Abstract
The four encapsidated RNAs of brome mosaic virus (BMV; B1, B2, B3, and B4) contain a highly conserved 3′ 200-nucleotide (nt) region encompassing the tRNA-like structure (TLS) which is required for packaging in vitro (Y. G. Choi, T. W. Dreher, and A. L. N. Rao, Proc. Natl. Acad. Sci. USA 99:655-660, 2002). To validate these observations in vivo, we performed packaging assays using Agrobacterium-mediated transient expression of RNAs and coat protein (CP) (P. Annamalai and A. L. N. Rao, Virology 338:96-111, 2005). Coexpression of TLS-less constructs of B1 or B2 or B3 and CP mRNAs in Nicotiana benthamiana leaves resulted in packaging of TLS-less B1 and B2 but not B3, suggesting that packaging of B3 requires the TLS in cis. This conjecture was confirmed by the efficient packaging of a B3 chimera in which the viral TLS was replaced with a cellular tRNATyr. When N. benthamiana leaves were infiltrated with a mixture of transformants containing wild-type B1 (wtB1) plus wtB2 plus a TLS-less B3 (wtB1+wtB2+TLS-lessB3), the 3′ end of progeny B3 was restored by heterologous recombination with that of either B1 or B2. This intrinsic cis-requirement of TLS in promoting B3 packaging was further confirmed when a mixture containing agrotransformants of TLS-less B1+B2+B3 was supplemented with either wtB4 or a 3′ 200-nt or 3′ 336-nt untranslated region (UTR) of B3. Northern blot analysis followed by sequencing of B3 progeny revealed that replication of TLS-less B3, but not TLS-less B1 or B2, was fully restored due to recombination with TLS from transiently expressed wtB4 or the B3 3′ UTR. Collectively, these observations suggested that the requirement of a cis-acting TLS is distinct for B3 compared with B1 or B2.
Purified virions of brome mosaic virus (BMV), the prototype of the plant virus family Bromoviridae (38), contain four single-stand, positive-sense RNAs. Among these, the largest three RNAs are genomic (gRNA) and the fourth is subgenomic (sgRNA). BMV replication is dependent on efficient interaction between two nonstructural proteins, 1a and 2a, encoded by monocistronic gRNA1 (B1) and gRNA2 (B2), respectively (28). Nonstructural protein 1a (109 kDa) contains a methyltransferase/guanyltransferase domain in its N-terminal half and a helicase domain in its C-terminal half, while nonstructural protein 2a (94 kDa) contains a central polymerase-like domain (28). The third gRNA3 (B3) encodes a 5′ nonstructural movement protein (MP) of 32 kDa and a 3′ capsid protein (CP) of 20 kDa. Only the 5′ proximal MP gene, not the 3′ proximal CP gene, is translated directly from B3. Instead, the CP is translated from an sgRNA (sgB4), which is synthesized by internal initiation on minus-strand progeny of B3 (31). The two gene products encoded by the dicistronic B3 are dispensable for viral replication but are required for infection of whole plants (33, 47). All four RNAs contain a highly conserved 3′ 200-nucleotide (nt) sequence and assume a tRNA-like structure (TLS) (19, 21). The TLS has been shown to be multifunctional since it not only harbors recognition signals for viral replicase to initiate minus-strand synthesis (20) but also mimics several tRNA-associated activities such as aminoacylation or interaction with nucleotidyltransferase (21). In addition, the TLS also serves as a 3′ telomere by recruiting the tRNA-specific host CCA nucleotidyltransferase to maintain intact 3′ CCA termini (27, 41).
BMV is a T=3 icosahedral virus (29) composed of 180 identical protein subunits (29, 49). Based on physical and biochemical analysis of BMV virions, it was hypothesized that genomic B1 and B2 are packaged independently into two virions whereas genomic B3 and the sgB4 are copackaged into a third virion (39). These three virion populations are physically and morphologically indistinguishable. The mechanism by which the CPs regulate this balanced distribution of four BMV RNAs into three individual virions remains elusive. However, the ability to reassemble virions in vitro from the dissociated CP and RNA allowed the identification of RNA sequences required for packaging. A series of in vitro assembly assays performed in our laboratory (13) demonstrated (i) that BMV genomic and sgRNAs lacking the 3′ TLS failed to assemble in vitro into virions and this defective assembly could be rescued by the addition of a 3′ 201-nt sequence encompassing the viral TLS to the assembly mixture; (ii) that tRNAs of wheat germ and yeast supplied in trans were similarly active in promoting the assembly of truncated BMV RNAs into virions; and (iii) that virions assembled from truncated BMV RNAs in the presence of cellular tRNAs or viral TLS did not incorporate the latter molecules. Based on these observations, it was hypothesized that the highly conserved 3′ TLS serves as a chaperone, in a transient association with virion CP, functioning as nucleating element (NE) to initiate the assembly of viral RNA into BMV virions.
In order to validate these in vitro findings in vivo and to analyze the extent to which the tRNAs of cellular and viral origin contribute toward the packaging of the four BMV RNAs, in this study we employed an efficient in vivo transient expression system that has proven to be ideal for dissecting replication from packaging (5). The results of this study indicated that a cis-acting TLS element of viral origin is obligatory for packaging B3 but not B1 or B2. The importance of this differential requirement by the tRNA elements in RNA packaging is discussed.
MATERIALS AND METHODS
Wild-type BMV transformants for agroinfiltration.
The construction and characterization of transfer DNA (T-DNA)-based plasmids for transient expression of full-length BMV genomic RNAs 1 (35S-B1), 2 (35S-B2), and 3 (35S-B3) and the CP mRNA (35S-B4.1) were described previously (5). Each plasmid contains, in sequential order, a double 35S promoter, cDNA complementary to the respective full-length BMV RNAs, ribozyme (RZ) sequence, and 35S terminator (Fig. 1A).
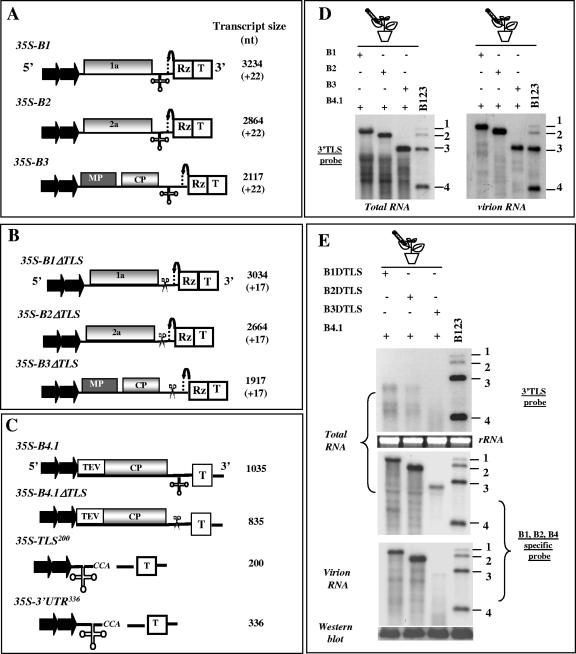
FIG. 1.
(A) Characteristics of T-DNA plasmids (agrotransformants) harboring BMV genomic RNAs used for Agrobacterium-mediated transient expression in plants. The 35S-B1, 35S-B2, and 35S-B3 constructs contain full-length cDNA copies of BMV genomic RNAs 1 (B1), 2 (B2), and 3 (B3), respectively (5). Filled arrows at the 5′ end represent the double 35S promoter (35S) whereas open boxes at the 3′ indicating RZ and T, indicate, respectively, the ribozyme sequence cassette derived from satellite tobacco ring spot virus and the 35S-polyadenylation terminator signals. The bent arrow represents the ribozyme cleavage site. The lengths of wt BMV RNAs and the number of nonviral nucleotides left after self-cleavage by ribozyme (shown in bracket) are indicated. (B) Characteristics of agrotransformants of BMV genomic RNAs lacking the 3′ TLS (ΔTLS). All other features are same those described for panel A. (C) Characteristic features of sgB4 agrotransformants of wild-type (35S-B4.1) and TLS-less (35S-B4.1/ΔTLS) constructs used for transient expression studies. Agrotransformants 35S-B3/TLS and 35S-B3/3′UTR were designed, respectively, to express either the 3′ 200-nt region or the entire 3′ UTR. All other features are same those described for panel A. (D) Transient expression and packaging in plants of individual BMV RNAs. N. benthamiana leaves were infiltrated with indicated Agrobacterium cultures, and the total and virion RNAs recovered 4 days postinfiltration were subjected to Northern blot hybridization. Approximately 5 μg of total nucleic acid preparations from agroinfiltrated leaves or 0.5 μg of virion RNA was denatured with formamide-formaldehyde and subjected to 1.2% agarose electrophoresis prior to vacuum blotting to a nylon membrane. The blot was then hybridized with 32P-labeled riboprobes complementary to the homologous 3′ TLS present on all four BMV RNAs (3′ TLS probe). The position of all four BMV RNAs is shown to the right of each panel. Marker lane B123 represents wt BMV virion RNA recovered from leaves infiltrated with a mixture containing all three wt BMV RNAs. (E) Transient expression and packaging of TLS-less BMV RNAs. The conditions of agroinfiltration and Northern blot analysis of total and virion RNAs are as described above. The blots were hybridized with either a 32P-labeled 3′ TLS probe or a mixture of riboprobes specific for each of the three genomic RNAs (B1-, B2-, and B4-specific RNAs) (15). Note that since the entire sequence of B4 is present in the 3′ half of B3, riboprobe complementary to the CP ORF sequences (referred to as B4 probe) would detect both B3 and B4. Conditions of hybridization are as described previously (5). The position of four BMV RNAs is shown to the right. wt BMV virion RNA was used as a size marker. For Western blot analysis, total protein extracts from leaves agroinfiltrated independently with the indicated mixture of agrotransformants were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane, and probed with antibodies prepared against purified BMV (35).
Construction of TLS-less BMV genomic and sgRNA agrotransformants.
To construct TLS-less agrotransformants of B1 (B1/ΔTLS), B2 (B2/ΔTLS), and B3 (B3/ΔTLS) (Fig. 1B), sequence cassettes lacking the 3′ TLS sequence were amplified by PCR with the 5′ primers d(CGACTCACTGCAGTAGACCACGGAACGAGG) (PstI site is underlined), d(CGACTCACTTACGTAAACCACGGAACGAGG) (SnaBI site is underlined), and d(CGACTCACTTACGTAAAATACCAACTAATT) (SnaBI site is underlined), respectively, for B1, B2, and B3, and the 3′ reverse primer d(GTTATAGCACGGATCCAGCTAGTCTTG) (BamHI site is underlined) for B1 and d(TATCAGTTATTGTACGGATCCAACAAGC) (BamHI site is underlined) for B2 and B3. The resulting products were digested with either PstI (for B1) or SnaB1 (for B2 and B3), treated with T4 DNA polymerase to create blunt-ended products, digested with a commonly shared BamHI, and finally subcloned into pCASS4-RZ vector as SnaBI-BamHI fragments (5). For constructing TLS-less B4.1 (B4.1/ΔTLS), a 200-nt region (encompassing the 3′ TLS) located between restriction sites HindIII and BamHI was excised by digesting the 35S-B4.1 (5) with these two enzymes, and the gel-purified largest fragment was blunt-ended with T4 DNA polymerase, ligated, and transformed. The resulting construct 35S-B4.1ΔTLS contains, in sequential order, a double 35S promoter, tobacco etch virus enhancer element, BMV RNA4 sequence, and a 35S terminator (Fig. 1C).
Construction of plasmid 35S-B4δ.
A variant of pCASS4-RZ (5) was constructed to substitute the tobacco ringspot ribozyme with hepatitis delta virus ribozyme in PCR using a forward primer d(AGGGAATTCGGTACCCCATGGGGTCGGCATGGCATCTC) (boldface and underline sequences, respectively, represent KpnI and NcoI sites) and reverse primer d(GCTCGGTACGAGCTCTCCTGGCTCTCCCTTAGC) (SacI site is underlined). The resulting fragment was digested with KpnI and SacI and ligated into similarly treated pCASS4-RZ, yielding pCASS4-HDVδ. Full-length wild-type (wt) B4 sequence was amplified in a PCR using a forward primer d(TGTCCTAATTCTACGTATTAATAATGTC) (SnaBI is underlined) and reverse primer d(ACACACACCATATGGTCTCTTTTAGAG) (NdeI site is underlined). The resulting product was digested with NdeI and blunt ended with mung bean nuclease, followed by digestion with SnaBI, and it was then subcloned into pCASS4-HDVδ previously digested with StuI/NcoI and blunt ended with mung bean nuclease.
Construction of plasmid 35S-TLS200 and 35S-UTR336.
Plasmid 35S-B4.1 was digested with either HindIII/BamHI to release the 3′ 200-nt region encompassing the TLS region or StuI/BamHI to release the 3′ 336-nt region encompassing the entire untranslated region (UTR). Each fragment was gel purified, blunt ended with T4 DNA polymerase, and ligated into similarly treated pCASS4-RZ vector as described above.
Construction of plasmid 35S-B3Tyr.
To replace the 3′ TLS with cellular tRNATyr in B3, a two-step PCR procedure was used. In the first-round PCR, 3′ TLS (200 nt) was precisely replaced with a 68-nt sequence complementary to cellular tRNATyr by amplifying a 585-nt fragment using the 5′ primer d(TGGGAGGCGTCTTCGGAC) (located between nucleotide positions 1410 to 1427 of the CP open reading frame [ORF]) and a 3′ reverse primer d(TTTTGGATCCTGGTCCGACCTACCGGATTCGAACCAGTGACCTAAGGATCTACAGTCCTCCGCTCTACCAACTGAGCTAAGGTCGGTGTCAACCACGACTATCAGT) (BamHI site is underlined and italicized bases denote sequence complementary to the CP ORF sequence located between positions 1941 to 1961, and other bases represent a complementary sequence to cellular tRNATyr). During a second-round PCR this 585-nt fragment and the cDNA of wt B3 clone (pT7B3) (22) were used as templates along with a forward 5′ primer for B3, d(CGACTCACTTACGTAAAATACCAACTAATT) (SnaBI site is underlined), and the 3′ reverse primer shown above. The resulting 1,985-nt fragment was digested with SnaBI and BamHI and ligated into StuI/BamHI-treated pCASS4-RZ vector (5). The authenticity of all genetically engineered clones was confirmed by restriction analysis followed by sequencing.
Agroinfiltration.
The agroinfiltration procedure was performed as previously described (2). Briefly, Agrobacterium strain EHA105 containing the desired transformant was initially streaked on an LB plate containing the antibiotics kanamycin (50 μg/ml) and rifampin (10 μg/ml) and incubated at 28°C. A single colony was inoculated into a 2 ml of LB medium with the above antibiotics and grown at 28°C for 48 h with vigorous shaking. One milliliter of the culture was transferred to 50 ml of LB medium containing the above antibiotics, 10 mM MES (morpholineethanesulfonic acid), pH 5.6, and 40 μM acetosyringone. After incubation at 28°C for 16 h with vigorous shaking, the optical density at 600 nm of the culture reached 1.0. The bacteria were spun down at 2,000 × g for 10 min, the pellet was resuspended in 50 ml of 10 mM MgCl2, and then 125 μl of 100 mM acetosyringone was added. To maintain a uniform concentration of Agrobacterium in coinfiltration experiments involving two or more Agrobacterium cultures, the optical density at 600 nm of each culture was adjusted to 1.0, and equal volumes of desired bacterial cultures were mixed. The mixture was kept at room temperature for at least 3 h without shaking. These mixtures were infiltrated into the abaxial surface of the fully expanded Nicotiana benthamiana leaves using a 1-ml syringe without a needle.
Progeny analysis and packaging assays.
Total RNA from agroinfiltrated leaves was extracted using the hot phenol method (5), and the RNA pellet was suspended in RNase-free water. RNase-resistant virions were purified from agroinfiltrated leaves as described previously (42). For Northern blot analysis, samples of virion RNA (0.5 μg) or plant total RNA (5 μg) were dried in a microcentrifuge tube, suspended in 10 μl of sample buffer (10× MOPS [morpholinepropanesulfonic acid] buffer-formamide-formaldehyde-H2O in a ratio of 1:1.8:5:2.2, respectively), heated at 65°C for 10 min, and electrophoresed in 1.2% agarose-formaldehyde gel (46). In all Northern blots, total RNA and virion RNA recovered from leaves infiltrated with a mixture containing agrotransformants of all three wt BMV RNAs (referred to as B123) were used as markers. Following a 3-h electrophoresis, fractionated RNA was transferred to a nylon membrane with a VacuGene XL blotting unit (Pharmacia Biotech). The blot was then processed for prehybridization and hybridization using 32P-labeled riboprobes corresponding to either the 3′ conserved region (41), the MP ORF, the CP ORF, or sequences specific for B1 and B2 as described previously (14). CP samples were analyzed by Western blotting as described previously (35). Reverse transcription-PCR (RT-PCR) was performed using either virion or total RNA progeny recovered from several independently infiltrated leaf samples and the desired set of forward and reverse primers complementary to the B3 sequence using an AccessQuick RT-PCR system according to the manufacturer instructions (Promega, Wisconsin), and the product was sequenced.
RESULTS
In vivo packaging of TLS-less BMV genomic RNAs.
In the absence of sustained replication, mechanical inoculations of individual BMV gRNA transcripts are hard to detect, mainly due to our inability to control the amount of inoculum that enters each cell. T-DNA vectors harboring viral genes were delivered into plant cells via Agrobacterium and remained transiently in the nucleus, leading to high-level expression of mRNAs for several days even in the absence of sustained replication (5, 26). Furthermore agroinfiltration offers synchronized delivery of multiple transformants to the same cell, a trait particularly useful for dissecting replication from packaging (5, 30). Consequently this approach allowed us to demonstrate that coexpression of CP mRNA and either B1, B2, or B3 in a replication-independent mode resulted in the assembly of RNA containing virions from transiently expressed CP subunits (5). Furthermore, we also demonstrated that transiently expressed B1ΔTLS and B2ΔTLS RNAs are competent for efficient packaging by the CP subunits translated from transiently expressed CP mRNA (5). Since packaging in vitro of B1 and B2 requires the TLS or cellular tRNAs functioning as nucleating agents (13), the efficient packaging of B1ΔTLS and B2ΔTLS in these in vivo assays can be attributed to trans complementation of the required function by either B4 TLS or cellular tRNAs (5).
To further unravel this issue and to compare the relative packaging efficiencies of B3 and B3ΔTLS, N. benthamiana leaves were coinfiltrated with Agrobacterium transformants of B4.1 and either B1ΔTLS, B2ΔTLS, or B3ΔTLS (Fig. 1B). Plants infiltrated with B4.1 and either B1, B2, or B3 served as controls. Total and virion RNA preparations isolated from infiltrated leaves were subjected to Northern hybridization. Efficient packaging into virions of individual wt BMV RNAs by the transiently expressed CP was observed in leaves infiltrated with control transformants (Fig. 1D). Similar analysis of leaves agroinfiltrated with transformants of B1ΔTLS, B2ΔTLS, or B3ΔTLS resulted in efficient expression and accumulation of the respective truncated RNAs, and the absence of the 3′ TLS in each RNA component was confirmed by hybridization with probes specific for B1, B2, or B3 but not with the 3′ TLS probe (Fig. 1E, top and middle blots). Despite efficient levels of CP expression in each sample (refer to Western blot shown in Fig. 1E), Northern blot analysis of virion RNA showed detectable levels of B1ΔTLS and B2ΔTLS but not B3ΔTLS (Fig. 1E, bottom blot).
Previous in vitro RNA packaging assays demonstrated that efficient assembly of BMV gRNAs into virions requires the 3′ TLS and that the defective packaging exhibited by TLS-less BMV gRNA can be complemented in trans by tRNAs of cellular origin (13). Consequently, the packaging of B1ΔTLS and B2ΔTLS observed in the above experiment as well as similar results from other studies (4, 5) could have been due to trans complementation with either cellular tRNAs or the TLS present on CP mRNA.
Effect of cis-acting cellular tRNATyr on B3 packaging.
Contrary to in vitro packaging assays where the defective packaging of B3ΔTLS could be rescued in trans by cellular tRNAs (13), transiently expressed B3ΔTLS failed to get encapsidated by the CP subunits (Fig. 1E). To verify whether B3 packaging in vivo requires the TLS in cis, a 68-nt sequence encompassing the cellular tRNATyr (Fig. 2A) was substituted for a 200-nt sequence of the 3′ TLS (Fig. 2A). Since BMV RNAs are specifically aminoacylated with tyrosine (21), cellular tRNATyr was selected. Furthermore, in order to produce B4 with authentic 5′ and 3′ ends, another agrotransformant referred to as 35S-B4δ (Fig. 1Β) was constructed. In this construct, a self-cleaving ribozyme from hepatitis delta virus (37) was engineered such that the de novo synthesized RNA transcripts terminate correctly with a natural 3′ CCA sequence. Northern blot analysis of N. benthamiana leaves infiltrated with 35S-B4δ revealed the accumulation of a single, discrete transcript of B4δ mRNA identical to wt B4 (Fig. 2C), and Western blot analysis (Fig. 2C) confirmed its ability to translate BMV CP subunits.
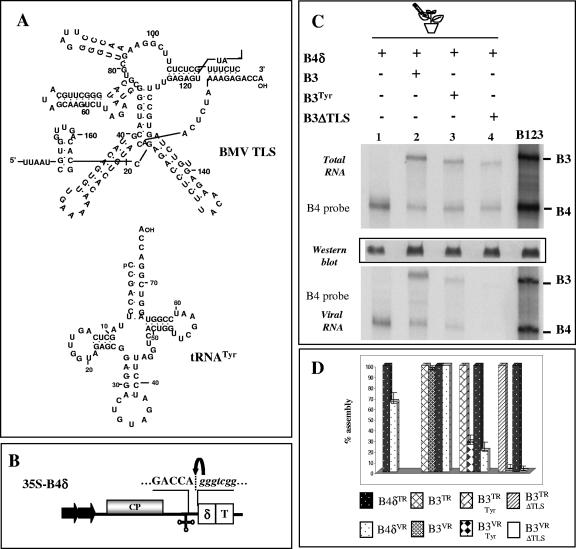
FIG. 2.
(A) Schematic representation of secondary structure of the 3′ 200-nt TLS of BMV RNA3 and 75 nt of cellular tRNATyr. (B) Characteristics of T-DNA construct of sgB4 (35S-B4δ) used for transient expression with authentic 5′ and 3′ termini. In 35S-B4δ, the position of the hepatitis delta ribozyme (δ) is shown. All other features are same those described in the legend of Fig. 1A. Viral and nonviral nucleotide sequences located at the 3′ end are shown by upper- and lowercase, respectively, and the bent arrow represents the ribozyme cleavage site. (C) Packaging competence in vivo of B3ΔTLS and B3Tyr by transiently expressed CP subunits. N. benthamiana leaves were infiltrated with indicated mixtures of Agrobacterium cultures, and the total and virion RNAs recovered were subjected to Northern hybridization with the indicated riboprobes as described in the legend of Fig. 1D. Marker lane B123 represents wt BMV virion RNA recovered from leaves infiltrated with a mixture containing all three wt BMV RNAs. Western blot analysis of viral CP was performed as described in the legend of Fig. 1D. The positions of B3 and B4 are shown to the right. (D) Packaging efficiency of B3ΔTLS and B3Tyr. The Northern blots shown in panel C were subjected to quantitation by a Typhoon 9410 PhosphoImager using wt B3 and B4 as internal standards. Histograms represent the relative accumulation of respective total RNA (TR) and virion RNA (VR) for either wt B4 (B4δ), wt B3 (B3), B3Tyr, or B3ΔTLS.
N. benthamiana leaves were infiltrated with agrobacterium cultures containing a mixture of B4δ and B3Tyr. Leaves infiltrated with B4δ or with B4δ and B3 (B4δ+B3) or B4δ+B3ΔTLS served as controls. Results of Northern blot hybridization of total and virion RNAs recovered from agroinfiltrated leaves are shown in Fig. 2C, and the relative packaging efficiency of each B3 variant is quantitatively shown in the histogram (Fig. 2D). In leaves infiltrated with either B4δ alone or B4δ+B3, most of the transiently expressed RNAs were packaged (Fig. 2C, lanes 1 and 2). However, in leaves infiltrated with B4δ+B3Tyr, packaging of B3Tyr and B4 was partially restored (Fig. 2C, lane 3). By contrast, in leaves infiltrated with B4δ+B3ΔTLS, none of the transiently expressed RNAs were found to be packaged. Collectively, these observations suggested that (i) TLS when present in cis has profound influence on packaging and (ii) efficient packaging prefers homologous TLS over cellular TLS.
Requirement of a cis-acting, homologous TLS for packaging selects for biologically active B3 recombinants.
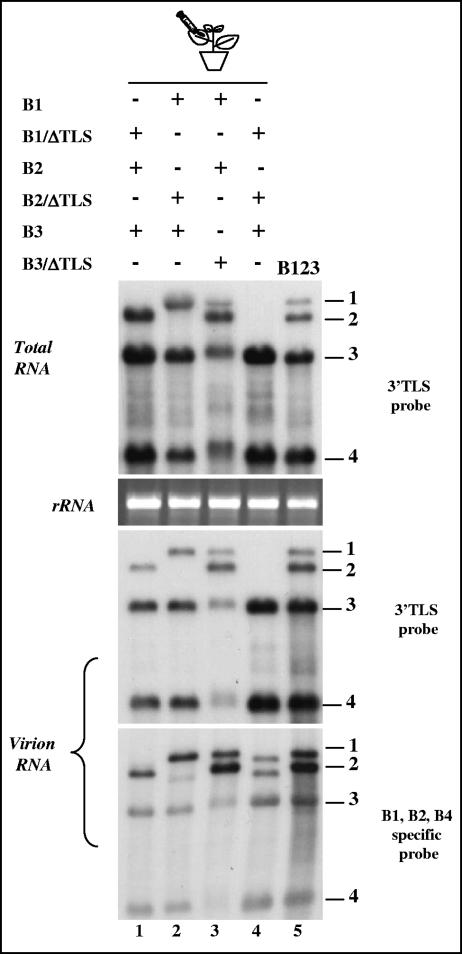
From the results of encapsidation assays of three TLS-less BMV RNAs by the CP subunits expressed in trans (Fig. 1 and 2), it is evident that, unlike B1 and B2, B3 requires a TLS in cis. To further address this issue, two independent agroinfiltration assays were performed. In the first assay, each agrotransformant of B1ΔTLS, B2ΔTLS, or B3ΔTLS was mixed with wild-type counterparts and infiltrated into N. benthamiana leaves. The results of Northern hybridization analysis are summarized in Fig. 3. In leaves infiltrated with either B1ΔTLS (Fig. 3, lanes 1 and 4), B2ΔTLS (Fig. 3, lanes 2 and 4), or B1ΔTLS and B2ΔTLS (Fig. 3, lane 4) along with the wt counterparts, the engineered deletion was conserved in each case (Fig. 3). Deletion of the TLS either in B1 or B2 could still provide a sufficient amount of viral replicase protein for the replication of B3 and transcription of B4 (Fig. 3, lanes 1, 2, and 4). By contrast, infiltration of the inoculum containing wt B1 and B2 and B3ΔTLS also resulted in the synthesis of sgB4, and restoration of the 3′ TLS in B3 was confirmed by hybridization with riboprobes specific for the 3′ region (Fig. 3, lane 3). Furthermore, the migration of the progeny B3 and sgB4 was slightly slower than the wild type. Since the 3′ TLS also functions as a minus-strand promoter (20), the origin of the TLS region of B3 must be either from B1 or B2 and involve homologous or heterologous recombination. To verify this assumption, the 3′ end of B3 progeny was subjected to RT-PCR, followed by sequencing. Two groups of recombinants were identified (Table 1). The first group was characterized by having 230 nt from B1 while the other had 306 nt from B2, based on the diagnostic nucleotide specific for each 3′ end (43). Within the 3′ 200-nt region, B3 can be distinguished from B2 by a single base substitution of G at position 43; B1 can be distinguished from B2 and B3 by 11 additional base substitutions (43). The nucleotide sequence in each BMV RNA, upstream to the 3′ TLS region, is heterologous. Since B3ΔTLS was characterized by precise deletion of a 3′ 200-nt region, recovery of B3 progeny with a 3′ sequence having an extra 30 nt and 106 nt in the first and second group of recombinants, respectively (Table 1), suggested that the restoration of the 3′ end involved heterologous recombination.
FIG. 3.
In vivo expression and packaging competence of each of the three replication-defective BMV RNAs. N. benthamiana leaves were infiltrated with indicated mixtures of Agrobacterium cultures (lanes 1 through 5), and the total and virion RNAs recovered were subjected to Northern hybridization with the indicated set of riboprobes. Marker lane B123 represents wt BMV virion RNA recovered from leaves infiltrated with a mixture containing all three wt BMV RNAs.
TABLE 1.
Characteristics of recombinant B3 progeny
| Inoculuma | No. clones sequenced | Event and comments |
|---|---|---|
| B1+B2+B3ΔTLS | 10 | Heterologous recombination. Five clones contained a 3′ 230-nt region of B1 and the remaining 5 clones contained a 3′ 306-nt region of B2. |
| B1ΔTLS+B2ΔTLS+ B3ΔTLS+B4.1 | 10 | Homologous recombination. All 10 clones contained a 3′ 191-nt region of B4.1. |
| B1ΔTLS+B2ΔTLS+B3ΔTLS + 3′ 200 nt | 12 | Homologous recombination. All 12 clones contained a 3′ 199-nt region of trans-complementing TLS. |
| B1ΔTLS+B2ΔTLS+B3ΔTLS + 3′ UTR | 12 | Homologous recombination. All 12 clones contained a 3′ 191-nt region of trans-complementing the 3′ UTR. |
| B1ΔTLS+B2ΔTLS+B3Tyr +B4.1 | 10 | Homologous recombination. All 12 clones contained a 3′ 200-nt region of B4.1. |
N. benthamiana leaves were infiltrated with the indicated mixture of agrotransformants. Four-day postinfiltration virions were purified, and B3 sequences were amplified by RT-PCR and sequenced as described in Materials and Methods.
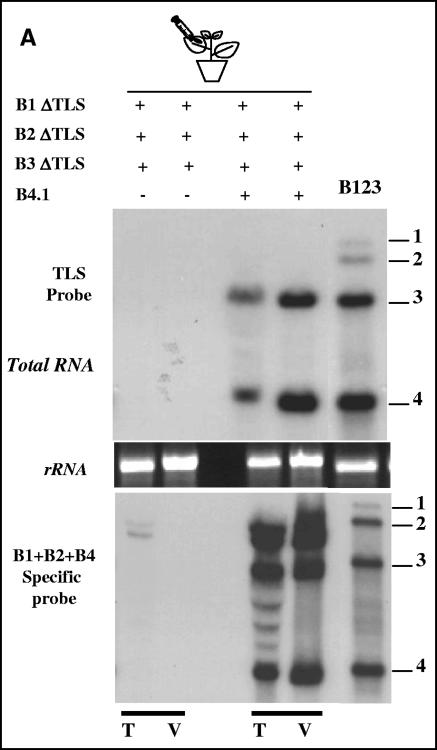
In the experiments described above B3 acquired TLS from replication-competent wt B1 and B2. To verify whether TLS provided in trans as a nonreplicated template can be used as a substrate for recombination, an inoculum containing a mixture of all three BMV RNAs lacking the 3′ TLS was infiltrated, and progeny were analyzed by Northern hybridization (Fig. 4). Hybridization with a riboprobe complementary to the 3′ TLS failed to detect any RNA, while hybridization with riboprobes specific for each of the three BMV RNAs detected, albeit at low levels, B1 and B2 but not B3 (Fig. 4). Interestingly, when an agrotransformant of sgB4 was complemented, accumulation of B3 and sgB4 RNAs in total nucleic acid preparations was evident in hybridization assays identical to those performed above (Fig. 4). These observations suggested that once again the 3′ end was restored for B3 but not for B1 or B2. Hybridization of virion RNA confirmed the encapsidation of B1ΔTLS, B2ΔTLS, and the newly generated progeny of B3 and sgB4 (Fig. 4). Unlike in the previous experiment (Fig. 4), neither B1 nor B2 had the 3′ TLS. Therefore, the only source to provide the 3′ TLS was the transiently expressed B4.1 mRNA. Sequence analysis confirmed that the progeny B3 acquired the 3′ TLS region from transiently expressed B4.1 mRNA by homologous recombination (Table 1). Since the entire sequence of sgB4 is contained within the 3′ half of gRNA3, it is difficult to determine the crossover sites involved in recombination. Agroinfiltration experiments similar to these were also performed with B3Tyr. Since B3Tyr cannot be recognized by the viral replicase, detection of sgRNA4 in leaves infiltrated with B1ΔTLS+B2ΔTLS+B3Tyr+B4.1 suggested that the 3′ end of B3Tyr was replaced by a functional 3′ TLS (data not shown). Subsequent cloning of progeny B3 and sequencing confirmed that homologous recombination with the 3′ end B4.1 (the only 3′ TLS sequences available) rendered a biologically active B3 (Table 1). Once again, because the 3′ halves B3 and B4 are identical, it is difficult to precisely map the crossover sites involved in recombination.
FIG. 4.
(A) Restoration of the deleted 3′ TLS of B3. N. benthamiana leaves were infiltrated with indicated mixtures of Agrobacterium cultures and the total (T) and virion (V) RNAs recovered were subjected to hybridization with the indicated set of riboprobes as described in the legend of Fig. 1D. Note that detection of sgB4 in total and virion RNAs indicates restoration of a functional 3′ end in B3. In each blot, marker lane B123 represents wt BMV virion RNA recovered from leaves infiltrated with a mixture containing all three wt BMV RNAs.
Transiently expressed 3′ TLS or 3′ UTR sequences serve as efficient substrates for recombination.
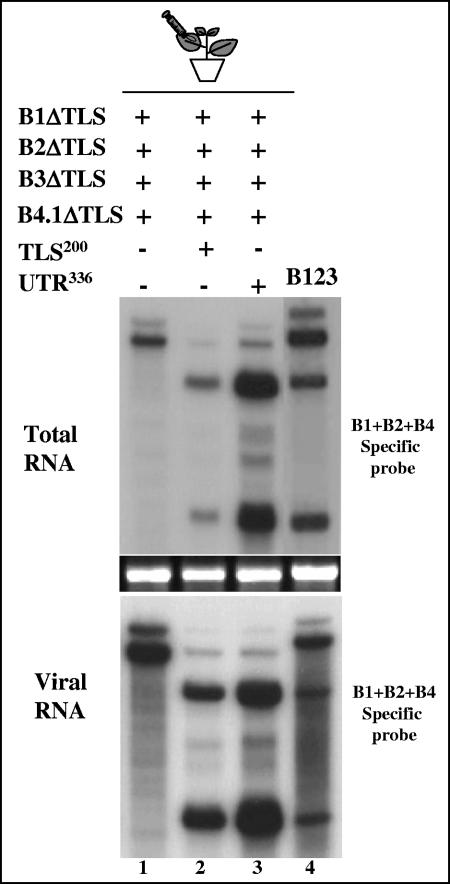
Results of the experiments presented above suggested that presence of a homologous TLS offers a convenient way to restore the biological activity of B3. However, in these experiments the homologous TLS was present either on B1 or B2 (Fig. 3) or on B4 (Fig. 4). The question that remains to be addressed is whether transiently expressed sequences encompassing only the 3′ TLS can function as efficient substrates for recombination. To address this question, two constructs amenable for transient expression of the 3′ TLS sequence were designed. Constructs 35S-TLS200 and 35S-UTR336 (Fig. 1C), respectively, direct the transcription of an RNA sequence encompassing the 3′ 200 nt and the 3′ 336 nt of the entire B3 3′ UTR.
A mixture of agrotransformants containing all three BMV RNAs lacking the 3′ TLS were complemented with either TLS200 or UTR336, infiltrated into the leaves of N. benthamiana, and the progeny in total and virion RNA preparations was analyzed by Northern hybridization (Fig. 5). Plants infiltrated with a mixture of all three BMV RNAs lacking the 3′ TLS served as negative controls. As observed in previous experiments (Fig. 1 to 4), the faster migration pattern of RNAs corresponding to B1 and B2 suggested that the engineered TLS deletion was conserved. By contrast, transcripts of B3 lacking the 3′ TLS were not detected. Since transcripts of B1ΔTLS and B2ΔTLS are packaged by the transiently expressed CP (Fig. 5, lane 1), they are protected from degradation by host nucleases whereas the transcripts of B3ΔTLS are not. This explains why the transcripts of B3ΔTLS are not detected (Fig. 5, lane 1). Similar observations were reported previously with nonreplicating or inefficiently replicating BMV RNA3 variants (1, 4). By contrast, efficient accumulation and detection of sgB4 in samples complemented with either TLS200 or UTR336 suggested that the 3′ end of the B3ΔTLS progeny was restored, permitting B3 to enter replication. This restoration of the cis-acting 3′ TLS allowed B3 to package efficiently (Fig. 5, lanes 2 and 3). Sequence analysis once again confirmed that transiently expressed RNAs of TLS200 or UTR336 served as substrates for viral replicase to mediate homologous recombination (Table 1).
FIG. 5.
Evidence for restoration of a functional viral TLS in B3 by recombination with transiently expressed 3′ end sequences. N. benthamiana leaves were infiltrated with the indicated mixtures of Agrobacterium cultures (lanes 1 through 4), and the total and virion RNAs recovered were subjected to Northern hybridization with a mixture of riboprobes specific for B1, B2, and B4 as described in the legend of Fig. 1D. Detection of sgB4 in total and virion RNAs indicates restoration of a functional 3′ end in B3. Note that in the first lane, the absence of the TLS in the inoculum failed to restore the 3′ end while packaging of B1ΔΤLS and B2ΔΤLS was not affected. In each blot, marker lane B123 represents wt BMV virion RNA recovered from leaves infiltrated with a mixture containing all three wt BMV RNAs.
DISCUSSION
The rationale for this study emerged from three sets of independent observations concerning BMV packaging. First, the highly conserved 3′ TLS of BMV RNA not only functions as a promoter to initiate minus-strand synthesis by viral replicase (20, 27) but is also obligatory for promoting packaging of all four BMV RNAs in vitro (13). Despite association with these two intrinsic processes, deletion of the 3′ TLS in B1 and B2 was preserved in agroinfiltrated leaves (5). Second, our recent observations that copackaging of sgB4, the mRNA for CP translation, is replication contingent implies that, unlike B1 and B2, B3 must preserve its 3′ end since synthesis of sgB4 occurs only if B3 enters the replication cycle (32). Results from a series of agroinfiltration experiments designed to clarify the discriminative role of viral TLS in this study revealed that the packaging requirement of the 3′ TLS for B3 is distinct from that of B1 and B2. Furthermore the need for a cis-acting TLS not only to function as a minus-strand promoter (20) but also to participate in packaging (16) selects for biologically functional B3 recombinants (Fig. 3 to 5). Taken together, the data indicate that a cis-acting TLS is obligatory for direct packaging of B3 but not B1 and B2.
Role of viral TLS in packaging BMV genomic RNAs.
Previous in vitro assembly assays showed that the 3′ TLS is obligatory for packaging of all four BMV RNAs, and the function can be complemented in trans either by viral TLS or cellular tRNAs (13). Despite this intrinsic requirement, TLS-less B1 and B2 are efficiently packaged in vivo (Fig. 1 and 3). This can be because of trans complementation by cellular tRNAs or because their packaging in vivo is independent of TLS. In contrast to B1 and B2, the requirement of TLS appears to be distinct for B3. A cis-acting TLS is required for B3 to perform two important roles. First, B3 functions as a minus-strand promoter. In BMV, synthesis of multifunctional CP is linked to replication of B3 since sgRNA is synthesized only from minus-strand B3 progeny by internal initiation (24, 48). Second, TLS plays an important role in RNA packaging. In vitro assembly assays performed with a series of B3 variants demonstrated (16, 17) that efficient packaging of B3 requires a bipartite signal: a 3′ TLS functioning as an NE and a 187-nt sequence located in the MP functioning as a packaging element; the function of TLS as NE can be complemented in trans (16). However, recent in vivo experimental evidence suggested that efficient packaging of BMV sgRNA is functionally coupled to replication-dependent transcription and translation of CP. Therefore, in order to perform the dual role, a strong selection exists to acquire viral TLS during replication either by homologous (9) or heterologous recombination (Table 1). These observations explain why the requirement of the 3′ TLS is distinct from that or B1 or B2.
Restoration of a defective 3′ end in BMV RNAs by homologous or heterologous recombination by a copy choice mechanism has been extensively documented (10, 11, 43, 44). Previous in vivo experiments demonstrated that a requirement for a functional 3′ end of B1 or B2 readily selected for functional recombinants (11, 40, 44, 45). By contrast, in this study restoration of a defective 3′ end by recombination was observed only for B3 and not for either B1 or B2 (Fig. 3 and Table 1). The reason for this observed variation between two independent studies is not immediately obvious. It is likely that the experimental approaches and host systems used could have a profound influence on the outcome, as suggested recently (26).
The mechanistic role of TLS in packaging of other eukaryotic RNA viruses.
The genomes of several groups of plant viruses and insect viruses of the family Tetraviridae have been shown to terminate in a TLS (19). In all these viruses the critical involvement of the TLS in replication is well established (19) while the role in packaging is not known. However, cellular tRNAs have in some instances been reported in the icosahedral capsids of RNA viruses. Two to three molecules of RNA with the properties of tRNALys, with smaller amounts of other tRNAs, were reported in the top component particles of a tymovirus eggplant mosaic virus (7). Virion preparations from other tymoviruses also contain RNAs (presumably tRNAs) capable of accepting a range of amino acids other than the valine bound by the viral RNA (50). However, it is not known in these cases whether these tRNAs are accidentally copackaged or whether their presence reflects active selection by the viral CP. Recent experimental evidence suggests that the requirement of TLS in packaging is distinct between viruses of the same taxonomic group. For example, in vitro assembly assays demonstrated that, unlike BMV, genomic RNA transcripts of another bromovirus, cowpea chlorotic mottle virus, lacking the 3′ TLS are efficiently packaged by CP subunits (3). Similarly, packaging of the genomic RNA of turnip yellow mosaic virus is independent of the 3′ TLS (12). However, in these two instances the possibility that host tRNAs function as nucleating agents of CP subunits during the assembly process cannot be excluded. Another interesting scenario with regard to the role of the TLS in packaging has been suggested for cucumber mosaic virus (CMV) belonging to Cucumovirus genus of family Bromoviridae (36). In the subgroup II of CMV, a 3′ 301-nt noncoding region encompassing the TLS is generated as a discrete RNA fragment (RNA5) during replication by an unknown mechanism (6). Since this RNA5 is efficiently packaged into virions, a potential role for its involvement in packaging has been suggested (18). However, this scenario differs from the one we have described here for BMV. Unlike CMV RNA5, trans complementing sequences encompassing the viral TLS or host tRNAs are not found in the assembled virions (13).
A scenario reminiscent of cis-acting tRNA-dependent replication and packaging was also observed in Sindbis virus (SIN) whose genome does not terminate in a TLS. However, a biologically active naturally occurring Sindbis virus defective interfering RNA (SIN DI RNA) having a cellular tRNAAsp sequence at the 5′ end has been isolated and characterized (34). Subsequent in vivo packaging studies revealed that SIN DI RNA having the 5′ tRNAAsp sequence replicated and packaged at higher levels than viruses containing the authentic 5′ terminus (8, 23, 25). Clearly, additional studies addressing whether the role of viral TLS in promoting RNA packaging is universally conserved or restricted to only some viruses such as BMV are needed to provide valuable insight into the mechanism of RNA packaging in viruses having icosahedral symmetry.
Acknowledgments
We thank Shou-Wei Ding for the generous gift of pCass4 plasmid and Melissanne de Wispelaere for helpful discussions.
Research in this laboratory was supported by a grant from National Institutes of Health (GM 064465-01A2).
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Allison, R., C. Thompson, and P. Ahlquist. 1990. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl. Acad. Sci. USA 87:1820-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annamalai, P., and A. L. Rao. 2006. Delivery and Expression of functional viral RNA genomes in planta by agroinfiltration, p. 16B.2.1-2.15. In T. Downey (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- 3.Annamalai, P., and A. L. Rao. 2005. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology 332:650-658. [DOI] [PubMed] [Google Scholar]
- 4.Annamalai, P., and A. L. Rao. 2006. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 80:10096-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96-111. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard, C. L., P. M. Boyce, and B. J. Anderson. 1996. Cucumber mosaic virus RNA 5 is a mixed population derived from the conserved 3′-terminal regions of genomic RNAs 2 and 3. Virology 217:598-601. [DOI] [PubMed] [Google Scholar]
- 7.Bouley, J., J. Briand, M. Genevaux, M. Pinck, and J. Witz. 1976. The structure of eggplant mosaic virus: evidence for the presence of low molecular weight RNA in top component. Virology 69:775-781. [DOI] [PubMed] [Google Scholar]
- 8.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujarski, J. J., and P. Kaesberg. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321:528-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bujarski, J. J., P. D. Nagy, and S. Flasinski. 1994. Molecular studies of genetic RNA-RNA recombination in brome mosaic virus. Adv. Virus Res. 43:275-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, T. J., and T. W. Dreher. 28. August 2006. Encapsidation of genomic but not subgenomic turnip yellow mosaic virus RNA by coat protein provided in trans. Virology doi: 10.1016/j.virol.2006.06.038. [DOI] [PubMed]
- 13.Choi, Y. G., T. W. Dreher, and A. L. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, Y. G., G. L. Grantham, and A. L. Rao. 2000. Molecular studies on bromovirus capsid protein. Virology 270:377-385. [DOI] [PubMed] [Google Scholar]
- 15.Choi, Y. G., and A. L. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residuals. Virology 275:207-217. [DOI] [PubMed] [Google Scholar]
- 16.Choi, Y. G., and A. L. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 77:9750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damayanti, T. A., S. Tsukaguchi, K. Mise, and T. Okuno. 2003. cis-Acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 77:9979-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies, C., and R. H. Symons. 1988. Further implications for the evolutionary relationships between tripartite plant viruses based on cucumber mosaic virus RNA 3. Virology 165:216-224. [DOI] [PubMed] [Google Scholar]
- 19.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 20.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201:31-40. [DOI] [PubMed] [Google Scholar]
- 21.Dreher, T. W., and T. C. Hall. 1988. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3′-adenylation. J. Mol. Biol. 201:41-55. [DOI] [PubMed] [Google Scholar]
- 22.Dreher, T. W., A. L. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425-438. [DOI] [PubMed] [Google Scholar]
- 23.Fayzulin, R., R. Gorchakov, O. Petrakova, E. Volkova, and I. Frolov. 2005. Sindbis virus with a tricomponent genome. J. Virol. 79:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopinath, K., B. Dragnea, and C. Kao. 2005. Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 79:14222-14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hema, M., and C. C. Kao. 2004. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 29.Lucas, R. W., S. B. Larson, and A. McPherson. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317:95-108. [DOI] [PubMed] [Google Scholar]
- 30.Marillonnet, S., A. Giritch, M. Gils, R. Kandzia, V. Klimyuk, and Y. Gleba. 2004. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 101:6852-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537-546. [DOI] [PubMed] [Google Scholar]
- 32.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 33.Mise, K., R. F. Allison, M. Janda, and P. Ahlquist. 1993. Bromovirus movement protein genes play a crucial role in host specificity. J. Virol. 67:2815-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monroe, S. S., and S. Schlesinger. 1983. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc. Natl. Acad. Sci. USA 80:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osman, F., G. L. Grantham, and A. L. Rao. 1997. Molecular studies on bromovirus capsid protein. IV. Coat protein exchanges between brome mosaic and cowpea chlorotic mottle viruses exhibit neutral effects in heterologous hosts. Virology 238:452-459. [DOI] [PubMed] [Google Scholar]
- 36.Palukaitis, P., and F. Garcia-Arenal. 2003. Cucumoviruses. Adv. Virus Res. 62:241-323. [DOI] [PubMed] [Google Scholar]
- 37.Perrotta, A. T., O. Nikiforova, and M. D. Been. 1999. A conserved bulged adenosine in a peripheral duplex of the antigenomic HDV self-cleaving RNA reduces kinetic trapping of inactive conformations. Nucleic Acids Res. 27:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao, A. L. 2001. Bromoviruses, p. 155-158. In O. C. Maloy and T. D. Murry (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Mississauga, Ontario, Canada.
- 39.Rao, A. L. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 44:61-87. [DOI] [PubMed] [Google Scholar]
- 40.Rao, A. L. 2005. Sensitivity of brome mosaic virus RNA1 replication to mutations in the 3′ tRNA-like structure implies a requirement for sustained synthesis of replicase protein 1a. Arch. Virol. 151:721-733. [DOI] [PubMed] [Google Scholar]
- 41.Rao, A. L., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao, A. L., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 43.Rao, A. L., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao, A. L., and T. C. Hall. 1990. Requirement for a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J. Virol. 64:2437-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao, A. L., B. P. Sullivan, and T. C. Hall. 1990. Use of Chenopodium hybridum facilitates isolation of brome mosaic virus RNA recombinants. J. Gen. Virol. 71:1403-1407. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. L. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Schmitz, I., and A. L. Rao. 1996. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology 226:281-293. [DOI] [PubMed] [Google Scholar]
- 48.Sivakumaran, K., S. K. Choi, M. Hema, and C. C. Kao. 2004. Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 78:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speir, J. A., S. Munshi, G. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Belkum, A., J. Bingkun, K. Rietveld, C. W. A. Pleij, and L. Bosch. 1987. Structural similarities among valine-accepting tRNA-like structures in tymoviral RNAs and elongator tRNAs. Biochemistry 26:1144-1151. [Google Scholar]