Abstract
The final assembly of herpes simplex virus (HSV) involves binding of tegument-coated capsids to viral glycoprotein-enriched regions of the trans-Golgi network (TGN) as enveloped virions bud into TGN membranes. We previously demonstrated that HSV glycoproteins gE/gI and gD, acting in a redundant fashion, are essential for this secondary envelopment. To define regions of the cytoplasmic (CT) domain of gE required for secondary envelopment, HSVs lacking gD and expressing truncated gE molecules were constructed. A central region (amino acids 470 to 495) of the gE CT domain was important for secondary envelopment, although more C-terminal residues also contributed. Tandem affinity purification (TAP) proteins including fragments of the gE CT domain were used to identify tegument proteins VP22 and UL11 as binding partners, and gE CT residues 470 to 495 were important in this binding. VP22 and UL11 were precipitated from HSV-infected cells in conjunction with full-length gE and gE molecules with more-C-terminal residues of the CT domain. gD also bound VP22 and UL11. Expression of VP22 and gD or gE/gI in cells by use of adenovirus (Ad) vectors provided evidence that other viral proteins were not necessary for tegument/glycoprotein interactions. Substantial quantities of VP22 and UL11 bound nonspecifically onto or were precipitated with gE and gD molecules lacking all CT sequences, something that is very unlikely in vivo. VP16 was precipitated equally whether gE/gI or gD was present in extracts or not. These observations illustrated important properties of tegument proteins. VP22, UL11, and VP16 are highly prone to binding nonspecifically to other proteins, and this did not represent insolubility during our assays. Rather, it likely reflects an inherent “stickiness” related to the formation of tegument. Nevertheless, assays involving TAP proteins and viral proteins expressed by HSV and Ad vectors supported the conclusion that VP22 and UL11 interact specifically with the CT domains of gD and gE.
Herpesvirus capsids cross nuclear membranes by envelopment at the inner nuclear membrane followed by fusion or deenvelopment at the outer nuclear membrane, delivering nucleocapsids into the cytoplasm (reviewed in references 35 and 46 to 48). Secondary envelopment occurs as herpesvirus tegument-coated capsids bind onto viral glycoprotein-enriched regions of the Golgi apparatus, trans-Golgi network (TGN), or endosomes. This delivers enveloped virions into cytoplasmic vesicles, which are subsequently trafficked to cell surfaces.
How tegument-coated nucleocapsids interact with membranes to promote herpesvirus budding is not well understood. Recent studies have suggested that redundant interactions between tegument proteins and the cytoplasmic (CT) domains of specific viral glycoproteins are required. Herpes simplex virus (HSV) produces as many as 12 membrane glycoproteins as well as other nonglycosylated membrane proteins (54). Any one of these membrane proteins can be deleted without substantially reducing the numbers of enveloped virions produced. However, HSV mutants lacking both gD and gE produce few enveloped particles (22). Instead, these mutants produced aggregates, including thousands of tegument-coated capsids, in the cytoplasm. gE forms a heterodimer with a second HSV glycoprotein, gI, and the majority of both glycoproteins are found in this complex in infected cells (34). Deletion mutants lacking gD and gI displayed more subtle defects in assembly, while mutants lacking gD, gE, and gI displayed more profound defects than gD−/gE− mutants (22). There were only minor (two- to threefold more) defects in secondary envelopment when either gD or gE was individually deleted (22, 36). Another alphaherpesvirus, the pig pseudorabies virus (PRV), exhibited a different requirement for this process. PRV mutants lacking both gM and gE failed to produce enveloped particles and accumulated capsids in the cytoplasm (6, 7). By contrast, an HSV mutant lacking gE and gM was not compromised for secondary envelopment (9). It has become an important paradigm that herpesviruses utilize viral glycoproteins, and likely tegument proteins, in redundant interactions for assembly.
HSV glycoproteins gE/gI and gD function in other facets of virus replication and spread. gD is essential for entry and spread in all cells tested to date, and mutants lacking gD are propagated on complementing cells (41). An HSV mutant lacking the gD CT domain could enter cells normally but produced smaller plaques and reduced yields of infectious virus (24), and in retrospect, this likely represented minor defects in assembly. HSV gE/gI complexes are required for HSV cell-to-cell spread in both neuronal and epithelial cells (15-17, 33). We proposed that gE/gI functions in HSV cell-to-cell spread by promoting the movement of newly assembled virions to epithelial cell junctions (reviewed in reference 35). The CT domains of gE/gI allow the glycoprotein to interact with TGN sorting machinery, and specifically, the gE CT domain is necessary for TGN accumulation as well as cell-to-cell spread (23, 36, 45, 58, 59). Coupled with observations that the HSV gE CT domain functions in secondary envelopment, this observation suggested that gE/gI promotes envelopment into specific subdomains of epithelial TGN in which sorting occurs so that nascent virions move specifically to cell junctions (35, 36). In polarized cells, the TGN is the major site for basolateral versus apical sorting. HSV mutants lacking the gE CT domain were mislocalized in epithelial cells and transported apically rather than to basolateral surfaces and cell junctions (36). We recently demonstrated that N-terminal, juxtamembrane regions of the gE CT domain promote TGN localization at early times of infection, while sequences nearer the C terminus promote trafficking to cell junctions late (23, 59). Both segments of the gE CT domain were required for gE/gI-mediated cell-to-cell spread.
There are also numerous (15 or more) HSV tegument proteins that form a complex lattice divided into at least two coats: (i) an inner layer including VP1/2 and UL37, which associate directly with capsids, and (ii) a more peripheral layer composed of proteins that interact with the envelope (reviewed in references 47 and 48). Among the peripheral tegument proteins are three polypeptides (VP22 [UL49], VP16 [UL48], and UL11), which have been implicated in secondary envelopment. VP22, which is one of the most abundant HSV tegument proteins (31), associates with cellular membranes and localizes to the TGN and endosomes when expressed without other HSV proteins (8). A mutant HSV lacking VP22 was able to assemble enveloped virions that reached extracellular compartments. This mutant produced virions with reduced immediate-early proteins (ICP0 and ICP4) and displayed delayed viral protein synthesis (19). Another VP22− mutant described recently displayed defects in reaching extracellular compartments and in cell-to-cell spread (18). Thus, VP22 plays some role in the final assembly of HSV, although likely in a redundant fashion with other tegument proteins.
VP16 is also an abundant component of the tegument (31) and interacts with VP22 (20). HSV and PRV VP16 mutants display increased numbers of cytoplasmic unenveloped capsids and few or no enveloped capsids (25, 49, 57). VP16− mutants accumulate capsids in the cytoplasm, but these are not highly aggregated, as in the case of gD−/gE− mutants. This implies that, without VP16, tegument proteins responsible for aggregation do not assemble around these capsids.
The HSV UL11 protein is modified with myristate and palmitate and contains a TGN/endosomal-sorting acidic cluster that promotes localization to the cytoplasmic surfaces of the Golgi apparatus and TGN (1, 42, 43). An HSV UL11− mutant exhibited fewer enveloped virions and a two- to threefold increase in unenveloped cytoplasmic capsids (2). Similarly, a PRV UL11− mutant displayed defects in secondary envelopment (40) and a UL11−/gM− double mutant displayed large cytoplasmic capsid aggregates (39). Consistent with the notion that UL11 functions in an important fashion in secondary envelopment with other herpesvirus families, a mutant human cytomegalovirus unable to express the UL11 homologue UL99 accumulated large numbers of tegument-coated, cytoplasmic capsids (53).
Interactions between tegument proteins and glycoproteins have been described by biochemical analyses. Early studies involving affinity purification and cross-linking indicated that VP16 interacts with glycoproteins gB, gD, and gH (37, 60). Two-hybrid analyses have indicated that PRV VP22 interacts with the CT domains of gE and gM (26). In vitro experiments indicated that the HSV gD CT domain fused to glutathione S-transferase interacts with VP22 as well as with tegument-coated nucleocapsids and that VP22 was immunoprecipitated with gD from HSV-infected cells (11). Moreover, VP16 can interact with glycoprotein H (gH) in vivo and by coimmunoprecipitation (28, 38). Genetic confirmation for some of these interactions has been previously reported. HSV VP22-null mutants incorporate reduced amounts of gB, gD, and gE into the virion envelope (18, 19). Moreover, a mutant PRV lacking both gE/gI and gM incorporated less VP22 into virions (26).
In order to better understand the final assembly of HSV, we attempted to define regions within the relatively large gE CT domain that are essential for secondary envelopment. A region in the middle of the gE CT domain including residues 470 to 495 played a key role in envelopment, although other residues nearer the C terminus also contributed. Using tandem affinity purification (TAP) fusion proteins containing the full-length gE CT domain, we identified HSV tegument proteins UL11 and VP22 as binding to the CT domain, and there was less UL11 and VP22 bound to truncated gE CT molecules. VP22 and UL11 were precipitated in conjunction with full-length gE/gI and also with gD from HSV-infected cells. However, substantial fractions of VP22 and UL11 bound nonspecifically to glycoproteins devoid of their CT domains.
MATERIALS AND METHODS
Cells and viruses.
HEC-1A cells (3) were grown in RPMI medium (BioWhittaker, Inc., Walkersville, MD) supplemented with 10% fetal bovine serum (FBS). HaCaT cells (5) and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) (BioWhittaker) supplemented with 10% FBS. VD60 cells, derived from Vero cells and capable of expressing gD (41), were maintained in DMEM lacking histidine and supplemented with 10% FBS and 0.5 mM histidinol (Sigma, St. Louis, MO). 293 M and 293-Cre4 cells (50) were obtained from Microbix (Toronto, Ontario, Canada) and maintained in Eagle's minimal essential medium supplemented with 10% FBS. Wild-type HSV-1 strain F (originally from Pat Spear, Northwestern Medical School) and the F-dl2 (24), F-gEΔCT (58), F-BAC HSV-1 (32), and F-BAC gE CT domain mutants (23) were propagated and their titers determined using Vero cells. vRR1097, a mutant lacking gD-coding sequences (51); F-BAC gD−, an HSV-1 derived from a bacterial artificial chromosome (BAC) lacking gD sequences; and F-BAC gD−/gE−, lacking gD and gE sequences, were propagated and their titers determined using VD60 cells. Adenoviruses (Ads) expressing gD [AdgD1(E1−)] (10), gE [Ad(E1−) gE], gI [Ad(E1−)gI] (17), VP22, or TAP fusion proteins were propagated and their titers determined using 293 cells.
Construction of F-BAC mutants.
HSV-1 mutants containing single and double mutations in gE and gD were constructed as previously described (23), using a modification of a protocol developed by Datsenko and Wanner (14). Briefly, bacteria containing BACs with wild-type HSV-1 strain F and mutants with gE− or gE truncations were further mutagenized to remove gD-coding sequences. PCR primers which included the first or last 18 nucleotides of the gD-coding sequences as well as sequences homologous to the kanamycin gene flanked by FRT (FLP recognition target) sites in plasmid pKD4 (14) (ATGGGGGGGGCTGCCGCCGTGTAGGTCGGAGCTGCTTC and CTAGTAAAACAAGGGCTGCATATGAATATCCTCCTTAG) were used to produce a DNA fragment in which the majority of the gD-coding sequences were replaced by a kanamycin cassette. This DNA was transformed into RR1 bacteria containing HSV BACs (containing wild-type HSV sequences or mutants with gE− or gE truncations) and pKD46, a plasmid encoding ampicillin resistance and the red recombinase. Selection for recombinant BACs was performed as previously described (23). BACs containing mutant HSV-1 genomes were sequenced across, upstream of, and downstream of the mutation site. BAC DNA was produced and transfected into Vero or VD60 cells as previously described (23).
Construction of recombinant Ad expression vectors.
Plasmid pAdTet7 contains Tet-responsive enhancer sequences, the simian virus 40 late poly(A) cassette, adenovirus E1A, and a single loxP site to increase recombination frequency (55). The sense oligonucleotide UL49F (GAATTCATGTGATTCCGTGTTCGTGGAACCATGA) and the antisense oligonucleotide UL49R (GGATCCTTAGTGGATCCGTTGGTGCTTTATTGTCT) were used to amplify the VP22 sequence from HSV-1 strain F by using Pfu DNA polymerase (Stratagene) and nucleotides from Promega. The product of this amplification reaction was cloned into pAdTet7, and the resulting construct, pAdTet7-VP22, was sequenced to verify the VP22 coding sequences. Plasmid pAdTet7-NTAP, a gift from Dan Streblow (Vaccine and Gene Therapy Institute, Portland, OR), was identical to pAdTet7 except that there is a TAP domain cloned upstream of the multiple cloning site of pAdTet7. The CT domains of various gE truncation mutants were amplified as described previously (23) and subcloned in frame at the C terminus of the TAP domain in pAdTet7-NTAP. Recombinant adenoviruses were produced by cotransfection of pAdTet7-VP22 or pAdTet7-NTAP/gE plasmids with adenovirus DNA (Ad5-psi5) into 294-Cre4 cells, which express Cre recombinase (50). Recombinant adenoviruses were expanded with four passages on Cre-4 cells to remove Ad5-psi5, and the titers were determined using 293 M cells.
Antibodies.
3104, a gI-specific HSV monoclonal antibody (MAb), and 3114, a gE-specific MAb, were gifts from Anne Cross and Nigel Stow (Institute of Virology, Glasgow, United Kingdom). DL6, a MAb specific for HSV-1 gD, was a gift from Gary Cohen and Roselyn Eisenberg (University of Pennsylvania, Philadelphia). The rabbit polyclonal anti-VP22 antibody AGV 030 was a gift from Gillian Elliott (Marie Curie Research Institute, Surrey, United Kingdom) (21), the rabbit polyclonal anti-UL11 antibody Rbt #73 was a gift from John W. Wills (College of Medicine, Pennsylvania State University, Hershey), and a rabbit polyclonal anti-VP16 antibody (catalog no. 3844-1) was purchased from BD Biosciences (San Jose, CA). An anti-calmodulin binding domain (CBD) antibody was obtained from Upstate Cell Signaling (Lake Placid, NY).
Electron microscopy.
HEC-1A cells were infected with wild-type F-BAC HSV-1 or F-BAC mutants for 16 h, washed with 0.1 M sodium cacodylate buffer (pH 7.2), and fixed in Ito and Karnovsky's fixative for 30 min at room temperature (1.6% paraformaldehyde, 2.5% glutaraldehyde, and 0.5% picric acid in 0.1 M sodium cacodylate). The samples were postfixed in 1.5% osmium tetroxide, rinsed, and then postfixed in 4% paraformaldehyde. The samples were dehydrated in a graded acetone series and embedded in epoxy resin, and ultrathin sections were double stained in uranyl acetate and lead citrate and viewed with a Philips EM 300 electron microscope.
Immunoprecipitation of radiolabeled HSV proteins.
Vero cells were infected with HSV-1 (using 10 PFU/cell) in DMEM containing 1% FBS for 2 h, and then fresh medium was added for an additional 6 to 7 h. The cells were washed twice with DMEM lacking methionine and cysteine and containing 1% dialyzed FBS and then labeled in the same medium with added [35S]methionine-cysteine (150 μCi/ml; NEN) for a further 3 h. The cells were lysed in Nonidet P-40 (NP-40)-deoxycholate extraction buffer (1% NP-40, 0.5% deoxycholate, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 1 mM phenylmethylsulfonyl fluoride and stored at −70°C. The cell extracts were thawed and centrifuged at 50,000 × g for 45 min, and anti-gE antibody (MAb 3114), anti-gD antibody (MAb DL6), or anti-gB antibody (MAb 15βB2) was added for 1 to 2 h at 4°C, followed by incubation with protein A-Sepharose. Immunoprecipitated proteins were subjected to electrophoresis on polyacrylamide gels followed by analysis by autoradiography (58).
Immunoprecipitation of radiolabeled or unlabeled TAP constructs.
HaCaT cells were initially coinfected with various Ad vectors expressing TAP/gE fusion proteins by using 50 PFU (defined using 293 cells)/cell and simultaneously with Adtet-trans by using 10 PFU/cell in DMEM supplemented with 1% FBS for 18 h. The cells were subsequently infected with F-gEΔCT or left uninfected for a further 7.5 h. For production of radiolabeled viral proteins, the cells were washed twice with a medium lacking methionine and cysteine and containing 1% dialyzed FBS and then labeled for 3 h with [35S]methionine-cysteine as described above. Cells were harvested, briefly washed, and incubated either in 0.5% NP-40 lysis buffer (0.5% NP-40, 50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 2 mM dithiothreitol, and protease inhibitor tablets [Roche Diagnostics]) containing either 100 mM or 500 mM NaCl or in 1% digitonin lysis buffer (1% digitonin, 50 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 2 mM dithiothreitol, and complete protease inhibitor tablets) containing either 100 or 500 mM NaCl for 20 min. Cell extracts were centrifuged at 60,000 × g for 30 min and incubated with immunoglobulin G (IgG)-Sepharose beads (Amersham-BioSciences, Piscataway, NJ) for 1 h at 4°C. Sepharose beads were pelleted at low speed (200 to 500 × g) for ≈30 s and washed four times in dilute wash buffers (0.1% NP-40 or 0.1% digitonin, 50 mM Tris-HCl) containing 150 mM or 500 mM NaCl, and bound proteins were released by boiling them in buffer containing 2% sodium dodecyl sulfate and 2% β-mercaptoethanol before electrophoresis on sodium dodecyl sulfate-polyacrylamide gels (58) and detection of proteins by autoradiography. In other cases, unlabeled cells infected as described above were extracted using the same buffers, centrifuged at 25,000 × g for 20 min, and subsequently incubated with IgG-Sepharose for 1 h at 4°C. The beads were washed four times in dilute NP-40 or digitonin wash buffers, pelleted at 200 to 500 × g for ≈30 s, and subjected to electrophoresis on polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, and then the membranes were immunoblotted with anti-tegument antibodies as described previously (58).
Immunoprecipitation of gD and gE from HSV-1- or Ad-infected cells followed by blotting for tegument proteins.
Confluent HaCaT cells were infected with HSV-1 for 12 h or with Ad vectors expressing gE/gI or gD and VP22 for 24 h. The cells were harvested, briefly washed, and incubated in 0.5% NP-40 lysis buffer containing 150 mM NaCl on ice for 20 min. The cell extracts were centrifuged at 25,000 × g for 20 min, and then the supernatants were precleared by incubation with protein A-Sepharose for 15 min. These beads were removed by centrifugation and anti-gE (3114) or anti-gD (DL6) MAb, and protein A-Sepharose was added to the extracts for an additional 20 min at 37°C. The beads were washed in a dilute NP-40 buffer and subjected to electrophoresis on polyacrylamide gels, and then the proteins were transferred to PVDF membranes and immunoblotted as described previously (58).
RESULTS
Construction of gD and gE mutant viruses.
We previously demonstrated that HSV-1 gD and gE/gI act in a redundant fashion to promote secondary envelopment (22). Tegument proteins likely interact with the CT domains of these glycoproteins in order to assemble an envelope around capsids as virions bud into the TGN. The gE CT domain is large, extending from residue 445 to residue 550, whereas the gD CT domain is 30 amino acids in length. The N-terminal half of the gE CT domain (residues 448 to 495) contains numerous recognizable TGN sorting motifs, and we previously demonstrated that these sequences were necessary for gE/gI complexes to localize to the TGN in early stages of virus replication (23). Residues nearer the C terminus of the CT domain (residues 495 to 519) were required for redistribution of gE/gI complexes to cell junctions, a process that occurs at late times coincident with the movement of virions to cell junctions.
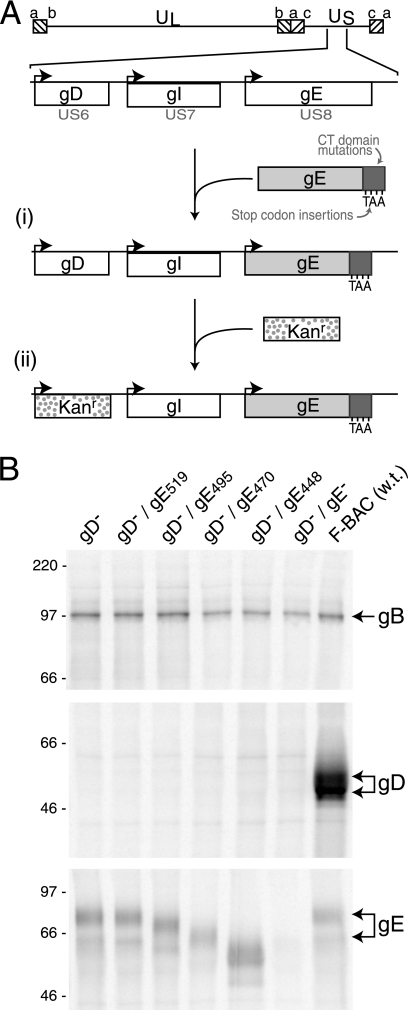
Here, we sought to determine which gE CT domain sequences were important for secondary envelopment. For these studies, it was necessary to construct HSVs expressing gE truncation mutations unable to express gD. This was done by beginning with several HSV-1 (strain F) genomes containing previously described gE truncations that had been cloned into BACs (23). For each of these HSV/BAC genomes, gD (US6) sequences were replaced with a kanamycin gene cassette (Fig. 1A). Additionally, two other BACs were produced as controls: (i) BAC gD−, a BAC in which the kanamycin gene cassette replaced the gD sequences, and (ii) BAC gD−/gE−, derived from BAC gE− (23) and lacking both gE-coding sequences and gD sequences. These BACs were transfected into cells to produce the following viruses: F-BAC gD−/gE519, expressing gE truncated after residue 519; F-BAC gD−/gE495, truncated after gE residue 495; F-BAC gD−/gE470, truncated after gE residue 470; F-BAC gD−/gE448, truncated after gE residue 448 (with only three residues of the CT domain); F-BAC gD−, expressing wild-type gE and lacking gD; and F-BAC gD−/gE−, lacking both gD and gE. Because gD is essential for entry, these BAC DNAs were transfected into VD60 cells which express gD (41).
FIG. 1.
Construction of recombinant viruses. (A) The genome of HSV-1, including the US6 (gD), US7 (gI), and US8 (gE) genes, is depicted (44). (i) The insertion of stop codons in the gE CT domain after residues 448, 470, 495, and 519 was previously described (23). (ii) A kanamycin resistance gene cassette was inserted into the coding sequences of gD, and this was transferred into BACs containing truncated versions of gE. (B) Vero cells were infected with F-BAC gD−, F-BAC gD−/gE519, F-BAC gD−/gE495, F-BAC gD−/gE470, F-BAC gD−/gE448, F-BAC gD−/gE−, or wild-type F-BAC HSV-1 (w.t.). The cells were labeled with [35S]methionine-cysteine, and gB, gD, or gE was immunoprecipitated from cell extracts by using MAb 15βB2, DL6, or 3114, respectively. The positions of gB, gD, and gE are indicated on the right side of the panel.
Mutant viruses were characterized for expression of gD, gB, and gE by infecting Vero cells, radiolabeling the cells with [35S]methionine-cysteine, and immunoprecipitating gD with MAb DL6, gE with MAb 3114, or gB with MAb 15βB2. Wild-type F-BAC expressed similar immature (51-kDa) and mature (56-kDa) forms of protein gD (Fig. 1B). F-BAC gD−, F-BAC gD−/gE519, F-BAC gD−/gE495, F-BAC gD−/gE470, F-BAC gD−/gE448, and F-BAC gD−/gE− did not express gD and expressed gE molecules of the sizes predicted from previous work (23). All viruses produced similar quantities of gB.
Secondary envelopment of F-BAC gD−/gE CT mutants.
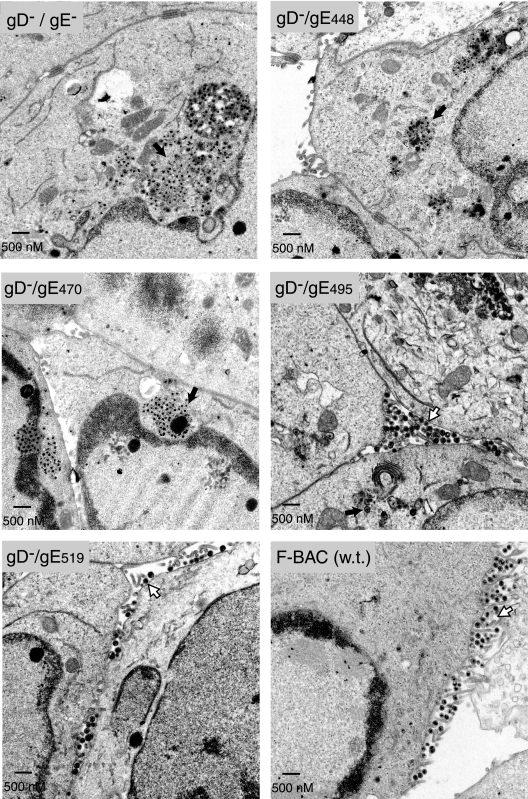
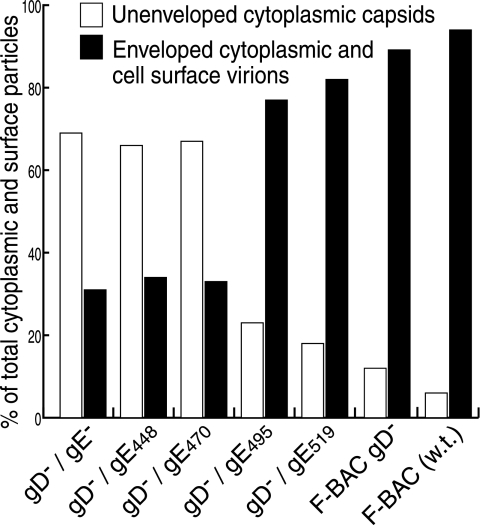
Viruses deleted for both gD and gE undergo final envelopment poorly and, instead, accumulate large numbers of nucleocapsids in aggregates in the cytoplasm (22). HEC-1A cells infected with F-BAC gD−/gE− accumulated large aggregates of nonenveloped capsids in the cytoplasm (Fig. 2), as had been described for other gD−/gE− and gD−/gE−/gI− mutants (22). As before, these aggregates appeared to be immersed in an electron-dense material, presumably containing tegument proteins. These observations were quantified by counting approximately 1,000 virus particles in multiple cell sections for each mutant. Particles were categorized as unenveloped capsids in the nucleus or cytoplasm or as enveloped particles in the perinuclear space, in the cytoplasm, or on the cell surface. We previously showed that gE−/gD− viruses do not differ in nuclear events, and this was the case in the present studies. Thus, Fig. 3 compares unenveloped capsids in the cytoplasm with enveloped particles in the cytoplasm combined with enveloped particles on the cell surface. There were 12- to 14-fold increases in the numbers of unenveloped nucleocapsids observed in comparing F-BAC gD−/gE− with wild-type F-BAC (Fig. 3). F-BAC gD−/gE448, missing the entire gE CT domain (gE448), also exhibited large numbers of unenveloped capsids in aggregates (Fig. 2), confirming that the gE CT domain was responsible for this defect. F-BAC gD−/gE470, which expresses a gE with only 25 residues of the CT domain, also displayed aggregates of unenveloped capsids, and fewer enveloped virions were observed (Fig. 2 and 3). F-BAC gD−/gE495 and F-BAC gD−/gE519 primarily produced enveloped virions in the cytoplasm and on the cell surface (Fig. 2), and unenveloped capsids in the cytoplasm were less common, as was the case with wild-type F-BAC. However, there were obvious differences between F-BAC gD−/gE495 and F-BAC gD−, which expresses wild-type gE; more unenveloped particles were observed with F-BAC gD−/gE495 (Fig. 2 and 3). F-BACgD−/gE515 was intermediate between F-BAC gD−/gE495 and F-BAC gD− (Fig. 2 and 3). These observations supported a role for the C-terminal sequences (residues 495 to 550) as a contributing factor in secondary envelopment. We also observed relatively minor defects in assembly with F-BAC gD− compared to those observed with F-BAC, as had been described before (22, 24). We concluded that a region of the gE CT domain between amino acids 470 and 495 is important for secondary envelopment, although sequences between residues 495 and 550 significantly influence this process.
FIG. 2.
Electron micrographs of cells infected with F-BAC gD−/gE CT mutants. Human HEC-1A epithelial cells were infected with F-BAC gD−/gE−, F-BAC gD−/gE448, F-BAC gD−/gE470, F-BAC gD−/gE495, F-BAC gD−/gE519, or F-BAC (w.t.) for 16 h. The cells were fixed and processed for electron microscopy. Black arrows point to aggregates of unenveloped capsids, while white arrows point to enveloped virions on the surfaces of cells.
FIG. 3.
Distributions of virus particles in cells infected by gD−/gE CT mutants. Randomly selected sections of HSV-infected HEC-1A cells were characterized by electron microscopy, and unenveloped nucleocapsids in the nucleus and cytoplasm and enveloped virions in the perinuclear space, in the cytoplasm, and on the cell surface were counted. In the figure, values for unenveloped capsids in the cytoplasm (white bars) are compared to those for enveloped virions in the cytoplasm combined with virions on cell surfaces (black bars).
Construction of gE CT domain/TAP fusions.
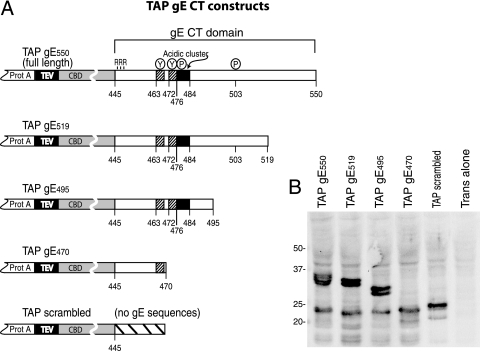
The CT domain of gE functions in three related process: TGN accumulation, secondary envelopment, and promotion of HSV cell-to-cell spread (reviewed in reference 35). As a method to identify both cellular and viral proteins involved in these processes, we fused the gE CT domain onto a TAP domain. This TAP domain included two protein A (IgG-binding) sequences separated by a tobacco etch virus protease site from a calmodulin binding domain (Fig. 4A), allowing sequential purification by using IgG and calmodulin affixed to Sepharose (52). Truncated versions of the gE CT domain (gE-519, -495, and -470) were fused onto the C terminus of the TAP domain as well as a control, TAP scrambled, which contains no gE sequences but instead has a scrambled sequence of amino acids (Fig. 4A). To achieve various levels of expression in different cells, the TAP-tagged proteins were expressed using replication-defective Ad vectors under the control of a tetracycline transactivator-inducible promoter (12, 29, 55). Cells can be coinfected with a second, nonreplicating Ad vector, Adtet-trans, which expresses the tetracycline transactivator protein (27, 55), in order to turn on the expression of gE/TAP fusion proteins. While it might have been reasonable to express the TAP fusion proteins in the context of the HSV genome, using Ad vectors had the benefit of enabling identification of interacting cellular proteins in future studies.
FIG. 4.
Construction of TAP-gE fusion proteins. (A) The TAP domain was composed of two tandem protein A (IgG-binding) domains separated from a CBD by a tobacco etch virus protease cleavage site. The TAP domain was fused N-terminal of the entire gE CT domain, beginning with three arginine residues that are adjacent to the gE transmembrane domain. Other TAP fusion proteins included truncated versions of the gE CT domain: gE519, gE495, and gE470. Also shown is a construct designated TAP scrambled, which contains 25 random amino acids unrelated to the gE CT domain. (B) Vero cells were coinfected with Ad vectors expressing TAP/gE550, TAP/gE519, TAP/gE495, TAP/gE470, or TAP scrambled and Adtet-trans or with Adtet-trans alone. Cell extracts were subjected to electrophoresis on polyacrylamide gels, and then proteins were transferred to the PVDF membrane and probed with an anti-CBD rabbit antibody. Molecular mass markers of 50, 37, 25, and 20 kDa are indicated.
To assess the expression of the TAP/gE fusions, Vero cells were coinfected with Ad vectors expressing TAP/gE proteins and Adtet-trans, and Western blot analyses were performed on cell extracts by using an anti-CBD antibody. Proteins of the expected sizes were expressed (Fig. 4B). Adtet TAP/gE550, Adtet TAP/gE519, and Adtet TAP/gE495 each produced a protein doublet, likely related to phosphorylation events that occur in the gE CT domain (58). There were also faster-migrating species of approximately 20 to 25 kDa observed with all the TAP fusion proteins, including TAP scrambled, which may consist of degradation products lacking the protein A domains, as these bands were not observed with IgG-Sepharose (not shown).
Binding of tegument proteins to the TAP gE CT domain.
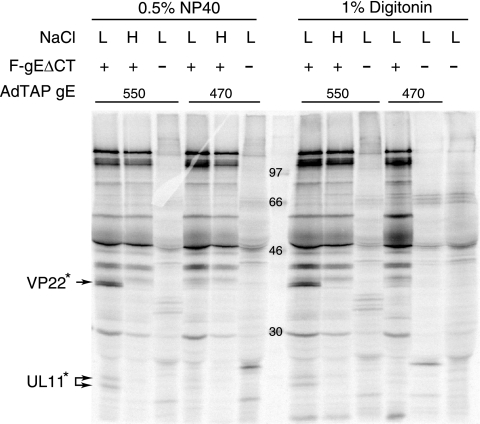
To characterize the binding of viral proteins to gE CT, the human keratinocyte cell line HaCaT was chosen because these cells display the most profound phenotype with gE-null mutants (58). HaCaT cells were infected with Ad vectors expressing TAP/gE550 (full-length gE CT) or TAP/gE470, which included only 25 gE CT residues (a domain insufficient for secondary envelopment). After 18 h, these cells were infected with an HSV gE-null mutant, F-gEΔCT (58), for an additional 7 to 8 h or not infected with HSV and then radiolabeled with [35S]methionine-cysteine for 3 h. Under these conditions, there was HSV-induced host protein shutoff and radiolabeled proteins were predominately of HSV origin, although the TAP/gE proteins expressed by the Ad vectors continued to be expressed at high levels (not shown). Cell extracts were made using mild nonionic detergents: 0.5% NP-40 or 1% digitonin and two concentrations of NaCl (100 mM or 500 mM NaCl) were used. The extracts were centrifuged at 60,000 × g to remove insoluble or aggregated material and then mixed with IgG-Sepharose, which binds the protein A domains of TAP proteins. The extracts were centrifuged at low speeds (200 to 500 × g) for ≈30 s to pellet IgG-Sepharose, under which conditions no significant quantities of viral proteins were pelleted (not shown). TAP/gE550 pulled down two sets of protein bands that were not observed, or were observed in lower quantities, with TAP/gE470 (Fig. 5). These proteins were observed with 0.5% NP-40 and 1% digitonin lysis buffers and were similar in size to tegument proteins VP22 and UL11. Thus, these proteins were designated VP22* and UL11* at this point. VP22* and UL11* were not observed with high salt concentrations or when the cells were not infected with F-gEΔCT (Fig. 5).
FIG. 5.
Interactions between TAP/gE fusion proteins and HSV proteins. Human HaCaT keratinocytes were infected with Ad expressing either TAP/gE550 or TAP/gE470 for 18 h and then infected with F-gEΔCT (a gE-null mutant) for an additional 7 to 8 h. The cells were radiolabeled with [35S]methionine-cysteine for 3 h, and then cell extracts were made using 0.5% NP-40 lysis buffer or 1% digitonin lysis buffer and low salt (L) (100 mM NaCl) or high salt (H) (500 mM NaCl). Extracts were centrifuged at 60,000 × g and supernatants incubated with IgG-Sepharose at 4°C. Proteins were eluted and subjected to electrophoresis. Two sets of bands, designated VP22* and UL11*, were observed when cells were infected with TAP/gE550 and F-gEΔCT but not with TAP/gE470 and F-gEΔCT and not without F-gEΔCT. Molecular mass markers of 97, 66, 46 and 30 kDa are indicated.
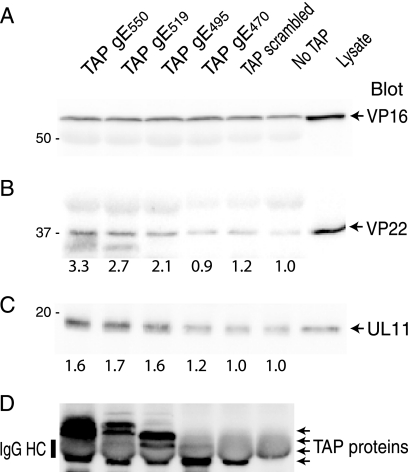
To identify the viral proteins observed in Fig. 5, HaCaT cells were infected with Ad vectors expressing TAP/gE fusions or TAP scrambled and then superinfected with HSV F-gEΔCT; then, TAP proteins were purified from 0.5% NP-40 cell extracts by using IgG-Sepharose before blotting with anti-tegument proteins. VP16 was observed in similar quantities whether a TAP/gE fusion protein, TAP scrambled, or no TAP protein was expressed in the cells (Fig. 6A). Thus, it was impossible to determine any specific binding of VP16 to these TAP proteins, although this nonspecific interaction provided a positive control for HSV infection in subsequent experiments. Binding of VP22 to TAP/gE550 was observed, and there was approximately one-third as much binding each to TAP/gE470 and TAP scrambled (Fig. 6B). Levels for TAP/gE519 and TAP/gE495 were intermediate. The nonspecific component of VP22 binding was illustrated by the levels of VP22 observed with TAP scrambled and where no TAP protein was expressed, which likely reflect nonspecific binding to IgG-Sepharose. UL11 also bound to TAP/gE550 and approximately as well to TAP/gE519 and TAP/gE495, and there was less binding to TAP/gE470, to TAP scrambled, and where no TAP protein was expressed. Thus, again, the nonspecific component of this binding was high. The expression of the TAP proteins was measured by blotting with anti-protein A antibodies (Fig. 6D). Here, there was some bleeding of IgG from the IgG-Sepharose that altered the mobilities of TAP proteins and secondary antibodies cross-reacted with this IgG to some extent. Still, it was clear that TAP/gE470, TAP/gE495, and TAP/gE519 were expressed at similar levels in these cells.
FIG. 6.
Coimmunoprecipitation of VP16, VP22, or UL11 with TAP/gE fusion proteins. HaCaT cells were coinfected with Ad vectors expressing TAP/gE550, TAP/gE519, TAP/gE495, TAP/gE470, TAP scrambled, and AdTet trans or were not infected with an Ad vector (No TAP) for 18 h. The cells were subsequently infected with HSV F-gEΔCT and harvested 12 h later in 0.5% NP-40 lysis buffer. Cell extracts were incubated with IgG-Sepharose and washed, and proteins were eluted and subjected to electrophoresis before transfer to PVDF membranes. Membranes were probed with rabbit polyclonal antibodies specific for VP16 (A), VP22 (B), UL11 (C), or anti-protein A antibodies to detect TAP proteins (D). Note that IgG eluted from IgG-Sepharose and the heavy chain comigrated with TAP proteins, as indicated in panel D. VP22 and UL11 levels were quantified using IP Lab Gel software and compared to immunoprecipitation levels for extracts containing no TAP proteins (No TAP), which were set at 1.
In other experiments, we assessed how well VP22, VP16, and UL11 were solubilized under these extraction conditions by using Western blot analyses to probe pelleted material versus protein in supernatants after centrifugation at 60,000 × g. Approximately 35% of the VP22 in cells was soluble, i.e., present in the supernatant fraction, with both 1% digitonin and 0.5% NP-40 extraction buffers containing 100 mM NaCl. Similarly, ≈25% each of UL11 and VP16 was solubilized with both of these buffers. In all these assays, and in the subsequent experiments described below, cell extracts were not frozen and experiments were carried out quickly under conditions in which tegument proteins did not aggregate or pellet under the low centrifugal forces (200 to 500 × g) used to harvest Sepharose beads (not shown). Therefore, insolubility during our assays cannot account for the tegument proteins observed. Instead, the nonspecific component of binding with these proteins must be caused by binding to these TAP domains or to IgG-Sepharose. The results for other experiments in which proteins associated with these TAP fusion proteins were immunoblotted with antibodies specific for VP1/2, vhs (UL41), and UL24 were consistent with very low or no binding for these tegument proteins (data not shown). VP1/2, vhs, and UL24 were partially solubilized in these cell extracts (not shown). Together, these results indicated that the gE CT domain can interact with VP22 and UL11 in the context of HSV-infected cells. Furthermore, the region of the gE CT domain most important for envelopment (residues 470 to 495), as well as more-C-terminal domains, contributed to VP22 and UL11 binding.
Binding of VP22 and UL11 to gE expressed by HSV.
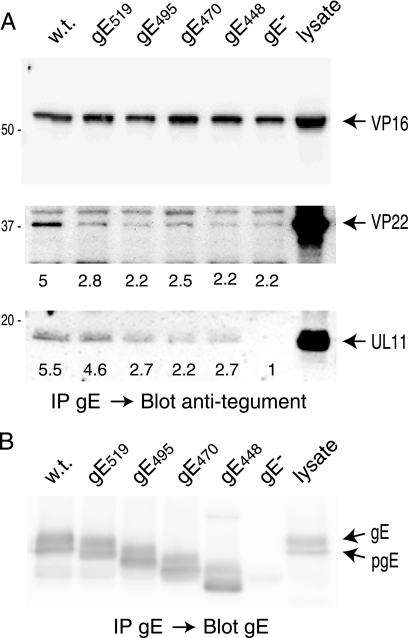
We next attempted to determine whether VP22 and UL11 could also interact in a specific fashion with wild-type gE, and less extensively with truncated gE, in HSV-infected cells. HaCaT cells were infected with F-BAC or HSV expressing truncated gE molecules, the cells were extracted with 0.5% NP-40, and the extracts were centrifuged at 25,000 × g and then preincubated with protein A-Sepharose beads, which were removed by low-speed centrifugation. The cell extracts were then incubated with anti-gE MAb 3114 and protein A-Sepharose for 1 h; then, the immunoprecipitated proteins were subjected to electrophoresis and transferred to PVDF membranes, and blots were probed with antibodies specific to VP16, VP22, or UL11. Again, VP16 was nonspecifically precipitated with antibody and protein A-Sepharose, i.e., in samples from cells that did not express gE (Fig. 7A, top panel). VP22 was observed with wild-type gE, and there were lesser quantities of VP22 observed with gE519, gE495, gE448 (lacking CT domain), and a gE-null mutant (Fig. 7A, middle panel). The expression levels of the different gE molecules were similar in each case (Fig. 7B). Therefore, a substantial fraction of the specific binding of VP22 to gE was to a region including residues 519 to 550 at the extreme C terminus of gE. UL11 also bound specifically to the full-length gE CT domain; there was approximately fivefold more UL11 bound to full-length gE than to a gE-null mutant and approximately twofold more UL11 bound to full-length gE than to gE448 (Fig. 7A, lower panel). In this case, the specific binding component of UL11 primarily involved residues near the C terminus of the gE CT domain between residues 495 to 550. There was also substantial nonspecific binding with gE448, which lacks most of the CT domain. This nonspecific binding was not due to insolubility during the assay, because these extracts had been centrifuged at high speeds for longer periods and then preincubated with protein A-Sepharose and centrifuged at low speeds (200 to 500 × g) before the pull-down assays, which involved low-speed centrifugation for ≈30 s. Instead, these nonspecific interactions involved binding of tegument protein to regions of gE other than the CT domain, with IgG or with protein A-Sepharose. Binding to the gE extracellular domain is illustrated by the levels of UL11 observed for gE448 compared with those observed for a gE-null mutant (Fig. 7A). Although nonspecific binding was observed, there was also clearly specific binding of VP22 and UL11 to gE, which required the gE CT domain and specifically sequences in the C-terminal half of the gE CT (residues 495 to 550), which contributes to secondary envelopment. Importantly, binding was assessed in a fashion that more accurately reflects that in vivo. Higher-order structures with other tegument proteins are likely preserved to some extent, and these may contribute to nonspecific binding.
FIG. 7.
Coimmunoprecipitation of VP16, VP22, or UL11 with gE from HSV-infected cells. HaCaT cells were infected with HSV F-BAC (w.t.) expressing wild-type gE, F-BAC gE519, F-BAC gE495, F-BAC gE470, F-BAC gE448, or F-BAC gE− for 12 h. Cell extracts were made using 0.5% NP-40 lysis buffer and gE immunoprecipitated with anti-gE MAb 3114. Precipitated proteins and a sample representing 20% of the cell lysate were subjected to electrophoresis, and proteins were transferred to membranes and then Western blotted with rabbit polyclonal antibodies specific for anti-VP16 (panel A, top), anti-VP22 (panel A, middle), anti-UL11 (panel A, bottom), or gE (B). VP22 and UL11 were quantified using IP Lab Gel software, with the background value (set at 1.0) determined by counting the pixels in the blot regions containing no obvious proteins.
Binding of VP22 and UL11 to the CT domain of HSV gD.
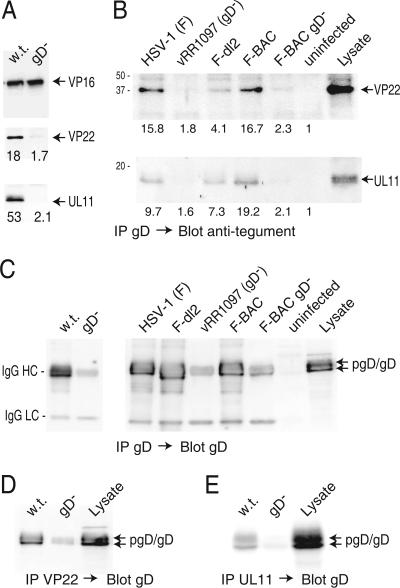
HSV gD and gE apparently interact with multiple tegument proteins where the glycoproteins and the tegument proteins serve redundant functions in secondary envelopment. We extended our analyses to gD by immunoprecipitating gD and blotting with anti-tegument antibodies. VP22 was precipitated extensively with gD, and much less (≈6%) VP22 was observed when gD was absent (Fig. 8A). Additionally, UL11 was precipitated from extracts of HSV-infected cells with gD antibodies, and less (≈4%) was detected when gD was absent (Fig. 8A). Again, VP16 was precipitated by anti-gD MAb whether cell extracts contained or did not contain gD.
FIG. 8.
Coimmunoprecipitation of VP22 and UL11 with gD from HSV-infected cells. (A) HaCaT cells were infected with F-BAC (w.t.) or F-BAC gD− for 12 h, and then cell extracts were made using 0.5% NP-40 lysis buffer. gD was immunoprecipitated using MAb DL6, proteins were subjected to electrophoresis and then transferred to membranes, and the membranes were probed with rabbit polyclonal anti-VP16 (upper panel), anti-VP22 (middle panel), or anti-UL11 (lower panel) antibodies. (B) HaCaT cells were infected with wild-type HSV-1 strain F, vRR1097 (a gD-null mutant), F-dl2 (expressing a gD lacking the CT domain), F-BAC, or F-BAC gD− for 12 h or were left uninfected. Cell extracts were made using 0.5% NP-40 lysis buffer, and gD was immunoprecipitated using anti-gD MAb DL6. Precipitated proteins and a sample representing 20% of the cell extract (Lysate) were subjected to electrophoresis, transferred to membranes, and probed with anti-VP22 or anti-UL11 rabbit antibodies. The numbers shown in panels A and B were derived as described in the legend to Fig. 7, with the background value (set at 1) corresponding to the blot regions with no obvious proteins. (C) Samples immunoprecipitated as described for panels A and B were also probed with rabbit anti-gD antibodies. The IgG-heavy and -light chains (indicated by HC and LC, respectively) derived from mouse MAb DL6 used to precipitate gD were detected through cross-reaction with secondary antibodies and were most obvious in samples from cells lacking gD. (D and E) VP22 or UL11 was precipitated using rabbit polyclonal antibodies, and samples were blotted with mouse anti-gD MAb DL6. In panel D, some IgG-heavy chains were detected by the secondary antibodies, most obviously in samples from the gD− mutant.
Given our observations that VP22 and UL11 were precipitated in substantial quantities with gE lacking the CT domain (Fig. 7), we also compared the binding of these tegument proteins to gD molecules with and without the CT domain. F-dl2 expresses a gD molecule lacking the CT domain and produces slightly smaller plaques than wild-type HSV-1 (24). HaCaT cells were infected with F-dl2, HSV-1 strain F (the parental virus), or vRR1097, a gD-null virus derived from F (51). gD was immunoprecipitated, and precipitated proteins were blotted with VP22-specific or UL11-specific antibodies. There was significantly less (11 to 16%) VP22 and UL11 precipitated from extracts of the gD-null mutant vRR1097 than from wild-type HSV expressing gD (Fig. 8B). However, substantially more VP22 and UL11 were observed when gD was immunoprecipitated from F-dl2-infected cells than when the gD-null mutant was immunoprecipitated. Binding of VP22 to gD lacking a CT domain (F-dl2) was reduced to 26% of that observed with wild-type gD, and binding of UL11 was reduced to only 76% of that observed with wild-type gD. Expression of wild-type and F-dl2 gD was characterized by Western blot analysis using a rabbit anti-gD serum. Unfortunately, anti-rabbit secondary antibodies cross-reacted with the mouse MAb DL6 (used to precipitate gD) and IgG-heavy and -light chains were observed (Fig. 8B). Nevertheless, it was clear that F-dl2 gD, a smaller molecule, was expressed at levels similar to those for wild-type gD.
To confirm interactions between VP22 and UL11 and gE and gD, reciprocal pull-down experiments were performed. It was impossible to perform these experiments with gE/gI because gE/gI is an IgG Fc receptor and binds rabbit IgG. The only available anti-tegument antibodies are rabbit antibodies which precipitate gE/gI. However, precipitation of VP22 from extracts of wild-type-HSV-infected cells followed by blotting with anti-gD MAb DL6 produced a strong band corresponding to gD (Fig. 8D). There was a fainter, faster-migrating band with extracts from a gD-null mutant, which resulted from the low-level cross-reactivity of the anti-mouse IgG secondary antibody. Consistent with this, the IgG light chain was also detected in this blot (not shown). Moreover, gD was detected in precipitates involving anti-UL11 antibodies from an extract of wild-type-HSV-infected cells but not with a gD-null virus (Fig. 8E). We concluded that VP22 interacts substantially and in a specific fashion with gD, interactions that require the CT domain of gD. There was more-limited specific binding of UL11 to gD. In our assays, as much as 50 to 75% of the binding of UL11 to gD occurred with a mutant lacking the CT domain, a process that is highly unlikely to occur in vivo. These results underscore the sticky or nonspecific qualities of these tegument proteins.
Binding of VP22 to gD and gE in the absence of other HSV proteins.
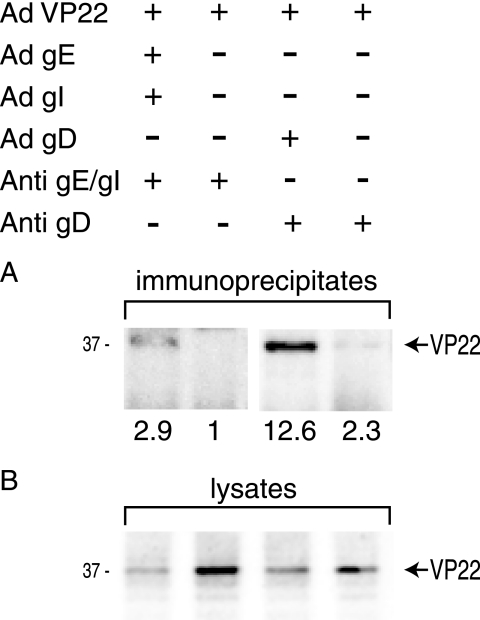
Given that tegument proteins exist in a matrix with other viral proteins in infected cells, it was of interest to determine whether VP22 could bind to gD and gE/gI in the absence of other HSV proteins. A nonreplicating Ad vector expressing VP22 was constructed as described previously (55), and Ad vectors expressing gE/gI and gD have been previously described (10). VP22, gE/gI, and gD were expressed in HaCaT cells by using the Ad vectors, and then gE/gI or gD was immunoprecipitated from cell extracts. Precipitated proteins were subjected to electrophoresis and probed with VP22-specific antibodies. Approximately threefold more VP22 was precipitated from extracts of cells expressing gE/gI than from those of cells not expressing gE/gI (Fig. 9, upper panel). This was observed despite the lower levels of expression of VP22 in cells that were also expressing gE/gI (Fig. 9, lower panel). Similarly, there was ≈5-fold more VP22 precipitated from cells expressing gD than from those not expressing gD when extracts were immunoprecipitated with an anti-gD antibody. The lower levels of VP22 expressed in cells coinfected with Ad vectors expressing HSV proteins relate to competition for transactivator proteins, an occurrence we previously observed with various Ad vectors expressing foreign genes (30). We concluded that VP22 can interact with gE/gI and gD without the requirement of other viral proteins.
FIG. 9.
VP22 binds to gE/gI and gD in the absence of other HSV proteins. HaCaT cells were infected for 24 h with nonreplicating Ad vectors expressing VP22, gE/gI, and gD in conjunction with AdTet-trans, as indicated in the upper panel. Cell extracts were made using 0.5% NP-40 lysis buffer. (A) gE/gI immunoprecipitated with pooled gE-specific MAb 3114 and gI-specific MAb 3104 (Anti-gE/gI) or gD precipitated with anti-gD MAb DL6 (Anti-gD), as indicated. Immunoprecipitated proteins were probed using antibodies specific to VP22. (B) Approximately 5% of the cell extract was subjected to electrophoresis, transferred to membranes, and then blotted with anti-VP22 antibodies.
DISCUSSION
The final stages of HSV assembly begin with the targeting of certain membrane glycoproteins and tegument proteins to the TGN/endosomes. Prominent among the viral membrane proteins that accumulate in the TGN when expressed without other viral proteins are gE/gI, gB, and gM/gN. gM/gN causes gD and gH/gL to relocalize from the plasma membrane to the TGN and can relocalize certain cellular proteins to the TGN (13). HSV gE/gI and gB localize to the TGN in early phases of virus replication but then redistribute to epithelial cell surfaces and at late times to cell junctions (45, 59). The CT domains of gE/gI, gB, and gM/gN contain TGN sorting motifs that cause them to accumulate at sites of virus assembly (4, 13, 45, 58). Apparently, there are other HSV proteins that then cause the redistribution of viral proteins and cellular components of the TGN to cell surfaces at late times of infection, apparently to promote egress of assembled virions (59). The CT domains of gE/gI and gD, acting in a redundant fashion, are necessary to tether tegument-coated capsids onto TGN membranes, so that envelopment occurs (22). Tegument proteins, such as UL11, VP16, and VP22, also accumulate at TGN assembly sites, either by directly interacting with TGN sorting machinery through acidic domains or other motifs, by interacting with other tegument proteins, or by interacting with membrane proteins, e.g., gD, gE/gI, or gM/gN. Presumably, interactions between tegument proteins and the CT domains of gE and gD drive the wrapping of a virion envelope around capsids as HSV particles bud into TGN-derived vesicles. Given the requirement for gE/gI or gD in this process and the phenotypes of HSV tegument mutants, it is very likely that tegument proteins interacting with gE/gI and gD also act in a redundant fashion in secondary envelopment.
In order to try to understand this assembly better, we characterized sequences in the gE CT domain required for secondary envelopment. We predicted that gE CT sequences closer to the C terminus, which were required for HSV spread but not for TGN localization (23), might play a role in envelopment. First, we established that a mutant lacking the entire gE CT domain and gD failed to undergo secondary envelopment. Consistent with the notion that N-terminal sequences were not sufficient for envelopment, F-BAC gD−/gE470 produced largely unenveloped capsids. Thus, sequences in the gE CT that are sufficient for TGN localization cannot promote envelopment. F-BAC gD−/gE495 exhibited a substantial improvement in secondary envelopment compared with F-BAC gD−/gE470, with largely enveloped instead of unenveloped capsids. However, sequences nearer the C terminus of the CT domain also contributed significantly more enveloped virions, and fewer unenveloped capsids were observed with gE519 and with gE550 (full-length gE) than with gE495. We concluded that a region of the gE CT domain between residues 470 and 495 was important for secondary envelopment, while more-C-terminal sequences also contributed. Interestingly, the sequences important for envelopment are similar to those required for relocalization of gE/gI to cell surfaces (23, 59).
To identify tegument proteins that bound to the gE CT domain, full-length and truncated CT domains were fused onto a TAP domain and expressed using Ad vectors in HSV-infected cells. This protocol allows viral proteins to interact with TAP constructs within HSV-infected cells and has advantages over the use of glutathione S-transferase proteins added after extraction of cells (11, 28, 38) and yeast two-hybrid analyses (26, 56). TAP/gE550 bound VP22 and UL11, and much less of these tegument proteins was observed with TAP/gE470 (Fig. 5). Other tegument proteins could have been missed either because they interact with the first 25 residues of the gE CT or because interactions were disrupted with detergent. Coinciding with our observations that a region between residues 470 and 495 was important in secondary envelopment, TAP/gE495 bound more UL11 and VP22 than TAP/gE470 (Fig. 6). However, increased binding of VP22 was also observed with TAP/gE550 and TAP/gE519 compared to that observed with TAP/gE495, suggesting that the C-terminal half of the gE CT domain (residues 495 to 550) contributes to this binding. Background binding of tegument proteins to TAP scrambled or where no TAP protein was expressed was substantial.
We extended the analysis of VP22 and UL11 binding to the gE CT domain with pull-down experiments involving HSV-infected cells expressing gE truncations immunoprecipitated with anti-gE MAb and blotted with anti-VP22 or anti-UL11 antibodies. There are obviously major advantages, as well as disadvantages (as discussed below), to studying these interactions in the context of HSV-infected cells where tegument and the virion envelope are formed. gE470 and gE495 bound approximately as much VP22 and UL11 as did gE448, which lacks all but three residues of the gE CT domain. Substantially more UL11 was observed with gE519 than with full-length gE470 and gE495. VP22 binding was highest with gE550 and lower with all the other truncation mutants. Thus, in these experiments, the binding of VP22 and UL11 to gE was specific but depended primarily on the more-C-terminal residues (amino acids 495 to 550). It is important to note that these residues contribute significantly to secondary envelopment. Thus, both assays for binding of tegument protein to the gE CT domain pointed to more-C-terminal residues, TAP assays pointed to residues 470 to 495, and pull-down assays pointed to residues 495 to 550. These differences may relate to differences in subcellular localization; gE/gI is membrane associated and incorporated into virions, while this is unlikely to be the case with TAP fusion proteins. Furthermore, gE interacts with gI, which contributes to secondary envelopment (22) and may influence binding of tegument proteins, and it is unlikely that TAP/gE proteins form a complex with gI (12). Whatever the origin of these differences in the two assays, it is clear that the C-terminal sequences of the gE CT domain are critical for both secondary envelopment and binding VP22 and UL11.
We also precipitated VP22 and UL11 in conjunction with gD from HSV-infected cells. These results supported and extended previous experiments in which VP22 was immunoprecipitated from HSV-infected cells with gD (11). gD appeared to bind more of both the VP22 and the UL11 proteins than gE/gI. Of course, there may be more gD in HSV-infected cells, the affinities of anti-gD and anti-gE MAb may differ, and detergents may affect these interactions differently, so this comparison may not accurately reflect the situation in cells. Compared with what was found for wild-type gD, less (≈25%) VP22 was precipitated with gD lacking the CT domain and ≈11% VP22 was precipitated with a gD-null mutant. Thus, there was clearly specific binding of VP22 that required the CT domain of gD. UL11 also bound to gD, although as much as 75% of the binding observed with wild-type gD was observed with the gD mutant lacking the CT domain. The amount of UL11 observed when gD was not present in extracts was much smaller. We concluded that both VP22 and UL11 can bind specifically to the CT domain of gD, but again, there was a substantial nonspecific component in these assays. Consistent with the notion that there was specificity for the binding of VP22 and UL11 to gE and gD, other viral proteins (VP1/2, UL24, and vhs [UL41]) were partially solubilized but did not substantially bind to either gE/gI or gD in parallel pull-down experiments.
It was important to study tegument/glycoprotein interactions in HSV-infected cells, but one of the principal disadvantages was that tegument proteins form tegument, a complex lattice of viral proteins. This can lead to two related problems in measuring the binding of tegument proteins to other viral proteins. First, tegument proteins can be largely insoluble and, indeed, UL11, VP22, and VP16 tegument proteins (65 to 75%) were insoluble (pelleted at 60,000 × g) with either 0.5% NP-40 or 1% digitonin buffer. Stronger detergents or salts would be expected to perturb protein-protein interactions. However, it is crucial to note that the soluble fractions of VP22, UL11, and VP16 did not aggregate and pellet during the course of our pull-down assays. Extracts were centrifuged at high speed, not frozen, and immediately precleared by incubation with Sepharose beads, a process involving low-speed centrifugation followed by incubation with antibody and protein A-Sepharose and then centrifugation at low speed (200 to 500 × g) for ≈30 s. Therefore, the fractions of UL11 and VP22 that were solubilized with NP-40 or digitonin remained soluble during our assays and nonspecific binding cannot be explained by insolubility.
Second, assembly into tegument probably increases the capacity of these proteins to stick to other viral proteins. Tegument is a matrix of numerous proteins that function to bridge capsids onto the envelope, favoring extensive protein-protein interactions. The relative amounts of nonspecific binding of VP22 and UL11 to gE and gD, or to IgG present in our assays, were often substantial, especially with extracts of HSV-infected cells. Based on our observations with gE and gD lacking the CT domains, which bound 20 to 75% of the VP22 and UL11 observed with wild-type gE or gD, there was apparently binding to the extracellular domains of gE/gI and gD. It is difficult to see how this could occur in vivo. More likely, this binding occurred after detergent extraction. Often, these types of controls involving tailless glycoproteins or other mutants have not been used. It is possible that UL11 and VP22 form higher-order structures, perhaps involving other tegument proteins in HSV-infected cells, and it is this material (still soluble after centrifugation at 60,000 × g) that interacts with glycoproteins. Such structures would likely be different in transfected cells or cells transduced with virus vectors. Our results, including those for VP16, provide a warning that efforts to characterize interactions between tegument proteins and other viral proteins require extreme caution and numerous controls.
One might argue that two- to threefold-increased binding of tegument proteins to gD or gE/gI compared with levels of binding to tailless glycoproteins does not represent a significant interaction. However, these observations can be viewed in two ways. First, the results underscore the notion that these tegument proteins are very sticky, i.e., readily adherent onto other viral or cellular proteins. This fits with the function of these proteins and their ability to form extensive protein-protein interactions during assembly. Second, our in vitro assays involving detergent solubilization of infected cells may not accurately predict the in vivo affinities of gD and gE/gI for VP22 and UL11. It is likely that there are very numerous interactions between VP22, UL11, gE/gI, and gD during the assembly of the virion envelope, so individual protein-protein interactions (as might be observed in detergents) may be of low affinity.
Nevertheless, our studies also strongly support the conclusion that VP22 and UL11 bind specifically onto the CT domains of gE and gD. These interactions were observed in three different assays, involving (i) TAP-tagged constructs, (ii) HSV-infected cells, and (iii) the use of Ad vectors to express HSV proteins. Binding of VP22 to gD was the better example of tegument/glycoprotein interaction. However, binding of VP22 and UL11 to gE/gI, although perhaps weaker or less extensive than with gD, should not be dismissed. Either gD or gE/gI can suffice in infected cells to allow the production of near-wild-type levels of assembly.
gD and gE are apparently the major membrane proteins that link the envelope onto tegument-coated capsids. Thus, the interactions described here further support the notion that VP22 and UL11 function in an important, but redundant, fashion in secondary envelopment. VP22 is highly conserved among alphaherpesviruses although not among other herpesvirus families. Although VP22-null mutants produce relatively normal numbers of enveloped virions, there is reduced movement of virus particles into extracellular compartments and reduced virus spread in cultured cells and in the cornea (18, 19). A significant point, related to our observations, is that VP22− mutants exhibit defects in the incorporation of gB, gD, and gE into virions. Moreover, a PRV gE/gI/gM-null mutant failed to incorporate VP22 into virions (26). Together, these observations provide strong support for the hypothesis that VP22 interacts with HSV gD and gE/gI to promote secondary envelopment. However, enveloped virions are produced in normal quantities with VP22− mutants (18, 19), which is likely related to the involvement of another tegument, as has been observed with gD and gE/gI. Our studies support the hypothesis that UL11 contributes to secondary envelopment. HSV and PRV UL11 mutants display defects in secondary envelopment (2, 40), although these defects are not as profound as those of gE−/gD− HSV. A human cytomegalovirus mutant lacking the UL11 homologue UL99 produces few or no enveloped particles (53), suggesting that UL11 may be more important in other herpesvirus families. To date, there are no reports of HSV VP22−/UL11− double mutants to test the hypothesis that these two proteins function in a redundant fashion to bridge capsids onto the envelope. Other tegument proteins may also bind to gE and gD, and there is the potential that other HSV membrane proteins may be involved in secondary envelopment.
Acknowledgments
We are grateful to Michael Webb of the OHSU Electron Microscopy Core for technical assistance and to Tiffani Howard for preparing graphics and cartoons. We thank John Wills and Gillian Elliott for antibodies and Dan Streblow for help in constructing Ad vectors. We are also indebted to Henry Mattison for his patience throughout this work.
These studies were supported by grants from the National Institute of Allergy and Infectious Diseases (AI-73996) and the National Eye Institute (EY11245).
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, J. M., Z. Moldoveanu, L. R. Melsen, P. A. Kozlowski, S. Jackson, M. J. Mulligan, J. F. Mestecky, and R. W. Compans. 1995. A polarized human endometrial cell line that binds and transports polymeric IgA. In Vitro Cell. Dev. Biol. Anim. 31:196-206. [DOI] [PubMed] [Google Scholar]
- 4.Beitia Ortiz de Zarate, I., K. Kaelin, and F. Rozenberg. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 78:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browne, H., S. Bell, and T. Minson. 2004. Analysis of the requirement for glycoprotein m in herpes simplex virus type 1 morphogenesis. J. Virol. 78:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunetti, C. R., K. S. Dingwell, C. Wale, F. L. Graham, and D. C. Johnson. 1998. Herpes simplex virus gD and virions accumulate in endosomes by mannose 6-phosphate-dependent and -independent mechanisms. J. Virol. 72:3330-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 12.Collins, W. J., and D. C. Johnson. 2003. Herpes simplex virus gE/gI expressed in epithelial cells interferes with cell-to-cell spread. J. Virol. 77:2686-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwell, K. S., L. C. Doering, and D. C. Johnson. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy, C., J. H. Lavail, A. N. Tauscher, E. G. Wills, J. A. Blaho, and J. D. Baines. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 80:8664-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 22.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farnsworth, A., and D. C. Johnson. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 64:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 29.Hardy, S., M. Kitamura, T. Harris-Stansil, Y. Dai, and M. L. Phipps. 1997. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde, N. R., C. Dunn, D. M. Lewinsohn, M. A. Jarvis, J. A. Nelson, and D. C. Johnson. 2005. Endogenous human cytomegalovirus gB is presented efficiently by MHC class II molecules to CD4+ CTL. J. Exp. Med. 202:1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, D. C., and V. Feenstra. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 61:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, D. C., M. C. Frame, M. W. Ligas, A. M. Cross, and N. D. Stow. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, D. C., M. Wittels, and P. G. Spear. 1984. Binding to cells of virosomes containing herpes simplex virus type 1 glycoproteins and evidence for fusion. J. Virol. 52:238-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamen, D. E., S. T. Gross, M. E. Girvin, and D. W. Wilson. 2005. Structural basis for the physiological temperature dependence of the association of VP16 with the cytoplasmic tail of herpes simplex virus glycoprotein H. J. Virol. 79:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 44.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 45.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 47.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mettenleiter, T. C., T. Minson, and P. Wild. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng, P., R. J. Parks, D. T. Cummings, C. M. Evelegh, U. Sankar, and F. L. Graham. 1999. A high-efficiency Cre/loxP-based system for construction of adenoviral vectors. Hum. Gene Ther. 10:2667-2672. [DOI] [PubMed] [Google Scholar]
- 51.Rauch, D. A., N. Rodriguez, and R. J. Roller. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J. Virol. 74:11437-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 53.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomazin, R., J. Boname, N. R. Hegde, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1043. [DOI] [PubMed] [Google Scholar]
- 56.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wisner, T., C. Brunetti, K. Dingwell, and D. C. Johnson. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wisner, T. W., and D. C. Johnson. 2004. Redistribution of cellular and herpes simplex virus proteins from the trans-Golgi network to cell junctions without enveloped capsids. J. Virol. 78:11519-11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]