strongly suggests that the failure of TME to replace DME in N2-fixing bacteroids is unrelated to differences in their PTA-like C-terminal domains.
DME has Kms of 9.4 mM for l-malate and 89 μM for NAD+, while TME has Kms for malate and NADP+ of 12.5 mM and 33 μM, respectively (39). The TME Vmax of 53 μmol/min/mg protein is close to the DME Vmax of 60 μmol/min/mg protein. Overall, the kinetic parameters for DME and TME are similar, and therefore it appears unlikely that these account for the different symbiotic properties of the two enzymes. However, unlike the NADP+-dependent malic enzyme, the S. meliloti NAD+ malic enzyme is allosterically regulated, with acetyl-CoA inhibiting and succinate and fumarate activating activity (39). Since kinetic results in our laboratory have revealed that DMEΔC is no longer allosterically regulated (unpublished results), and since DMEΔC functions in place of DME in nodules, we can conclude that the allosteric nature of DME is not essential to its function in bacteroids.
The E. coli NAD+ malic enzyme was characterized by several groups, and Km values for the substrate l-malate have been reported in the range from 0.1 to 0.4 mM, while the Km for the NAD+ cofactor has been estimated to be 23 and 55 μM (17, 33, 36, 41). As in the case of DME, the E. coli NAD+ malic enzyme is allosterically regulated, with inhibition of activity by acetyl-CoA and activation by malate (9, 28, 32, 39). Thus, except for a higher affinity for l-malate, the E. coli NAD+ malic enzyme compares favorably with DME, and it is not surprising that the E. coli enzyme can function in place of DME. However, as the two enzymes share little amino acid similarity, it would appear that the ability of the E. coli enzyme to function in bacteroids simply reflects its ability to catalyze the NAD+-dependent conversion of malate to pyruvate.
Given that neither differences in the catalytic rate nor the affinity for l-malate is sufficient to account for the failure of TME to act in place of DME in N2-fixing bacteroids, we hypothesize that there is a high ratio of NADPH+ + H+/NADP+ in bacteroids and that this prevents TME from functioning in nodules. The ratios of NAD(P)H+ + H+/NAD(P)+ in bacteroids from alfalfa nodules have not been determined; however, the levels in soybean bacteroids have been reported in two studies. Tezuka and Murayama (38) reported NAD/NADH and NADP/NADPH ratios for soybean bacteroids of 5.56 and 3.26, respectively. These values are higher than those reported by Tajima and Kouzai (37), which were 2.17 and 0.37, respectively. Possible reasons for the different results, such as methods used in the extraction and assay of pyridine nucleotides in bacteroids, have been discussed (38). Nevertheless, the very different values for the NADP/NADPH ratios (3.26 and 0.37) do not allow us to draw inferences; interpretations are also complicated by the fact that the properties of the Bradyrhizobium japonicum NAD+ and NADP+ malic enzymes appear to differ from those of the S. meliloti DME and TME enzymes, and it has been suggested that the physiological roles of the enzymes are different in S. meliloti and B. japonicum (4, 5).
The hypothesis that there is a high ratio of NADPH+ + H+ to NADP+ in bacteroids is difficult to reconcile with the observations that S. meliloti isocitrate dehydrogenase (Idh) is an NADP+-dependent enzyme and that mutants deficient for this activity are unable to support symbiotic nitrogen fixation, as determined by McDermott and Kahn (25). Isocitrate dehydrogenase has been purified from S. meliloti and has a Km of 15 μM for NADP+ (3), which is half that of TME (39). This low Km may be a reason why isocitrate dehydrogenase is capable of functioning under conditions of restricted NADP+ while TME cannot. Interestingly, an NAD+-dependent isocitrate dehydrogenase has been reported for rhizobia infecting Lotus pedunculatus; however, this does not appear to be a widespread phenomenon among prokaryotes (27). To directly address the issue of cofactor dependence, we wish to alter the cofactor specificity of TME from NADP+ to NAD+ to determine whether such an enzyme would support N2 fixation by a dme mutant strain.
Microbial genome sequences have revealed the presence of malic enzyme-like proteins, presumably the NAD+ and NADP+ enzymes, in many bacteria. In E. coli, the presence of two enzymes has been known for some time; however, the distinct roles played by these enzymes have not been clearly defined (17, 28, 33, 41). In Corynebacterium glutamicum the malE-encoded NADP-dependent malic enzyme plays an important role in routing carbon flux between glycolysis and gluconeogenesis (15, 29). Aymerich and colleagues recently identified and characterized four malic enzyme isoforms from Bacillus subtilis (6, 23). Disruption of the ytsJ gene resulted in a markedly reduced growth on malate despite the presence of the other enzymes. YtsJ is the sole NADP-dependent malic enzyme, and the mutant phenotype showed that it plays a role distinct from those of the other malic enzymes in B. subtilis. Paradoxically, while overexpression of the E. coli NADP-dependent malic enzyme in B. subtilis did not suppress the malate growth phenotype of the ytsJ mutant, overexpression of the E. coli transdehydrogenase gene (udhA) did partially suppress this phenotype. Those authors concluded that YtsJ may play an additional physiological role beyond the conversion of malate to pyruvate. It is clear that much remains to be established regarding the physiological roles of malic enzyme, and accordingly the analysis of the Sinorhizobium DME and TME enzymes and the establishment of their roles should be informative with respect to general microbial metabolism.
Supplementary Material
Acknowledgments
We express our appreciation to M. Donnely for providing the sfcA construct and R. T. Voegele and S. Vlaar for analyzing the biochemical characteristics of DMEΔC. We thank Punita Anjea and Rahat Zaheer for critical reading of the manuscript.
T.M.F. gratefully acknowledges the financial support from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 27 October 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allaway, D., E. M. Lodwig, L. A. Crompton, M. Wood, R. Parsons, T. R. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. [DOI] [PubMed] [Google Scholar]
- 2.Boles, E., P. Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandrasekharan Nambiar, P. T., and Y. I. Shethna. 1976. Purification and properties of an NADP+-specific isocitrate dehydrogenase from Rhizobium meliloti. Antonie Leeuwenhoek 42:471-482. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., Y. Okabe, K. Osano, and S. Tajima. 1997. Purification and characterization of the NADP-malic enzyme from Bradyrhizobium japonicum A1017. Biosci. Biotechnol. Biochem. 61:384-386. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., Y. Okabe, K. Osano, and S. Tajima. 1998. Purification and characterization of an NAD-malic enzyme from Bradyrhizobium japonicum A1017. Appl. Environ. Microbiol. 64:4073-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doan, T., P. Servant, S. Tojo, H. Yamaguchi, G. Lerondel, K. Yoshida, Y. Fujita, and S. Aymerich. 2003. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology 149:2331-2343. [DOI] [PubMed] [Google Scholar]
- 7.Driscoll, B. T., and T. M. Finan. 1993. NAD+-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol. Microbiol. 7:865-873. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll, B. T., and T. M. Finan. 1996. NADP+-dependent malic enzyme of Rhizobium meliloti. J. Bacteriol. 178:2224-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Driscoll, B. T., and T. M. Finan. 1997. Properties of NAD+- and NADP+-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology 143:489-498. [DOI] [PubMed] [Google Scholar]
- 10.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finan, T. M., B. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finan, T. M., I. Oresnik, and A. Bottacin. 1988. Mutants of Rhizobium meliloti defective in succinate metabolism. J. Bacteriol. 170:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finan, T. M., J. M. Wood, and D. C. Jordan. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J. Bacteriol. 154:1403-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gourdon, P., M. F. Baucher, N. D. Lindley, and A. Guyonvarch. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66:2981-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, E. J., and E. Juni. 1975. Isolation of mutants of Escherichia coli lacking NAD- and NADP-linked malic. Biochem. Biophys. Res. Commun. 65:559-566. [DOI] [PubMed] [Google Scholar]
- 17.Katsuki, H., K. Takeo, K. Kameda, and S. Tanaka. 1967. Existence of two malic enzymes in Escherichia coli. Biochem. Biophys. Res. Commun. 27:331-336. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, S., H. Suzuki, K. Yamamoto, M. Inui, H. Yukawa, and H. Kumagai. 1996. Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing of its gene. Appl. Environ. Microbiol. 62:2692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, K., S. Doi, S. Negoro, I. Urabe, and H. Okada. 1989. Structure and properties of malic enzyme from Bacillus stearothermophilus. J. Biol. Chem. 264:3200-3205. [PubMed] [Google Scholar]
- 20.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., A. Bourdes, and P. Poole. 2005. De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J. Bacteriol. 187:5493-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 23.Lerondel, G., T. Doan, N. Zamboni, U. Sauer, and S. Aymerich. 2006. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis. J. Bacteriol. 188:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodwig, E., S. Kumar, D. Allaway, A. Bourdes, J. Prell, U. Priefer, and P. Poole. 2004. Regulation of l-alanine dehydrogenase in Rhizobium leguminosarum bv. viciae and its role in pea nodules. J. Bacteriol. 186:842-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott, T. R., and M. L. Kahn. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsch, M. J., R. T. Voegele, A. Cowie, M. Osteras, and T. M. Finan. 1998. Chimeric structure of the NAD(P)+- and NADP+-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti. J. Biol. Chem. 273:9330-9336. [DOI] [PubMed] [Google Scholar]
- 27.Moustafa, E., and C. K. Leong. 1975. Effect of adenine nucleotides on NAD-dependent isocitrate dehydrogenases in rhizobia and bacteroids of legume root nodules. Biochim. Biophys. Acta 391:9-14. [DOI] [PubMed] [Google Scholar]
- 28.Murai, T., M. Tokushige, J. Nagai, and H. Katsuki. 1971. Physiological functions of NAD- and NADP-linked malic enzymes in Escherichia coli. Biochem. Biophys. Res. Commun. 43:875-881. [DOI] [PubMed] [Google Scholar]
- 29.Netzer, R., M. Krause, D. Rittmann, P. G. Peters-Wendisch, L. Eggeling, V. F. Wendisch, and H. Sahm. 2004. Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch. Microbiol. 182:354-363. [DOI] [PubMed] [Google Scholar]
- 30.Poole, P., and D. Allaway. 2000. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43:117-163. [DOI] [PubMed] [Google Scholar]
- 31.Ronson, C. W., P. Lyttleton, and J. G. Robertson. 1981. C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. USA 78:4284-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanwal, B. D. 1970. Allosteric controls of amphibolic pathways in bacteria. Bacteriol. Rev. 34:20-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanwal, B. D. 1970. Regulatory characteristics of the diphosphopyridine nucleotide-specific malic enzyme of Escherichia coli. J. Biol. Chem. 245:1212-1216. [PubMed] [Google Scholar]
- 34.Schweizer, H. P. 2001. Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr. Opin. Biotechnol. 12:439-445. [DOI] [PubMed] [Google Scholar]
- 35.Scott, J. D., and R. A. Ludwig. 2004. Azorhizobium caulinodans electron-transferring flavoprotein N electrochemically couples pyruvate dehydrogenase complex activity to N2 fixation. Microbiology 150:117-126. [DOI] [PubMed] [Google Scholar]
- 36.Stols, L., and M. I. Donnelly. 1997. Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima, S., and K. Kouzai. 1989. Nucleotide pools in soybean nodule tissue, a survey of NAD(P)/NAD(P)H ratios and energy charge. Plant Cell Physiol. 30:589-593. [Google Scholar]
- 38.Tezuka, T., and Y. Murayama. 2002. Formation of pyridine nucleotides under symbiotic and non-symbiotic conditions between soybean nodules and free-living rhizobia. Phytochemistry 61:637-644. [DOI] [PubMed] [Google Scholar]
- 39.Voegele, R. T., M. J. Mitsch, and T. M. Finan. 1999. Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1432:275-285. [DOI] [PubMed] [Google Scholar]
- 40.Waters, J. K., B. L. Hughes, L. C. Purcell, K. O. Gerhardt, T. P. Mawhinney, and D. W. Emerich. 1998. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc. Natl. Acad. Sci. USA 95:12038-12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, M., M. Tokushige, and H. Katsuki. 1973. Studies on regulatory functions of malic enzymes. II. Purification and molecular properties of nicotinamide adenine dinucleotide-linked malic enzyme from Escherichia coli. J. Biochem. (Tokyo) 73:169-180. [PubMed] [Google Scholar]
- 42.Yarosh, O. K., T. C. Charles, and T. M. Finan. 1989. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol. Microbiol. 3:813-823. [DOI] [PubMed] [Google Scholar]
- 43.Yurgel, S. N., and M. L. Kahn. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28:489-501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.