Abstract
The primer for reverse transcription of the human immunodeficiency virus type 1 (HIV-1) genome is tRNA3Lys. During assembly of HIV-1 particles, tRNA3Lys is taken up from the host cell along with lysyl-tRNA synthetase (LysRS), the tRNA binding protein that specifically aminoacylates the different tRNALys isoacceptors. In humans, the cytoplasmic and mitochondrial species of LysRS are encoded by a single gene by means of alternative splicing. Here, we show that polyclonal antibodies directed to the full-length cytoplasmic enzyme equally recognized the two enzyme species. We raised antibodies against synthetic peptides that allowed discrimination between the two enzymes and found that mitochondrial LysRS is the only cellular source of LysRS detected in the virions. These results open new routes for understanding the molecular mechanisms involved in the specific packaging of tRNA3Lys into viral particles.
Human immunodeficiency virus type 1 (HIV-1) is the etiological agent of AIDS. Many aspects of replication of this classical retrovirus are known (27). Current drug therapies are effective in blocking viral development but require multitarget approaches. Major advances in improving therapeutics require a better understanding of the virus life cycle to uncover new putative targets.
The virally encoded reverse transcriptase requires a host cell RNA primer to initiate replication of the HIV-1 RNA genome. In retroviruses, the RNA primer is a tRNA. In HIV-1, the primer binding site is complementary to the 3′-end 18 nucleotides of tRNA3Lys. The two RNA molecules form an extended template/primer complex, and in HIV virions, the tRNA is believed to be bound to the complementary primer binding site (14). Accordingly, tRNA3Lys is incorporated into HIV-1 particles (17). The other tRNALys isoacceptors, tRNA1,2Lys, are also found in the virions. Indeed, the specificity of tRNA packaging is believed to be due to the selective incorporation of host lysyl-tRNA synthetase (LysRS) within virions (4). Interestingly, decreasing cellular expression of LysRS with small interfering RNA reduced tRNALys packaging and reduced viral infectivity (12). The current model for the selective packaging of the LysRS-tRNALys complex involves the formation of a packaging complex with the two viral proteins Gag and Gag-Pol (16).
Hamster and human LysRS share 43% identical residues with bacterial LysRS (1, 26) but display N-terminal polypeptide extensions that provide the core synthetases with potent tRNA-binding capacities (9, 10). This N-terminal tRNA-binding domain is required for triggering tRNALys packaging into human immunodeficiency virus type I viral particles (3). However, in mammalian cells, aminoacylation of cytoplasmic tRNALys is accomplished by an enzyme that is exclusively recovered as 1 of the 11 polypeptide components of MARS, a stable multiaminoacyl-tRNA synthetase complex (6, 22). One puzzling observation was that the other components of this complex were not detected in HIV-1 virions along with LysRS (4). One possibility is that newly synthesized LysRS may interact with Gag before it associates with other components of the multisynthetase complex (13). However, the interaction between LysRS and Gag is not highly specific, which led to the suggestion that other factors may contribute to the specificity (18). Alternatively, another LysRS species might be targeted to the virion.
Interestingly, human cytoplasmic and mitochondrial LysRSs are encoded by a single gene by means of alternative splicing (30). The two LysRS species share 576 identical amino acid residues. Thus, polyclonal antibodies directed to the cytoplasmic protein and used to identify the presence of LysRS in HIV-1 extracts (4) are likely unable to discriminate between the two LysRS species. Only the very N-terminal sequence of 21 or 49 amino acid residues, respectively, for the cytoplasmic and mitochondrial LysRS is distinct (Fig. 1). To uncover the cellular source of viral LysRS, we raised antibodies against synthetic peptides specific for the two LysRS species and analyzed the LysRS content of HIV-1 extracts. The results show that the mitochondrial enzyme is found in the viral particles and suggest that the mitochondrion-specific sequences of LysRS may contribute to the selectivity of its packaging.
FIG. 1.
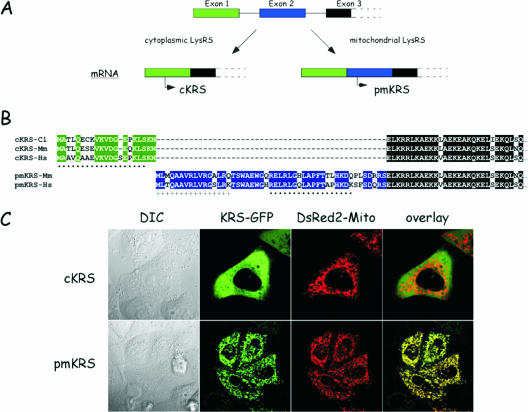
The cytoplasmic and mitochondrial species of LysRS are encoded by the same gene. (A) The three first exons of the KARS gene are shown. Two distinct mRNA are produced by alternative splicing. The mRNA encoding cytoplasmic LysRS (cKRS) lacks exon 2. Premitochondrial LysRS (pmKRS) is translated from an ATG codon located in exon 3. (B) Alignment of the N-terminal amino acid sequences of cKRS from hamster (Cl), mouse (Mm), or human (Hs) and of pmKRS from mouse and human. The N-terminal sequence of pmKRS putatively removed during mitochondrial import is indicated by a plus sign. The peptide sequences used to raise antibodies specific for the cytoplasmic and mitochondrial species of human LysRS are indicated by dots. (C) Localization by confocal microscopy of cKRS and pmKRS. HeLa cells were cotransfected with pEGFP/cKRS or pEGFP/pmKRS and with pDsRed2-Mito plasmids, and localization of the fusion proteins was analyzed by confocal microscopy. The overlay shows a perfect colocalization of pmKRS and of the mitochondrial marker. DIC, differential interference contrast.
MATERIALS AND METHODS
Antibodies and Western blot analysis.
Monoclonal antibodies directed to human cytochrome c were from PharMingen, and those directed to green fluorescent protein (GFP) were from BD Biosciences. Polyclonal anti-LysRS antibodies have been described previously (21). Rabbit anti-cytoplasmic KRS (anti-cKRS) and anti-mitochondrial KRS (anti-mKRS) antibodies were generated against synthetic peptides corresponding to residues 1 to 19 or 25 to 42 of the human cytoplasmic or mitochondrial form of LysRS (Neosystem, Strasbourg, France). Western blot analyses were conducted with goat antirabbit or goat antimouse secondary antibodies conjugated with peroxidase (Chemicon) and the ECL detection reagents (Amersham Biosciences).
Extracts of HIV-1 particles.
The purified particles of HIV-1 were obtained from Françoise Barré-Sinoussi (Institut Pasteur, Paris). The HIV-1 LAI strain was cultured on CEM cells (25). Viral particles were concentrated by polyethylene glycol precipitation, purified on a 20 to 50% sucrose gradient, and concentrated by ultracentrifugation (8). The protein concentration of the sample was 600 μg/ml. Total extracts were prepared by heating for 2 min at 100°C in 62.5 mM Tris-HCl (pH 7.5), 2% sodium dodecyl sulfate, 5 M urea, 100 mM dithiothreitol, and 0.002% bromophenol blue and kept frozen at −70°C. Aliquots of 10 μl were analyzed by Western blotting.
Isolation of mitochondria.
U937 cells were grown in suspension in RPMI medium supplemented with 10% fetal calf serum (FCS). Subcellular fractionation of U937 cell extracts was conducted essentially as described previously (29). U937 cells (100 × 106 cells) were harvested by centrifugation at 600 × g for 10 min at 4°C, washed once with 10 ml of ice-cold phosphate-buffered saline and resuspended in 2 ml of buffer MitoA (20 mM HEPES-KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, and 1 mM dithiothreitol). All subsequent steps were conducted at 4°C. After 10 min of incubation on ice, cells were pelleted at 600 × g, resuspended in 550 μl of buffer MitoA, and incubated for 10 min. Cells were lysed with 30 to 50 strokes of a Teflon homogenizer (Kontes) and diluted with 1 volume of buffer MitoA. The lysate was centrifuged at 750 × g for 10 min and then at 900 × g for 10 min to remove cell debris and nuclei. After another centrifugation at 5,500 × g for 10 min, the supernatant was recovered and centrifuged at 8,000 ×g for 10 min to remove residual mitochondria, and the resulting supernatant was referred to as the cytoplasmic fraction. The pellet from the centrifugation at 5,500 × g, containing mitochondria, was resuspended with 0.5 ml of buffer MitoA, centrifuged at 5,500 × g for 10 min, resuspended with 0.5 ml of buffer MitoA, and centrifuged at 8,000 × g for 10 min. The resulting pellet was referred to as the mitochondrial fraction.
Expression of human LysRS in yeast.
The cDNAs encoding the human cytoplasmic and mitochondrial forms of LysRS were introduced between the BglII and EcoRI sites of the yeast expression vector pYeDP10 (31) after PCR amplification with oligonucleotides GGGGAGATCTCATAATGGCGGCCGTGCAGGCGGCCGA, GGGGAGATCTCATAATGGCGACCT CCTGGGCAGAGTGGGGTCAC, and GGGGAATTCCTAGACAGAACTGCCAACTGTTG to give the recombinant plasmids pyHKc and pyHKm. Recombinant proteins are expressed under the control of the PGK promoter. Transformation of yeast and sporulation were performed as described previously (1).
Transient expression of human LysRS in HeLa cells.
The cDNAs encoding the human mitochondrial and premitochondrial forms of LysRS were introduced between the BamHI and EcoRV sites of the pTRE2hyg vector (BD Biosciences). HeLa Tet-Off cells were grown in F12 medium supplemented with 10% fetal calf serum and were transfected with Effectene (QIAGEN). After transfection, at the times indicated, crude extracts were prepared and analyzed by Western blotting.
Confocal imaging.
The cDNAs encoding the human cytoplasmic and premitochondrial forms of LysRS were introduced between the BglII and EcoRI sites of pEGFP-N1 (BD Biosciences) to give pEGFP/cKRS and pEGFP/pmKRS. HeLa cells were grown in F12 medium supplemented with 10% fetal calf serum. Cells were transfected with Effectene (QIAGEN). For localization experiments, cells were cotransfected with a pEGFP-N1 derivative and with pDsRed2-Mito (BD Biosciences). Cells were grown into eight-well Lab-Tek II chambers (Nalge Nunc International) and observed by confocal laser scanning microscopy using a Leica TCS SP2 confocal microscope.
RESULTS
Polyclonal antibodies directed to LysRS recognize the cytoplasmic and mitochondrial species with similar efficiencies.
In the human genome, a single gene (KARS) at position 16q23-q24 encodes LysRS. Two mRNA are produced by alternative splicing (Fig. 1A) and specify the two species of human LysRS, the cytoplasmic (cyto-LysRS) and the mitochondrial (mito-LysRS) enzymes (30). The human cytoplasmic enzyme displays a unique N-terminal sequence of 21 amino acid residues well conserved among other mammalian enzymes (Fig. 1B). The Asn residue at position 21, encoded by a codon spanning exons 1 and 3 (the codon is made of two nucleotides from exon 1 and one nucleotide from exon 3), was missing in the sequence published previously (30). When expressed as a fusion protein with GFP appended at the C-terminal extremity, cyto-LysRS had a purely cytoplasmic localization (Fig. 1C; note that it is also completely excluded from the nucleus). The human mitochondrial enzyme is translated from an initiator codon located in exon 2. Translation of this mRNA from the AUG codon located in exon 1 led to premature termination of translation at an in-frame stop codon provided by exon 2, upstream from the second AUG. Consequently, human mito-LysRS has a distinct N-terminal sequence made of 49 amino acid residues (Fig. 1B). This mito-LysRS-specific sequence is also well conserved among other species. When mito-LysRS is fused at the N terminus of GFP, a characteristic mitochondrial pattern is observed. The labeling with GFP exactly matched that observed with the DsRed fluorescent protein fused to the well-characterized mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase (Fig. 1C). These data are in agreement with previous observations (30). According to the program Mitoprot, a putative cleavage site could be predicted within the mitochondrial targeting sequence. The first 16 amino acid residues might be removed after import into the mitochondria. Accordingly, a fusion of mitochondrial LysRS with a deletion of this putative presequence to GFP gave cytoplasmic labeling similar to that observed with cyto-LysRS (result not shown). Hereafter, the mitochondrial LysRS translated from the initiator codon will be referred to as premitochondrial LysRS (premito-LysRS), and the mature species starting from Thr at position 17 will be referred to as mito-LysRS.
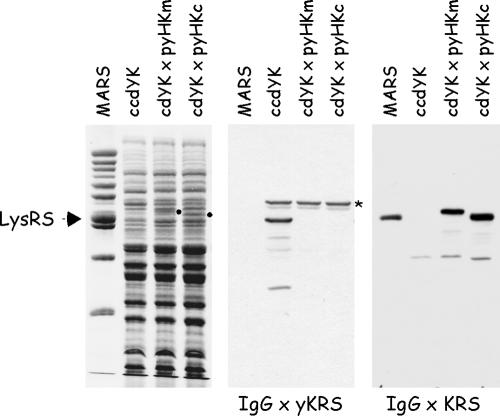
The two human LysRS species, cyto-LysRS and mito-LysRS, were expressed in yeast. The diploid yeast strain CCdYK01 contains a wild-type and a TRP1-disrupted KRS1 allele (in Saccharomyces cerevisiae, KRS1 is an essential gene that encodes the cytoplasmic form of LysRS (19). A second gene, MSK1, encodes the mitochondrial species (11)). After transformation of CCdYK01 with the 2μ plasmid carrying URA3 and expressing cyto-LysRS or mito-LysRS, and sporulation, spores with the Trp+ and Ura+ phenotype were isolated. These spores corresponded to haploid strains bearing a disrupted allele of the yeast KRS1 chromosomal gene rescued by plasmid-encoded human LysRS. As shown in Fig. 2, LysRS could not be identified in these strains by using antibodies directed to the yeast enzyme. In contrast, antibodies directed to the cytoplasmic form of mammalian LysRS recognized both human cytoplasmic and mitochondrial LysRS species in the corresponding haploid strains. Thus, human cyto-LysRS or mito-LysRS can functionally replace the cytoplasmic yeast enzyme. Because cyto-LysRS and mito-LysRS were expressed at similar levels (see the corresponding polypeptides stained by Coomassie blue) and were equally recognized by Western blotting, anti-KRS antibodies could not discriminate between the two forms of LysRS.
FIG. 2.
Expression of human cyto-LysRS and mito-LysRS in yeast. Total extracts from the yeast diploid strain ccdYK or from the yeast haploid strains cdYK×pyHKm and cdYK×pyHKc, lacking the yeast KRS1 gene but complemented with human mito-KRS or cyto-KRS, respectively, were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue (left panel) or subjected to Western blotting using antibodies directed to yeast LysRS (IgG × yKRS) or raised against the full-length cytoplasmic LysRS of mammalian origin (IgG × KRS). The polypeptides corresponding to mito-LysRS and cyto-LysRS, visualized by Coomassie blue staining, are indicated by dots. MARS, the multi-aminoacyl-tRNA synthetase complex purified from rabbit containing cyto-LysRS (indicated by an arrow). *, a polypeptide cross-reacting with anti-yKRS antibodies.
Mitochondrial LysRS accounts for less than 10% of total LysRS.
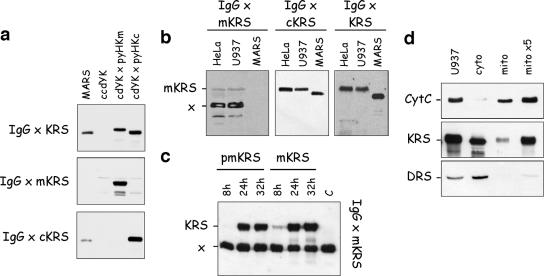
In order to be able to distinguish between cyto-LysRS and mito-LysRS by Western blotting on HIV extracts, we raised antibodies in rabbit against peptides specific for the cytoplasmic or mitochondrial form of LysRS. Peptides of 19 amino acid residues, located within the cytoplasm-specific sequence encoded by exon 1, and of 18 residues, located within the mitochondrion-specific sequence encoded by exon 2 and positioned 8 residues after the putative cleavage site of the import presequence (Fig. 1B), were synthesized, conjugated to ovalbumin, and used for immunization. As shown Fig. 3a, the rabbit antisera proved to be specific for the human cytoplasmic (anti-cKRS antibodies) or mitochondrial (anti-mKRS antibodies) LysRS expressed in yeast. As expected, the LysRS component of the multi-aminoacyl-tRNA synthetase complex isolated from rabbit liver could be revealed only with anti-cKRS antibodies (Fig. 3a). With anti-cKRS antibodies, a single polypeptide was revealed in total extracts of human HeLa and U937 cells in culture (Fig. 3b). With anti-mKRS antibodies, a faint band corresponding to the expected size for mito-LysRS (69.7 kDa) and a major band migrating as a polypeptide of 55 kDa were observed (Fig. 3b). This polypeptide of 55 kDa was not detected with polyclonal antibodies directed to full-length LysRS, which clearly established that it did not correspond to an alternative form of mito-LysRS (Fig. 3b). To further ascertain that the minor band did correspond to human mitochondrial LysRS, HeLa cells were transiently transfected with the pTRE2hyg vector, which expressed mito-LysRS or premito-LysRS (Fig. 3c). After 8, 24, and 32 h of transfection, cell extracts were analyzed by Western blotting using anti-mKRS antibodies. The intensity of the polypeptide of 69.7 kDa was severely increased after 24 or 32 h of transfection, whereas the intensity of the polypeptide of 55 kDa remained constant (Fig. 3c). Thus, the latter polypeptide is not related to mito-LysRS.
FIG. 3.
Analysis of the specificity of antibodies raised against N-terminal peptides of cyto-LysRS and mito-LysRS from human. (a) The yeast extracts described in Fig. 2 were analyzed by Western blotting using antibodies directed to full-length LysRS (IgG × KRS) or to synthetic peptides specific for the mitochondrial (IgG × mKRS) or cytoplasmic (IgG × cKRS) form of human LysRS. (b) Total extracts from HeLa or U937 cells were analyzed by Western blotting using antibodies directed to synthetic peptides specific for the mitochondrial (IgG × mKRS) or cytoplasmic (IgG × cKRS) form of human LysRS or antibodies directed to full-length LysRS (IgG × KRS). mKRS, polypeptide corresponding to mito-LysRS; x, a polypeptide cross-reacting with IgG × mKRS. (c) Total extracts of HeLa Tet-Off cells transfected with pTRE2hyg vectors that overexpressed the premitochondrial (pmKRS) or mitochondrial (mKRS) form of LysRS were analyzed by Western blotting using antibodies directed to the synthetic peptide specific for mito-LysRS (IgG × mKRS). The extracts were prepared 8 h, 24 h, or 32 h after transfection. C, a control extract of HeLa Tet-Off cells prepared before transfection. Note that the intensity of the cross-reacting polypeptide (x) is independent of transfection of HeLa cells with plasmids that express different forms of mito-LysRS. (d) A total extract of U937 cells (U937) or the cytoplasmic (cyto) or mitochondrial (mito) fraction obtained after subcellular fractionation of a U937 cell extract was analyzed by Western blotting using antibodies directed to full-length LysRS (KRS) or to cytochrome c (CytC) or AspRS (DRS), used as markers of the mitochondrial and cytoplasmic compartments, respectively. Lanes “cyto” and “mito” contain equivalent amounts of the initial extract of U937 cells. Lane “mito ×5” contains a fivefold excess of the “mito” fraction.
The polypeptide of 69.7 kDa, corresponding to mito-LysRS, is barely visible by Western blotting with anti-mKRS antibodies after short times of exposure of the blot (Fig. 3c, lane C). This suggested that the amount of mito-LysRS was quite low compared with that of cyto-LysRS, which was readily detected (Fig. 3b, middle panel) (data reported in Fig. 3a showed that anti-mKRS and anti-cKRS antibodies recognized mito-LysRS and cyto-LysRS, respectively, with similar efficiencies). The relative amounts of the cytoplasmic and mitochondrial species of LysRS were determined by Western blotting with anti-KRS antibodies (these antibodies recognize cyto-LysRS and mito-LysRS with similar efficiencies) (Fig. 2) after subcellular fractionation of a U937 cell extract. Antibodies to cytochrome c and to the cytoplasmic species of aspartyl-tRNA synthetase, representative of a mitochondrial protein and of a cytoplasmic protein, respectively, were used as markers of the subcellular fractions. The intensities of the signals obtained by Western blotting were quantified; the fraction of mito-LysRS was found to represent about 5 to 10% of total LysRS (Fig. 3d).
Extracts from HIV-1 contain the mitochondrial LysRS species.
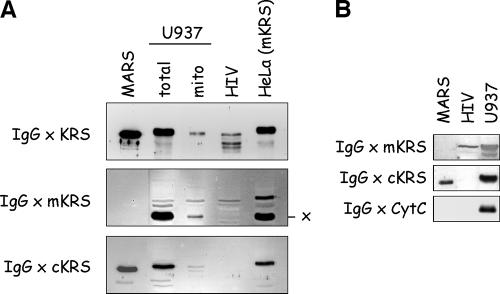
Human LysRS is incorporated into HIV-1 particles (4). We first verified that polyclonal antibodies directed to full-length cytoplasmic LysRS did cross-react with a polypeptide of appropriate size within an HIV-1 extract. As expected, a polypeptide similar in size to the LysRS component of MARS or to the polypeptides revealed in total cellular extracts of U937 or HeLa cells could be detected in the extract of purified HIV-1 particles (Fig. 4A, top panel). Polypeptides of smaller sizes were also observed and may correspond to degradation products of LysRS, an enzyme very sensitive to proteolysis (5). Earlier studies also reported the presence of LysRS species with various sizes in the viruses. These truncations also occurred in protease-negative viruses, suggesting that viral protease is not involved in this process (4). Because the cytoplasmic and mitochondrial species of LysRS observed after fractionation of U937 cell extract have very similar sizes, it was not possible to assign the signal obtained from the HIV-1 extract to either of these two forms.
FIG. 4.
Extracts from HIV-1 viral particles exclusively contain the mitochondrial LysRS species. (A) A total extract of purified HIV-1 particles (HIV) was analyzed by Western blotting using antibodies directed to full-length LysRS (IgG × KRS) or to synthetic peptides specific for the mitochondrial (IgG × mKRS) or cytoplasmic (IgG × cKRS) form of human LysRS. The HIV extract was analyzed along with MARS, with the total and mitochondrial fractions of a U937 cell extract, and with a total extract of HeLa cells that overexpressed mito-LysRS. x, the polypeptide cross-reacting with IgG × mKRS. (B) An HIV-1 extract was also probed with antibodies directed to cytochrome c (IgG × CytC).
When antibodies directed to the cyto-LysRS-specific peptide were used (Fig. 4A, bottom panel, IgG × cKRS), no cross-reacting polypeptide was identified in the sample from HIV-1 extract. By contrast, a polypeptide with the expected size was visualized when the HIV-1 extract was probed with antibodies directed to the synthetic peptide specific for the mitochondrial species of human LysRS (Fig. 4A, middle panel). Thus, only the mitochondrial form of LysRS can be detected in extracts from HIV-1 viral particles. It is noticeable that the degradation products of LysRS observed above with antibodies directed to full-length LysRS are not recognized by antibodies directed to the N-terminal peptide of mito-LysRS. This suggests that the truncated derivatives have lost the N-terminal region of mito-LysRS. To ascertain that the preparation of HIV-1 particles was not contaminated by mitochondria or mitochondrial proteins, the HIV-1 extract was also probed with antibodies directed to cytochrome c, one of the most abundant proteins found in the mitochondrial compartment. No signal for cytochrome c could be detected in the HIV-1 extract (Fig. 4B). As already published (4), no polypeptide cross-reacting with antibodies directed to the MetRS or ArgRS component of MARS could be detected in the HIV-1 fraction (results not shown). Altogether, these results suggest that the LysRS species that is specifically packaged within virions does not correspond to the cytoplasmic form of LysRS that is a component of MARS but does correspond to the mitochondrial LysRS species.
DISCUSSION
Our findings that the mitochondrial form but not the cytoplasmic form of LysRS is recovered in the HIV-1 viral particles is an essential step toward the understanding of the molecular mechanisms involved in packaging of tRNA3Lys, the primer RNA for reverse transcription of the HIV-1 genome. Earlier studies clearly showed that the cytoplasmic form of human LysRS, when overexpressed in COS7 cells, can be packaged in HIV-1 viral particles (3). These authors also established that amino acid residues located in the motif 1 region of LysRS are required for packaging in virus-like particles made of Gag only (16). The crystal structure of homologous LysRS from bacteria showed that this domain in LysRS is involved in the dimer interface (7, 24). In vitro studies also identified a domain of Gag that could be important for the interaction with LysRS (16). Recent studies revealed that interaction of cyto-LysRS with Gag is not highly specific (18). These authors suggested that another factor may contribute to the specificity of the interaction and thus to the selectivity of the packaging process. Obviously, most of the data obtained with cyto-LysRS most certainly hold true with mito-LysRS, which differs from cyto-LysRS only in its very N-terminal region. Thus, packaging of mito-LysRS is likely to involve its association with Gag via the domain conserved in cyto-LysRS, but the selectivity of this process might involve a specific contact between the mitochondrion-specific sequence of mito-LysRS with Gag or with another viral protein.
Our results also provide a rational explanation for the absence of other aminoacyl-tRNA synthetases within HIV-1 particles. Whereas cyto-LysRS is a component of MARS, a stable multisynthetase complex containing eight other aminoacyl-tRNA synthetases (20), mito-LysRS is directed to the mitochondria, where the other components of this complex are not present. Indeed, most of the other mitochondrial species of aminoacyl-tRNA synthetases are encoded by genes that are distinct from their cytoplasmic counterparts (2). Interestingly, mito-LysRS shares with cyto-LysRS the eukaryote-specific tRNA-binding domain, referred to as a tIF (tRNA-interacting factor), that provides eukaryotic LysRS with potent tRNA-binding properties compared with the bacterial enzyme (9, 10). Mutational analysis identified residues K19, K23, R24, and K27 from the N-terminal polypeptide extension of cyto-LysRS as important for the binding of the tRNA molecule (10). The amino acid residues K23, R24, and K27 are common to mito- and cyto-LysRS (Fig. 1). The residue K19 is encoded by exon 1 and is embedded in the cytoplasm-specific sequence peptide. However, in the corresponding position of mito-LysRS, this lysine residue is replaced by an amino acid with a side chain displaying similar physicochemical properties, an arginine residue. Thus, the tIF of mito-LysRS should be equivalent to the tIF of cyto-LysRS. The conservation of the tIF domain is especially important for incorporation of tRNALys into HIV-1 virions, since the deletion of this domain in cyto-LysRS is accompanied by a lower efficiency in tRNA packaging (3).
In human cells, three isoforms of LysRS are produced from a single gene. Two mRNAs are transcribed by means of alternative splicing and are translated in the cytoplasm to give the cytoplasmic species (cyto-LysRS) and the precursor of the mitochondrial form of LysRS (premito-LysRS). Cyto-LysRS associates with the other components of the multisynthetase complex, MARS (20). The matured form of mito-LysRS is produced during importation into the mitochondria by cleavage of the N-terminal targeting sequence of premito-LysRS. Whether premito- and mito-LysRS or one of these two LysRS species is the source of viral LysRS remains to be deciphered. Whereas association of premito-LysRS with viral components may explain its direct targeting to the viral particle during the packaging process, the selection of mito-LysRS would first require its release from the mitochondria. The viral protein Vpr, which has been shown to alter the permeability of the mitochondrial membrane (15, 23) and has been found to interact with LysRS (28), might be involved in this process.
The finding that mito (or premito)-LysRS is the molecular species of LysRS that is specifically packaged into HIV-1 viral particles opens new routes for understanding the function of Vpr in the HIV-1 life cycle. In addition, the role of Gag and Gag-Pol in the packaging process of LysRS should be reexamined. It is now possible to look for viral proteins that may discriminate between the cyto- and mito- forms of LysRS.
Acknowledgments
We thank Françoise Barré-Sinoussi (Institut Pasteur, Paris) for the gift of purified HIV-1 pellets. We also thank Spencer Brown and Susanne Bolte for access to the confocal microscope facility (ISV, Gif-sur-Yvette, France).
This work was supported by grants from the “Centre National de la Recherche Scientifique” (CNRS), the “Agence Nationale de Recherches sur le SIDA” (ANRS), the “Association pour la Recherche sur le Cancer” (ARC), and “La ligue.” M.K. was the recipient of postdoctoral fellowships from the “Fondation pour la Recherche Médicale” and “Association pour la Recherche sur le Cancer,” and V.S. was the recipient of a postdoctoral fellowship from the “Agence Nationale de Recherches sur le SIDA.”
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Agou, F., S. Quevillon, P. Kerjan, M. T. Latreille, and M. Mirande. 1996. Functional replacement of hamster lysyl-tRNA synthetase by the yeast enzyme requires cognate amino acid sequences for proper tRNA recognition. Biochemistry 35:15322-15331. [DOI] [PubMed] [Google Scholar]
- 2.Bonnefond, L., A. Fender, J. Rudinger-Thirion, R. Giegé, C. Florentz, and M. Sissler. 2005. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry 44:4805-4816. [DOI] [PubMed] [Google Scholar]
- 3.Cen, S., H. Javanbakht, M. J. Niu, and L. Kleiman. 2004. Ability of wild-type and mutant lysyl-tRNA synthetase to facilitate tRNALys incorporation into human immunodeficiency virus type 1. J. Virol. 78:1595-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirakoglu, B., and J. P. Waller. 1985. Leucyl-tRNA and lysyl-tRNA synthetases, derived from the high-Mr complex of sheep liver, are hydrophobic proteins. Eur. J. Biochem. 151:101-110. [DOI] [PubMed] [Google Scholar]
- 6.Cirakoglu, B., and J. P. Waller. 1985. Multiple forms of arginyl- and lysyl-tRNA synthetases in rat liver: a re-evaluation. Biochim. Biophys. Acta 829:173-179. [DOI] [PubMed] [Google Scholar]
- 7.Cusack, S., A. Yaremchuk, and M. Tukalo. 1996. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 15:6321-6334. [PMC free article] [PubMed] [Google Scholar]
- 8.Delfraissy, J. F., C. Wallon, F. Boue, F. Barre-Sinoussi, and P. Galanaud. 1991. Tumor necrosis factor-alpha inhibits the competence signal delivered by HIV to normal B cells. J. Immunol. 146:1516-1521. [PubMed] [Google Scholar]
- 9.Francin, M., M. Kaminska, P. Kerjan, and M. Mirande. 2002. The N-terminal domain of mammalian lysyl-tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 277:1762-1769. [DOI] [PubMed] [Google Scholar]
- 10.Francin, M., and M. Mirande. 2003. Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase. J. Biol. Chem. 278:1472-1479. [DOI] [PubMed] [Google Scholar]
- 11.Gatti, D. L., and A. Tzagoloff. 1991. Structure and evolution of a group of related aminoacyl-tRNA synthetases. J. Mol. Biol. 218:557-568. [DOI] [PubMed] [Google Scholar]
- 12.Guo, F., S. Cen, M. J. Niu, H. Javanbakht, and L. Kleiman. 2003. Specific inhibition of the synthesis of human lysyl-tRNA synthetase results in decreases in tRNALys incorporation, tRNA3Lys annealing to viral RNA, and viral infectivity in human immunodeficiency virus type 1. J. Virol. 77:9817-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halwani, R., S. Cen, H. Javanbakht, J. Saadatmand, S. Kim, K. Shiba, and L. Kleiman. 2004. Cellular distribution of lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 78:7553-7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isel, C., C. Ehresmann, G. Keith, B. Ehresmann, and R. Marquet. 1995. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA3Lys (template/primer) complex. J. Mol. Biol. 247:236-250. [DOI] [PubMed] [Google Scholar]
- 15.Jacotot, E., L. Ravagnan, M. Loeffler, K. F. Ferri, H. L. Vieira, N. Zamzami, P. Costantini, S. Druillennec, J. Hoebeke, J. P. Briand, T. Irinopoulou, E. Daugas, S. A. Susin, D. Cointe, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2000. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J. Exp. Med. 191:33-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javanbakht, H., R. Halwani, S. Cen, J. Saadatmand, K. Musier-Forsyth, H. Gottlinger, and L. Kleiman. 2003. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 278:27644-27651. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovaleski, B. J., R. Kennedy, M. K. Hong, S. A. Datta, L. Kleiman, A. Rein, and K. Musier-Forsyth. 2006. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J. Biol. Chem. 281:19449-19456. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, R., M. T. Latreille, and M. Mirande. 1991. A PMR2 tandem repeat with a modified C-terminus is located downstream from the KRS1 gene encoding lysyl-tRNA synthetase in Saccharomyces cerevisiae. Mol. Gen. Genet. 227:149-154. [DOI] [PubMed] [Google Scholar]
- 20.Mirande, M. 1991. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog. Nucleic Acid Res. Mol. Biol. 40:95-142. [DOI] [PubMed] [Google Scholar]
- 21.Mirande, M., B. Cirakoglu, and J. P. Waller. 1982. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. III. Assignment of aminoacyl-tRNA synthetase activities to the polypeptide components of the complexes. J. Biol. Chem. 257:11056-11063. [PubMed] [Google Scholar]
- 22.Mirande, M., D. Le Corre, and J. P. Waller. 1985. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur. J. Biochem. 147:281-289. [DOI] [PubMed] [Google Scholar]
- 23.Muthumani, K., D. S. Hwang, B. M. Desai, D. Zhang, N. Dayes, D. R. Green, and D. B. Weiner. 2002. HIV-1 Vpr induces apoptosis through caspase 9 in T cells and peripheral blood mononuclear cells. J. Biol. Chem. 277:37820-37831. [DOI] [PubMed] [Google Scholar]
- 24.Onesti, S., A. D. Miller, and P. Brick. 1995. The crystal structure of the lysyl-tRNA synthetase (LysU) from Escherichia coli. Structure 3:163-176. [DOI] [PubMed] [Google Scholar]
- 25.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 26.Shiba, K., T. Stello, H. Motegi, T. Noda, K. Musier-Forsyth, and P. Schimmel. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 272:22809-22816. [DOI] [PubMed] [Google Scholar]
- 27.Sierra, S., B. Kupfer, and R. Kaiser. 2005. Basics of the virology of HIV-1 and its replication. J. Clin. Virol. 34:233-244. [DOI] [PubMed] [Google Scholar]
- 28.Stark, L. A., and R. T. Hay. 1998. Human immunodeficiency virus type 1 (HIV-1) viral protein R (Vpr) interacts with Lys-tRNA synthetase: implications for priming of HIV-1 reverse transcription. J. Virol. 72:3037-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, Y., Y. Imai, H. Nakayama, K. Takahashi, K. Takio, and R. Takahashi. 2001. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8:613-621. [DOI] [PubMed] [Google Scholar]
- 30.Tolkunova, E., H. Park, J. Xia, M. P. King, and E. Davidson. 2000. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J. Biol. Chem. 275:35063-35069. [DOI] [PubMed] [Google Scholar]
- 31.Urban, P., C. Cullin, and D. Pompon. 1990. Maximizing the expression of mammalian cytochrome P-450 monooxygenase activities in yeast cells. Biochimie 72:463-472. [DOI] [PubMed] [Google Scholar]