Coronaviruses are a family of enveloped, plus-stranded RNA viruses with helical nucleocapsids and extraordinarily large genomes. The hallmark of coronavirus transcription is the production of multiple subgenomic mRNAs that contain sequences corresponding to both ends of the genome. (Transcription is defined as the process whereby subgenome-sized mRNAs are produced, and replication is the process whereby genome-sized RNA, which also functions as mRNA, is produced.) Thus, the generation of subgenomic mRNAs involves a process of discontinuous transcription. The aim of this minireview is to describe our current understanding of coronavirus replication and transcription. For more detailed information, the reader is directed to other recent reviews (25, 28, 44a, 49, 63, 74, 94).
The coronavirus genomic RNA of approximately 30,000 nucleotides encodes structural proteins of the virus, nonstructural proteins that have a critical role in viral RNA synthesis (which we will refer to as replicase-transcriptase proteins), and nonstructural proteins that are nonessential for virus replication in cell culture but appear to confer a selective advantage in vivo (which we will refer to as niche-specific proteins). At least one niche-specific protein, nonstructural protein 2 (nsp2), and one structural protein, the nucleocapsid protein (N), are involved in viral RNA synthesis (1, 30, 68).
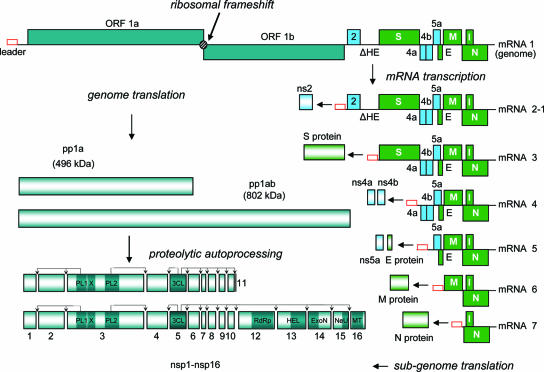
The expression of the coronavirus replicase-transcriptase protein genes is mediated by the translation of the genomic RNA. The replicase-transcriptase proteins are encoded in open-reading frame 1a (ORF1a) and ORF1b and are synthesized initially as two large polyproteins, pp1a and pp1ab. The synthesis of pp1ab involves programmed ribosomal frame shifting during translation of ORF1a (48). During or after synthesis, these polyproteins are cleaved by virus-encoded proteinases with papain-like (PLpro) and chymotrypsin-like folds into 16 proteins; nsp1 to nsp11 are encoded in ORF1a, and nsp12 to nsp16 are encoded in ORF1b (Fig. 1) (96). The replicase-transcriptase proteins, together with other viral proteins and, possibly, cellular proteins, assemble into membrane-bound replication-transcription complexes (RTC). (We will use the term RTC to describe complexes copying or producing genome- or subgenome-length RNA.) These complexes accumulate at perinuclear regions and are associated with double-membrane vesicles (15, 76). Hydrophobic transmembrane domains are present in nsp3, nsp4, and nsp6 and likely serve to anchor the nascent pp1a/pp1ab polyproteins to membranes during the first step of RTC formation.
FIG. 1.
Organization and expression of the MHV-A59 genome. The structural relationships of the MHV-A59 genome- and subgenome-length mRNAs are shown. The virus ORFs are depicted in teal (nsp1-nsp16 genes), blue (ns2, ns4a, ns4b, and ns5a genes), and green (S, M, E, N, and I structural protein genes). The ORFs are defined by the genomic sequence of MHV-A59 as published by Coley et al. (20). The open red box represents the common 59-leader sequence, and the barred circle represents the programmed (−1) frameshifting element. The translation products of the genome- and subgenome-length mRNAs are depicted, and the autoproteolytic processing of the ORF1a and ORF1a/ORF1b polyproteins into proteins nsp1 to nsp16 is shown. A number of confirmed and putative functional domains in the nsp proteins are also indicated. NeU, uridylate-specific endoribonuclease; PL1, papain-like protease 1; PL2, papain-like protease 2.
Subcellular fractionation, electron microscopic in situ hybridization, and immunofluorescence studies suggest that most, if not all, coronavirus nsp proteins are recruited to RTCs synthesizing both genome- and subgenome-length RNA (29, 32, 52, 76). At later times of infection, it appears that the nsp proteins encoded in ORF1a remain tightly bound to the RTC, while proteins encoded in ORF1b detach and diffuse to the cytosol (85). In addition to the nsp proteins, and consistent with it playing a role in RNA synthesis, the RTC contains the viral N protein (13, 85). A number of cellular proteins have been shown to interact with coronaviral RNA. These include heterogeneous nuclear ribonucleoprotein A1, polypyrimidine tract binding protein, poly(A)-binding protein, and mitochondrial aconitase (73), although there is no evidence that these proteins specifically colocalize with RTCs.
Sequence analysis of the nsp1 to -16 proteins predicts that they have at least eight enzymatic activities (75). Some of these activities, e.g., proteinase, RNA-dependent RNA polymerase (RdRp), and 5′-to-3′ helicase (HEL) activities, are common to RNA viruses, but others appear to be unique to coronaviruses or viruses closely related to them. Clearly, while most of these enzymatic functions are concerned with viral RNA synthesis, some may also have relevance to cellular processes. For example, nsp3, which contains the PLpro activity, is a deubiquitinating enzyme and is capable of reversing the conjugation of protein with ISG15 (interferon-stimulated gene 15), which may subvert cellular processes to facilitate viral replication (7, 42). Also, the ADP-ribose 1′′-phosphatase (ADRP) activity of nsp3 may act to influence levels of ADP-ribose, a key regulatory molecule in the cell.
CORONAVIRUS RNA REPLICATION AND TRANSCRIPTION
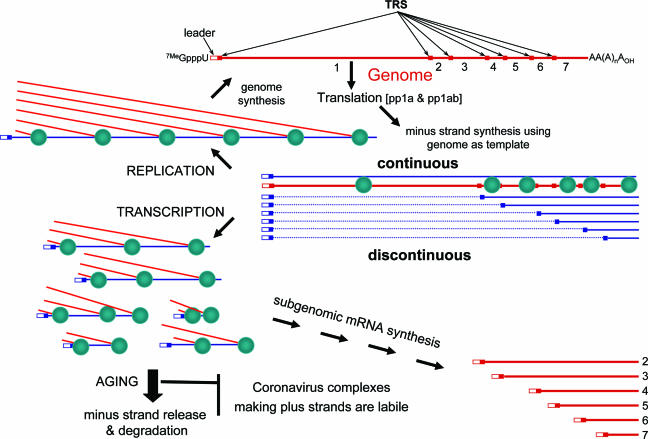
Model of discontinuous extension during subgenome-length minus-strand synthesis.
We have proposed a model of discontinuous extension during subgenome-length minus-strand synthesis to explain the generation of coronavirus mRNAs (65) (Fig. 2). Each subgenome-length mRNA contains a 5′ leader sequence corresponding to the 5′ end of the genome. This 5′ leader is joined to a mRNA “body,” which represents sequences from the 3′-poly(A) stretch to a position that is upstream of each genomic ORF encoding a structural or niche-specific protein. The junction of the leader and body elements in each mRNA can be identified by a characteristic short, AU-rich motif of about 10 nucleotides that is known as the transcription-regulating sequence (TRS). In the genome, functional TRS motifs are found at the 3′ end of the leader (leader TRS) and in front of each ORF that is destined to become 5′ proximal in one of the subgenome-length mRNAs (body TRSs).
FIG. 2.
Model for coronavirus replication-transcription. The ORF1 of genomic RNA (red) is translated to produce pp1a and pp1ab, which assemble into an RTC (teal oval) that recognizes cis-acting elements at the 5′ and 3′ ends of the genome. This RTC copies the genome either continuously into genome-length template or discontinuously into the various subgenome-length minus-strand templates. The minus strands (blue) are used as templates for genomic and subgenomic mRNA synthesis. Only genomes are used as templates for minus-strand synthesis, i.e., replication. The RTCs engaging in plus-strand synthesis age and release their minus-strand templates, which are then degraded specifically.
Our explanation of how the subgenomic mRNAs are generated has two central tenets: (i) that the process of discontinuous transcription occurs during the synthesis of minus-strand subgenome-length templates, and (ii) that the process of discontinuous transcription resembles the mechanism of similarity-assisted or high-frequency copy-choice RNA recombination.
Mechanistically, the process of discontinuous transcription during minus-strand synthesis can be viewed as a number of consecutive events. (i) The components of a functional RTC are recruited and minus-strand synthesis is initiated at the 3′ end of a genomic RNA. (ii) Elongation of nascent minus-strand RNA continues until the first functional body TRS motif is encountered. A fixed proportion of RTCs will either (iii) disregard the TRS motif and continue to elongate the nascent strand or (iv) stop synthesis of the nascent minus strand and relocate and complete its synthesis. (v) This relocation will be guided by complementarity between the 3′ end of the nascent minus strand and the leader TRS motif. The translocated minus strand will be extended by copying the 5′ end of a genome. The completed minus-strand RNA would then serve as a template for mRNA synthesis. The evidence to support this model is now extensive and has been discussed in detail in recent reviews (49, 63).
There are many unanswered questions that are central to the model.
First, we have only a vague idea of what constitutes the signal that stops, or attenuates, minus-strand synthesis at each of the body TRS motifs. Three parameters that are considered important are the stability of TRS base pairing between the template and nascent minus strand, the nature of sequences flanking the TRS motifs, and the location of the TRS motif relative to the promoter for minus-strand synthesis (21, 25, 77, 92a, 97). Also, it is often proposed that protein-to-protein interactions may be important for attenuation but, in our opinion, the lack of rigid sequence specificity in the TRS base-pairing interaction implies that protein cofactors will not be major players in TRS recognition. Rather, specific RNA motifs (cis-acting and higher-order RNA structures) and their accessibility may be of importance, along with TRS base-pairing potential. Further evidence for the extremely tight regulation of body TRS attenuation is provided by a series of experiments done with the Equine arteritis virus (EAV) system; EAV belongs to a family that is distantly related to coronaviruses, with similar, if not identical, mechanisms of subgenome-length mRNA synthesis (49). In this system, it has been shown that the functional inactivation of a 3′-proximal body TRS element by minimal mutagenesis does not, as might have been predicted, increase the frequency with which more 3′-distal body TRS elements are used (50). In other words, the TRS element still enforces attenuation, i.e., the “launching” of the RTC, even when the “landing” is prevented.
Second, there is no information on how the relocation of the 3′ end of the nascent minus strand to the 5′ end of the genomic template is mediated or how the realignment of complementary bases on the template is facilitated. One idea is that this relocation might be mediated by protein-to-protein interactions between the polymerase attached to the growing minus strand and a protein associated with the 5′ end of the genome. It also could occur by the polymerase binding directly to the sequences downstream of the leader RNA. A variation on this theme is that the 5′ end of the genomic RNA is actually bound to the polymerase attached to the growing minus strand and essentially scans the newly synthesized RNA for the synthesis of the complementary TRS sequence (25). This is an attractive idea, but it implies that, at any one time, each genomic RNA template engages in the synthesis of only one subgenome-length minus strand, and it is contradictory to the idea that each template generates concurrently a complete set of genome- and subgenome-length minus strands. The difference should be experimentally testable once sensitive methods are developed. With regard to the realignment of complementary bases on the template, it has been suggested that coronavirus polymerase might copy the template RNA in a fashion analogous to DNA-dependent RNA polymerases, where regulatory elements in the template and nascent RNA disrupt elongation (59, 89) and cause retraction at pause sites (40, 41) and where the polymerase remains associated with the growing nascent strand rather than with the template. This would endow the 3′ end of the nascent minus strand with primer capability. Of interest, a similar mechanism was recently proposed for proofreading by RNA polymerases (71), and several gene products of ORF1b of coronaviruses could plausibly have a role in this sort of process (see section below on proteins involved in coronavirus transcription).
Kinetics of plus-strand and minus-strand synthesis.
Coronaviruses resemble other plus-stranded RNA viruses and produce plus strands at 50- to 100-fold excess of their minus-strand templates. Based on studies with the A59 strain of Mouse hepatitis virus (MHV-A59), coronavirus RNA synthesis is detectable as early as 2 to 3 h postinfection when metabolic labeling ([32P]orthophosphate or [3H]uridine) is employed and by 90 min postinfection when reverse transcription-PCR is used (63; data not shown). Throughout infection, syntheses of the various species of plus strands remain constant, i.e., in a fixed molar ratio, relative to each other. In addition, the same ratio is retained under conditions where overall viral RNA synthesis declines late in infection, after recovery from the inhibition of protein synthesis by cycloheximide, or after transfer from restrictive to permissive temperatures in the case of temperature-sensitive (ts) mutants. The pattern of plus-strand synthesis reflects a basic principle of coronavirus transcription; namely, the amount of genome- and subgenome-length mRNA synthesis is determined at the level of minus-strand synthesis.
Minus-strand synthesis begins 75 to 90 min after infection. At this time, genome-length and subgenome-length minus strands and their plus-strand complements can be detected simultaneously by reverse transcription-PCR (S. Sawicki, unpublished data). Therefore, once RTCs form, they immediately start to produce minus-strand RNA. Minus strands accumulate exponentially, and their rate of synthesis peaks at 5 to 6 h postinfection; their synthesis then declines but does not cease (62). Why the assembly of RTCs is hindered beyond 6 h postinfection is not known.
Coronavirus RIs/TIs and RFs/TFs.
The templates in viral RTCs are recoverable after deproteinization of infected-cell lysates as a multistranded replication intermediate (RI) with genome-length templates or multistranded transcription intermediates (TIs) with templates corresponding in length to subgenome-length mRNAs and the similarly sized double-stranded replication form (RF) with genome-length templates and double-stranded transcription forms (TFs) with templates corresponding in length to subgenome-length mRNAs. The native RF/TF structures are essentially single-stranded, but they become largely double-stranded following deproteinization. The TIs and TFs are authentic transcription structures that are active in subgenomic mRNA synthesis (60, 64).
In MHV-infected cells, all of the RTCs making minus and plus strands are labile. The short-lived nature of MHV minus-strand-synthesizing complexes is a constant feature throughout infection. It is demonstrated by inhibiting the synthesis of nsp's with cycloheximide (62), which means that only newly made viral proteins function in minus-strand synthesis and suggests that polyprotein intermediates probably function in minus-strand synthesis. The instability of plus-strand-synthesizing RTCs is more difficult to observe but can be seen under certain conditions. For instance, at 7 to 8 h postinfection, plus-strand synthesis starts to decline, but it is difficult to study because with MHV-A59 cell fusion also occurs. However, by use of a mutant of MHV-A59 that does not cause cell fusion (61), RNA synthesis can be seen to decline to undetectable levels by 12 to 15 h postinfection. This occurs even though viral proteins, at least the structural proteins, are being produced at high levels. Another way to demonstrate the lability of plus-strand-synthesizing RTCs is to inhibit protein synthesis early in infection when plus-strand synthesis is increasing. Plus-strand synthesis stops increasing almost immediately, starts to decline an hour later, and disappears after 4 to 5 h. Under these circumstances, the synthesis of all species of viral plus strands is equally affected.
What is the basis of the instability shown by plus-strand-synthesizing RTCs? One clue is that RI/RF and TI/TF structures disappear coincidently in time with the loss of plus-strand synthesis (63). This could reflect activation of specific RNase activities targeting minus strands. Our hypothesis is that coronavirus RTCs age and lose their activity and then their minus-strand templates; the virus is then required to produce new RTCs throughout infection.
Biogenesis of minus strands.
The current model of coronavirus RNA synthesis proposes that minus strands arise from copying the genome continuously to form genome-length templates and discontinuously to form subgenome-length templates. While early experiments ruled out a role for splicing in the generation of viral subgenome-length mRNAs, it remained possible that the genome-length minus strands are subject to splicing, which would result in the formation of subgenome-length minus-strand templates with the same 5′ and 3′ ends. To test this possibility, we determined the sensitivity of MHV minus-strand synthesis to UV irradiation (data not shown). The kinetics of overall minus-strand sensitivity showed a dose dependency that was not linear and did not follow the inactivation of synthesis of any of the individual plus strands. Rather, it reflected the sum of their individual sensitivities. This result supports the argument that the subgenome-length minus-strand templates do not arise via splicing of genome-length minus strands. This conclusion is also supported by studies using replicons derived from the infectious clone of Human coronavirus, strain 229E (HCoV-229E) where, in the absence of N protein expression, the synthesis of a subgenome-length mRNA for green fluorescent protein (GFP) occurred even when there was little or no replication of the replicon-length RNA (68). Since the subgenome-length mRNA encoding GFP had a leader fused to its 5′ end and was produced in the absence of demonstrable replicon-length minus-strand synthesis, discontinuous synthesis rather than splicing was likely responsible for the generation of the minus-strand template.
Although they are central to any understanding of coronavirus transcription, fundamental issues regarding the mechanisms of the RTCs responsible for minus-strand synthesis remain to be addressed. The most obvious remaining question is the mechanism of discontinuous synthesis. However, it is also important to ask other questions. Can a single genome serve as a template for the synthesis of only one minus strand, or can it serve simultaneously for all species of minus strands? Is the ratio of each minus-strand product per genome 1 or greater than 1? We believe that the fixed ratio of different minus strands produced early and late during infection suggests that each genome serves as a template for the complete nested set of minus strands, with cis-active signals and the 3′ position relative to the genome influencing or determining the final abundance or molar ratios of each product. This prediction can be tested, however, only when a sensitive method is devised to analyze the templates and products of RTCs engaged in minus-strand synthesis.
Promoters for minus- and plus-strand synthesis.
It is difficult to reconcile experiments investigating the sequences and structures found at the 5′ and 3′ ends of coronavirus RNA with simple models for the initiation of plus- and minus-strand synthesis. Two regions of the 3′ untranslated region (UTR) have been suggested as containing cis-acting regulatory elements that play a role in coronavirus RNA synthesis. The first region of ∼150 nucleotides adjoins the poly(A) stretch and is predicted to form a number of different stem-loop structures (37, 57). One of these is known as the stem-loop II motif and has been identified in the Severe acute respiratory syndrome coronavirus (SARSCoV) genome (57). This region also contains the sequence 5′-GGAAGAGC-3′ that is present in all coronavirus genomes. The second region is upstream of the terminal 150 nucleotides and contains two structures, known as the bulged stem-loop and the hairpin-type pseudoknot. It is interesting that the pseudoknot structure involves nucleotides at the base of the bulged stem-loop structure, which means the structures are mutually exclusive. This may represent a form of “molecular switch” related in an as-yet-unknown way to different modes of RNA synthesis (27). Structures implicated in RNA synthesis have also been predicted in the 5′ UTR of the coronavirus genome (14, 54, 55). These include four higher-order structures, stem-loops I to IV, that may function as cis-acting elements in RNA replication and transcription. Stem-loops I and III demonstrate a cis-acting function in Bovine coronavirus (BCoV) defective interfering (DI) RNA replication. Stem-loop II harbors the leader TRS motif, and stem-loop IV may be involved in the initiation of minus-strand synthesis (55).
In a model analogous to the models for picornavirus replication-transcription (8), we propose that the 3′ and 5′ ends of the coronavirus genome interact, either directly (RNA to RNA) or indirectly (protein to RNA or protein to protein), to form the promoter for minus-strand synthesis. Only genomes containing a 5′ element downstream of the leader would be able to engage the 3′ end to serve as templates for minus-strand synthesis. The subgenome-length mRNAs would be missing the 5′ element (although they would all contain the 3′ element), and this would explain why they are not able to replicate (43, 54, 55). Similarly, the 3′ end of minus strands presumably must function as part of the promoter for plus-strand synthesis. Again, we would propose that the 3′ and 5′ ends of the minus strand might interact to circularize the template and create functional plus-strand promoters. DI minus strands are capable of replicating when introduced into helper virus-infected cells (38), and this indicates that minus-strand templates are capable of being recruited in trans into a functional RTC to produce plus strands. Thus, either the minus-strand templates of RTCs are released after completion of synthesis of the plus strand, allowing the RTC to initiate RNA synthesis on another minus-strand template, or newly assembled RTCs might recognize minus strands in trans and not be obliged to first create a minus-strand template by copying a genome.
PROTEINS INVOLVED IN CORONAVIRUS TRANSCRIPTION
As mentioned above, sequence analyses of the nsp proteins of MHV, SARSCoV, and other coronaviruses predict that they include domains associated with at least eight enzymatic activities (75). The enzymatic activities and characteristics of these protein domains are listed in Table 1. Crystallographic or nuclear magnetic resonance structures have been determined for the PLpro and ADRP (or X) domains of nsp3, nsp5, nsp7, nsp8, nsp9, and nsp10 (3, 39, 51, 56, 58, 82, 93). In addition, a number of other features, including domains with conserved cysteine and histidine residues (C/H domains in nsp3, nsp13, and nsp14), putative transmembrane domains (TM domains in nsp3, nsp4, and nsp6), and domains with conserved features (the A [acidic] and Y domains within nsp3) have also been identified in coronavirus nsp proteins (94).
TABLE 1.
Enzymatic activities and characteristics of coronavirus nsp protein domains
| Activity | Location and designation | Protein family | Architecture | Comments | References |
|---|---|---|---|---|---|
| Papain-like proteinase | One or two PLpro domains within nsp3 | Deubiquitinating enzyme family | Finger-palm-thumb | Fingertips contain zinc-binding domain | 5, 33, 56 |
| ADP-ribose 1"-phosphatase | ADRP (or X) domain within nsp3 | Macro-HS2A-fold family | Three-layered α/β/α | Also found in other plus-strand RNA viruses | 53, 58 |
| 3C-like cysteine proteinase | nsp5 (3CLpro or main protease, Mpro) | Two-β-barrel proteinase family | Twelve antiparallel β-strands | Extra, α-helical, carboxyl-terminal domain III | 3, 4, 95 |
| RNA-dependent RNA polymerase | RdRp domain within nsp12 | Viral RdRp family | Finger-palm-thumb (predicted) | RdRp motif VI signature GDD replaced with SDD | 19, 87 |
| 5′-to-3′ helicase (associated, NTPase, and RNA 5′-triphosphatase activities) | HEL domain within nsp13 | Superfamily 1 helicase | Modeled to Escherichia coli Rep helicase | Adjacent to amino-proximal, binuclear, zinc-binding domain | 9, 35, 36, 70 |
| 3′-to-5′ exonuclease | ExoN domain within nsp14 | DEDD superfamily | Hexahelical bundle (predicted) | Unable to cleave ribose 2′-O-methylated RNA substrates | 45, 75 |
| Uridylate-specific endoribonuclease | NendoU domain within nsp15 | XendoU family | Wing-body-wing (butterfly fold) | Active in hexameric form; RNA bound to internal cavity | 10, 11, 31, 34, 56a, 88 |
| S-adenosylmethionine-dependent 2′-O-methyltransferase | nsp16 (MT) | Rrmj methyltransferase family | Methyltransferase fold (predicted) | Activity yet to be determined | 75, 86 |
In addition to the proteins for which enzymatic functions have been predicted or demonstrated, the structures of four other coronavirus nsp proteins have been reported. The SARSCoV proteins nsp7 and nsp8 cocrystallize to produce a supercomplex that can be viewed as a hollow, cylinder-like structure assembled from eight copies of nsp8 and eight copies of nsp7. This complex has a central channel with positive electrostatic properties favorable for nucleic acid binding. It has been suggested that the role of this structure may be to confer processivity to the viral RdRp (51, 93). Second, the crystal structure of SARSCoV nsp9 has been reported as comprising a single β-barrel with a fold that resembles a carboxyl-extended oligonucleotide-oligosaccharide binding (OB) fold. The crystal structure suggests that the protein is dimeric, and gel shift assays show that it is able to bind single-stranded RNA (24, 82). And finally, recent studies have elucidated the crystal structure of the SARSCoV nsp10 protein (39). The structure predicts a single domain protein consisting of a pair of antiparallel amino-terminal helices stacked against an irregular β-sheet, together with a coil-rich carboxyl terminus and two zinc fingers. Twelve subunits assemble to form a unique dodecameric superstructure. The nsp10 protein is the first representative of a new family of zinc-finger proteins which, so far, are found exclusively in coronaviruses (39, 81).
Many review articles on coronavirus RNA synthesis include references to a nonstructural protein, ns2, which is encoded in the genome of a subset of group II coronaviruses, including MHV, BCoV, and HCoV-OC43. The ns2 gene lies immediately downstream of ORF1b, and the protein is translated from a subgenome-sized mRNA. This protein has been predicted to have cyclic phosphodiesterase (CPD) activity, although this has not yet been experimentally verified. It is tempting to envisage this activity, together with the ADRP activity of nsp3, as acting in the regulation of a pathway involving the processing of (viral) RNA by enzymes (for example, the NendoU activity of nsp15) that generate intermediates with terminal cyclic phosphate residues (78). It is worth remembering, however, that only a few coronaviruses encode this protein, that it is not essential for virus replication in cell culture (69), and that there is no evidence to suggest that it is recruited into the coronavirus RTC.
GENETICS OF CORONAVIRUS TRANSCRIPTION
Forward genetics.
The classical approach to the genetic analysis of coronavirus replication and transcription, i.e., the characterization of conditionally lethal, usually ts, virus mutants that are unable to synthesize RNA when the infection is initiated and maintained at the nonpermissive temperature, has been adopted in only a few laboratories. This is disappointing, because the essential feature of such mutants is that they are likely to be defective in different aspects of viral RNA synthesis, and a detailed characterization of their genotype and phenotype should provide insights into the mechanisms of RNA synthesis, the functions of individual viral replicase proteins, and the protein-RNA and protein-protein interactions that regulate the activity of the RTC. These conditional-lethal mutants may also be used in a cis-trans test to define the number of complementation groups, or cistrons, that contribute to a specific phenotype. This sort of analysis can provide valuable insight into the possible pathways that polyproteins must travel to assume functional configurations.
MHV-A59 mutants have been produced in a number of laboratories over a period of 20 years (67, 79, 80). In a recent analysis of 19 ts mutants that are unable to produce viral RNA at 39°C, the nonpermissive temperature, four complementation groups were identified using classical and molecular complementation assays (66). Most of the 19 mutants, including Alb ts6, Alb ts16, and LA ts6, were members of complementation group 1. The lesions in these three mutants were located in nsp4, nsp5, and nsp10. Thus, by definition, these individual nsp's are cis-acting, i.e., their ts defects cannot be complemented in trans by nsp's made by a second virus. This result suggests three possibilities: (i) nsp4-nsp10 function as a polyprotein before cleavage into individual polypeptides, (ii) nsp4-nsp10 first assemble into an RTC and are then cleaved, with a gain of function(s) expressed in individual polypeptides, or (iii) ts-induced alteration to the folding or interaction of protein domains within the nsp4-nsp10 region of pp1a/pp1ab interferes with polyprotein processing or function. In the same study, five further mutants could be assigned to three additional complementation groups. Alb ts22 (mutation in nsp12) comprises group 2, Alb ts17 and Wü ts38 (mutations in nsp14) comprise group 4, and Wü ts18 and Wü ts36 (mutation in nsp16) comprise group 6. Thus, each of these nsp's is trans-active and diffusible between pp1ab complexes. Taken together, the results of this analysis are consistent with the idea that nsp4 to nsp10 of pp1a act together as a complex, multidomain structure or scaffold onto which the trans-acting nsp's (e.g., nsp12, nsp14, and nsp16) and viral RNA associate. It will be interesting to discover whether other complementation groups can be assigned to nsp1, nsp2, nsp3, nsp13, or nsp15 and if any of the individual nsp's in nsp4 to -10 have later and separate functions, i.e., are trans-acting. This information will be especially interesting for nsp's in pp1a that are thought to function as multimers.
Another major advantage of using ts mutants is that they can be analyzed by temperature-shift protocols; i.e., by allowing them to gain function at the permissive temperature, shifting to the nonpermissive temperature, and then determining if the function is lost. Generally, their failure to lose function after the temperature shift indicates that the amino acid change in the mutant has affected the formation of the complex or activity. If they lose function, the mutation probably directly affects function. When this analysis was done for the mutants described above, we were surprised to find that MHV-A59 mutants from cistrons 2, 4, and 6 were of the latter type. In one case, Alb ts22, where the mutation lies in the virus RdRp, it is easy to rationalize this phenotype. However, we also found that mutations in nsp14 and nsp16 have an immediate effect on plus-strand RNA synthesis by preformed RTCs after the shift to 39°C, showing that the activity of the MHV-A59 RTC is dependent on a complex structure of interacting domains contributed by nsp's encoded in ORF1b. In contrast, mutations in nsp4, nsp5, and nsp10 (cistron 1, i.e., Alb ts6, Alb ts16, and LA ts6) did not inhibit ongoing plus-strand synthesis following the temperature shift. This result is consistent with the notion that the nsp4-to-nsp10 portion of pp1a is providing structural and not catalytic components of the complex (see Addendum in Proof).
An RNA-negative phenotype, i.e., the failure to synthesize viral RNA at the nonpermissive temperature, might be due to the inability to form a minus-strand RTC or the inability to convert the minus strand into a RTC-making plus strand (the so-called conversion phenotype) (22). A more detailed analysis of the cistron 1 mutants showed that mutant LA ts6 was defective in continuing minus-strand synthesis after the shift to the nonpermissive temperature. This result implicates nsp10 in minus-strand synthesis. In contrast, Alb ts16 appeared to have a conversion phenotype: Alb ts16-infected cells made minus strands but did not increase the rate of plus-strand synthesis after shift to 39°C. Alb ts16 has a mutation in the carboxyl-terminal domain of nsp5 (3C-like cysteine proteinase [3CLpro]), and it will be important to determine if this mutation affects the activity of the proteinase (72, 84). It might affect the folding of pp1a or pp1ab, or the nsp5 C-terminal domain itself could have a function in plus-strand RNA synthesis. Nevertheless, because minus-strand RNA synthesis ceased in Alb ts16-infected cells that were treated with cycloheximide to block translation of new nsps at the time of the shift to nonpermissive temperature (66), we propose that the Alb ts16 RTC does not retain artifactually its activity for minus-strand synthesis. Rather, it fails to gain plus-strand synthesis activity at the nonpermissive temperature. We favor a model where the activity that makes plus strands is gained at the expense or loss of the activity to make minus strands.
Reverse genetics.
The development of infectious cDNA constructs for coronaviruses is a recent event, and the use of reverse genetics in the analysis of coronavirus transcription is now gathering momentum. Already, a number of important findings have been made. First, reverse genetics, in particular the method of targeted RNA recombination (44), has been used to analyze cis-acting elements that regulate coronavirus transcription, as discussed in the above section on coronavirus RNA replication and transcription. Second, reverse genetics has been used to confirm the involvement of coronavirus nsp proteins in RNA synthesis and to discriminate between essential and nonessential functions (6, 26, 83). For example, mutation at the active site of the HCoV-229E nsp15 NendoU domain or the active site of the HCoV-229E nsp14 3′-to-5′ exonuclease (ExoN) domain suggests that these functions are essential for virus RNA synthesis in cell culture (34) although, in some cases, residual viral RNA synthesis was detected in cells transfected with mutant RNA. In contrast, mutation of the active site of the HCoV-229E nsp3 ADRP domain had no significant effect on virus RNA synthesis or virus titer, and no reversion to wild-type sequence was observed when the mutant virus was passed in cell culture (53).
Reverse genetics has been used to show that the MHV and SARSCoV nsp2 proteins are not essential for virus replication but that their deletion attenuates virus growth and virus RNA synthesis (30). In a similar fashion, MHV mutants that are incapable of liberating nsp1 from the nascent polyprotein (i.e., nsp1/nsp2 cleavage mutants) exhibit delayed replication, low titers, small plaque morphology, and reduced RNA synthesis compared to wild-type virus (23). A more extensive deletion and site-specific mutagenesis study of the MHV nsp1 has identified domains and residues that are important for polyprotein processing and virus RNA synthesis (16). Although informative, this sort of reverse genetic analysis must be approached with caution. The frequency of lethal mutation is likely to be high, and there is, as always, the possibility of indirect effects, such as phenotypes caused by the disruption of cis-acting RNA structures.
ROLE OF THE N PROTEIN
The coronavirus N protein has been implicated in virus RNA synthesis by three lines of evidence. First, the N protein was shown to bind specific RNA sequences, including the leader sequence, the TRS sequence, and sequences located at the 3′ end of the virus genome (73). Second, in addition to a cytoplasmic distribution in the host cell, at least a fraction of the N protein colocalizes with RTCs early in infection (13, 85). And third, there is clearly a requirement for sustained translation of the N protein in trans or in cis for optimal replication of BCoV DI RNA and Transmissible gastroenteritis virus-derived replicons (1, 18). The importance of the N protein was also emphasized when it was shown that the rescue of recombinant coronaviruses from cells transfected with infectious RNA was greatly enhanced by, if not dependent upon, the expression of N protein in the same cell (2, 17, 20, 90-92).
These observations were followed up by experiments showing that, in the absence of N protein, HCoV-229E replicons expressing GFP from a subgenomic mRNA produced low levels of GFP and little or no amplification of the replicon. In contrast, the expression of N in the same cells, in cis or in trans, significantly increased the number of transduced cells, the levels of GFP expression and, concomitantly, the amplification of the replicon (68). Our interpretation of these data is that the N protein has a role in determining the ratio of genome- to subgenome-length minus strands and, in the absence of N, this ratio is perturbed, with the underproduction of genome-length templates. In the virus-infected cell, there should always be sufficient N protein, either introduced with the nucleocapsid structure or synthesized within 90 min of infection (see the section on coronavirus RNA replication and transcription), to fulfill this role. Therefore, according to this interpretation, N does not function as a replication-transcription switch.
What could the role of the N protein be? One possibility is that it is acting as an RNA chaperone as proposed for the N protein of hantaviruses (46), where chaperone activity results in the transient dissociation of RNA structures, including RNA duplexes with adjoining single-stranded regions, which may be required for facilitating correct higher-order RNA structure. For coronaviruses, such chaperone activity could be important for the initiation of minus-strand synthesis or, perhaps, for suppressing attenuation and/or template switching at the TRS element during discontinuous synthesis (25). Second, coronaviruses possess helical nucleocapsids. This is not common among plus-stranded RNA viruses, Tobacco mosaic virus being another exception, but is typical of minus-stranded RNA viruses. It is possible that the role of the coronavirus N protein is to associate with the genomic RNA to produce a template that is “configured” to balance the ratio of RTCs engaged in either transcription or replication, as has been proposed for measles virus (12). In this respect, it is also worth noting that replication and transcription from the genome of EAV, a virus that has an icosahedral nucleocapsid structure, does not appear to involve N protein function (47).
WHY IS CORONAVIRUS TRANSCRIPTION SO COMPLEX?
Two reasons are most often given for the complexity of coronavirus transcription. (i) The generation of multiple subgenomic mRNAs and the process of discontinuous transcription during minus-strand synthesis demand complex replication-transcription machinery. (ii) The large size of the coronavirus genome demands unusual activities to maintain genetic stability.
With regard to the first reason, it is true that the generation of multiple minus-strand RNAs from a single template must be a complex process but, essentially, it is only the process of discontinuous transcription during minus-strand synthesis that is unique. The initiation of plus-strand and minus-strand synthesis, elongation, and termination are carried out by all RNA viruses, usually with a smaller number of replicase components than are found in the coronavirus RTC. Also, it is important to realize that arteriviruses, which, as already mentioned, are a family of viruses related to coronaviruses and have similar, if not identical, mechanisms of subgenome-length mRNA synthesis, do not encode proteins with functions analogous to the ExoN, S-adenosylmethionine-dependent 2′-O-methyl transferase (MT), ADRP, or CPD functions of coronaviruses (75). This implies that these functions, although they may be an integral part of the coronavirus RTC, are not needed to allow for discontinuous transcription. Biological advantages to discontinuous transcription include economies of coding and the ability to regulate individual mRNA abundance, but these do not seem to justify the extraordinary genetic investment that coronaviruses have made in replicase-transcriptase and niche-specific proteins.
An alternative view is proposed by Gorbalenya and colleagues (28). Basically, these authors argue that the acquisition of the coronavirus enzymatic activities may have improved the fidelity of RNA replication and transcription to allow for genome expansion. This, in turn, would provide coronaviruses with the opportunity, for example, to expand their host range and adapt rapidly to changing environmental conditions while maintaining genomic stability. Specifically, it is proposed that the HEL, ExoN, NendoU, and MT functions may provide RNA specificity and that the ADRP and CPD functions (when present) modulate the pace of a reaction in a common pathway, which could be part of an oligonucleotide-directed repair mechanism (75). The existence of such a repair mechanism in coronaviruses, with similarities to the “proofreading” or repair activities associated with DNA replication, would require a paradigm shift in our view of RNA virus replication.
ADDENDUM IN PROOF
It was recently shown that SARSCoV encodes a second, noncanonical RdRp residing in nsp8, and it was proposed that the nsp8 RdRp produces primers utilized by the primer-dependent nsp12 RdRp (I. Imbert, J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard, EMBO J. 25:4933-4942, 2006).
Acknowledgments
The Sawicki laboratory is supported by the NIAID, NIH. The Siddell laboratory is supported by the Wellcome Trust and the EU.
We thank Alexander Gorbalenya, Paul Lehmann, and the reviewers for helpful comments on the manuscript.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Almazán, F., C. Gálan, and L. Enjuanes. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 5.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baric, R. S., and A. C. Sims. 2005. Development of mouse hepatitis virus and SARS-CoV infectious cDNA constructs. Curr. Top. Microbiol. Immunol. 287:229-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 9.Bernini, A., O. Spiga, V. Venditti, F. Prischi, L. Bracci, J. Huang, J. A. Tanner, and N. Niccolai. 2006. Tertiary structure prediction of SARS coronavirus helicase. Biochem. Biophys. Res. Commun. 343:1101-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhardwaj, K., J. Sun, R. Sidhartharaja, A. Holzenburg, L. A. Guarino, and C. C. Kao. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361:243:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhella, D., A. Ralph, and R. P. Yeo. 2004. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340:319-331. [DOI] [PubMed] [Google Scholar]
- 13.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockway, S. M., and M. R. Denison. 2005. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 340:209-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, R. Y., and D. A. Brian. 1996. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 70:2201-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coley, S. E., E. Lavi, S. G. Sawicki, L. Fu, B. Schelle, N. Karl, S. G. Siddell, and V. Thiel. 2005. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 79:3097-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis, K. M., B. Yount, A. C. Sims, and R. S. Baric. 2004. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. J. Virol. 78:6061-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enjuanes, L., F. Almazán, I. Sola, and S. Zuñiga. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60:211-230. [DOI] [PubMed] [Google Scholar]
- 26.Enjuanes, L., I. Sola, S. Alonso, D. Escors, and S. Zuñiga. 2005. Coronavirus reverse genetics and development of vectors for gene expression. Curr. Top. Microbiol. Immunol. 287:161-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 79:13399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. Kao. 2005. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 353:1106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herold, J., S. G. Siddell, and A. E. Gorbalenya. 1999. A human RNA viral cysteine proteinase that depends upon a unique Zn2+-binding finger connecting the two domains of a papain-like fold. J. Biol. Chem. 274:14918-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78:7833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, R. F., M. Feng, P. Liu, J. J. Millership, B. Yount, R. S. Baric, and J. L. Leibowitz. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570-14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joo, M., S. Banerjee, and S. Makino. 1996. Replication of murine coronavirus defective interfering RNA from negative-strand transcripts. J. Virol. 70:5769-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, A. Brooun, M. Griffith, K. Moy, M. K. Yadav, J. Velasquez, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2006. Crystal structure of nonstructural protein 10 from the SARS coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 80:7894-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komissarova, N., and M. Kashlev. 1997. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 272:15329-15338. [DOI] [PubMed] [Google Scholar]
- 41.Komissarova, N., and M. Kashlev. 1997. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. USA 94:1755-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters, P. S., and P. J. M. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mir, M. A., and A. T. Panganiban. 2006. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 80:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 48.Namy, O., S. J. Moran, D. I. Stuart, R. J. Gilbert, and I. Brierley. 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 50.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. 2004. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol. 78:8102-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peti, W., M. A. Johnson, T. Herrmann, B. W. Neuman, M. J. Buchmeier, M. Nelson, J. Joseph, R. Page, R. C. Stevens, P. Kuhn, and K. Wüthrich. 2005. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 79:12905-12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1′′-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raman, S., P. Bouma, G. D. Williams, and D. A. Brian. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raman, S., and D. A. Brian. 2005. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434-12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA 103:11892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson, M. P., H. Igel, R. Baertsch, D. Haussler, M. Ares, Jr., and W. G. Scott. 2005. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure (Cambridge) 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santangelo, T. J., and J. W. Roberts. 2004. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell 14:117-126. [DOI] [PubMed] [Google Scholar]
- 60.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 61.Sawicki, S. G. 1987. Characterization of a small plaque mutant of the A59 strain of mouse hepatitis virus defective in cell fusion. Adv. Exp. Med. Biol. 218:169-174. [DOI] [PubMed] [Google Scholar]
- 62.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 66.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathogens 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaad, M. C., S. A. Stohlman, J. Egbert, K. Lum, K. Fu, T. Wei, Jr., and R. S. Baric. 1990. Genetics of mouse hepatitis virus transcription: identification of cistrons which may function in positive and negative strand RNA synthesis. Virology 177:634-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwarz, B., E. Routledge, and S. G. Siddell. 1990. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 64:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seybert, A., C. C. Posthuma, L. C. van Dinten, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr. 2005. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 79:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shaevitz, J. W., E. A. Abbondanzieri, R. Landick, and S. M. Block. 2003. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature 426:684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi, J., and J. Song. 2006. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 273:1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siddell, S., J. Ziebuhr, and E. J. Snijder. 2005. Coronaviruses, toroviruses, and arteriviruses, p. 823-856. In B. W. J. Mahy and V. ter Meulen (ed.), Topley & Wilson's microbiology and microbial infections: virology. Hodder Arnold, London, United Kingdom.
- 75.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. M. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sola, I., J. L. Moreno, S. Zuñiga, S. Alonso, and L. Enjuanes. 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 79:2506-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stalcup, R. P., R. S. Baric, and J. L. Leibowitz. 1998. Genetic complementation among three panels of mouse hepatitis virus gene 1 mutants. Virology 241:112-121. [DOI] [PubMed] [Google Scholar]
- 80.Sturman, L. S., C. Eastwood, M. F. Frana, C. Duchala, F. Baker, C. S. Ricard, S. G. Sawicki, and K. V. Holmes. 1987. Temperature-sensitive mutants of MHV-A59. Adv. Exp. Med. Biol. 218:159-168. [DOI] [PubMed] [Google Scholar]
- 81.Su, D., Z. Lou, F. Sun, Y. Zhai, H. Yang, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. Dodecameric structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 80:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thiel, V., and S. G. Siddell. 2005. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 287:199-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Aken, D., E. J. Snijder, and A. E. Gorbalenya. 2006. Mutagenesis analysis of the nsp4 main proteinase reveals determinants of arterivirus replicase polyprotein autoprocessing. J. Virol. 80:3428-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu, X., Y. Liu, S. Weiss, E. Arnold, S. G. Sarafianos, and J. Ding. 2003. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 31:7117-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu, X., Y. Zhai, F. Sun, Z. Lou, D. Su, Y. Xu, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 80:7909-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611-615. [DOI] [PubMed] [Google Scholar]
- 90.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92a.Yount, B., R. S. Roberts, L. Lindesmith, and R. S. Baric. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl. Acad. Sci. USA 103:12546-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ziebuhr, J., J. Herold, and S. G. Siddell. 1995. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 97.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]