Abstract
Hepatitis C virus (HCV) infection induces the α-chemokine interleukin-8 (CXCL-8), which is regulated at the levels of transcription and mRNA stability. In the current study, CXCL-8 regulation by double-stranded (ds)RNA pathways was analyzed in the context of HCV infection. A constitutively active mutant of the retinoic acid-inducible gene I (RIG-I), RIG-N, activated CXCL-8 transcription. Promoter mutagenesis experiments indicated that NF-κB and interferon (IFN)-stimulated response element (ISRE) binding sites were required for the RIG-N induction of CXCL-8 transcription. IFN-β promoter stimulator 1 (IPS-1) expression also activated CXCL-8 transcription, and mutations of the ISRE and NF-κB binding sites reduced and abrogated CXCL-8 transcription, respectively. In the presence of wild-type RIG-I, transfection of JFH-1 RNA or JFH-1 virus infection of Huh7.5.1 cells activated the CXCL-8 promoter. Expression of IFN regulatory factor 3 (IRF-3) stimulated transcription from both full-length and ISRE-driven CXCL-8 promoters. Chromatin immunoprecipitation assays demonstrated that IRF-3 and NF-κB bound directly to the CXCL-8 promoter in response to virus infection and dsRNA transfection. RIG-N stabilized CXCL-8 mRNA via the AU-rich element in the 3′ untranslated region of CXCL-8 mRNA, leading to an increase in its half-life following tumor necrosis factor alpha induction. The data indicate that HCV infection triggers dsRNA signaling pathways that induce CXCL-8 via transcriptional activation and mRNA stabilization and define a regulatory link between innate antiviral and inflammatory cellular responses to virus infection.
Hepatitis C virus (HCV) infects an estimated 3% or 170 million of the world's population and causes an estimated 476,000 deaths per year due to complications of end-stage liver disease (56, 62). In the United States, about 1.8% of the general population or 4 million persons are infected. Of those acutely infected with HCV, approximately 85% develop chronic infection, and about 70% of these patients develop histological evidence of chronic liver disease. Moreover, viremia is not cleared in about 50% of infected patients treated with pegylated interferon (IFN)-ribavirin therapy, the current standard of care. Compounding this issue are the predictions that in the next 20 years, HCV-related complications, including hepatic decompensation, hepatocellular carcinoma, and liver-related deaths, will increase by 106%, 81%, and 180%, respectively (12). Thus, chronic hepatitis C is a serious global medical problem necessitating effective treatment. Given the propensity of HCV for chronic infection, association with severe liver disease, and difficulty of treatment, many studies are focused on HCV-host interactions that contribute to HCV persistence and pathogenesis.
The inflammatory response to virus infection involves the regulated induction of cytokines and chemokines. The response is initiated within the infected cell by pathogen-associated molecular pattern (PAMP) recognition but soon thereafter affects neighboring cells and tissues due to the paracrine effects of cytokine and chemokine release. Cytokine and chemokine release occurs rapidly in response to virus infection, and its chief objectives are to recruit inflammatory leukocytes, limit virus replication and spread, and induce adaptive immunity. However, prolonged expression of chemokines in the context of chronic viral infections may be detrimental to the host. For example, patients with chronic hepatitis C have elevations in serum levels of α-chemokine interleukin-8 (CXCL-8), and patients who are nonresponsive to IFN therapy have high pretreatment levels of CXCL-8 (39, 51). When expressed in cell culture, the HCV NS5A protein induces CXCL-8, which is associated with the inhibition of the antiviral effects of IFN (4, 16, 50).
CXCL-8 is a 71-amino-acid chemokine belonging to the CXC family and is produced by many cell types, including monocytes, epithelial cells, fibroblasts, and hepatocytes (42). CXCL-8 elicits many effects, including neutrophil, T-lymphocyte and basophil chemotaxis and degranulation, oxidative burst, and lysosomal-enzyme release (42). CXCL-8 has been shown to be an important mediator of the inflammatory responses to many viruses and bacteria. For example, CXCL-8 is induced in response to the expression of the HCV NS5A protein (3, 4, 16, 50) and HCV replication (19). Moreover, CXCL-8 inhibits the antiviral actions of IFN-α (29), and recent studies indicate that CXCL-8 protein levels are associated with HCV replication (30).
CXCL-8 is induced by a variety of stimuli, including those from CXCL-1, tumor necrosis factor alpha (TNF-α), phorbol esters, lipopolysaccharide, and virus infection (42). The induction of CXCL-8 involves both the transcriptional activation of the CXCL-8 promoter and the stabilization of CXCL-8 mRNA (61). CXCL-8 mRNA stabilization involves the binding of regulatory proteins to AU-rich elements (AREs) in the 3′ untranslated region (3′UTR) of the mRNA (1, 6, 9, 53). The CXCL-8 promoter contains binding sites for NF-κB, NF-interleukin 6 (IL-6), AP-1 protein, and an IFN-stimulated response element (ISRE)-like element, which has been shown to bind proteins belonging to the IFN regulatory factor (IRF) family (8, 65). The mechanisms of CXCL-8 gene induction have been investigated using different stimuli and cell types, and deletion and mutational analyses of the promoter indicate that CXCL-8 is activated in a cell-type- and stimulus-specific manner (43, 45). It has previously been shown that HCV NS5A-induced transcriptional activation of the CXCL-8 promoter requires intact NF-κB and AP-1 binding sites (4, 50). However, the role of upstream regulatory elements, such as the ISRE site, is not known.
IRF proteins play pivotal roles in innate antiviral responses and bind to DNA sequences containing ISREs (2, 36). IRFs are activated by virus infection when viral PAMPs, such as double-stranded (ds)RNA, are sensed by Toll-like receptor 3 (TLR3) or by cytoplasmic dsRNA sensors such as the retinoic acid-inducible gene I (RIG-I) (66). IPS-1 (also known as the MAVS, Cardif, or VISA protein) (21) is an adaptor protein for RIG-I signaling (34). RNA binding to RIG-I and interaction with IPS-1 or TLR3 activate IRF-3 and NF-κB. IRF-3 and NF-κB translocate to the nucleus, interact with other transcription factors, and bind to their cognate promoter elements to induce gene expression, which classically includes IFN-α and IFN-β genes.
IRFs and NF-κB also regulate transcription of chemokine genes. For instance, IRF-1 is involved in the respiratory syncytial virus (RSV) and Helicobacter pylori induction of CXCL-8 by binding to one of several ISRE-like elements in the CXCL-8 promoter (8, 65). Similarly, IRF-3 induces the chemokine RANTES by binding to ISREs in the RANTES promoter (32). Moreover, NF-κB is the principal transcription factor responsible for CXCL-8 induction (42). Thus, the regulation of chemokines by NF-κB and especially by IRFs may represent a functional link between innate antiviral and inflammatory responses. In the current report, we examined the induction of CXCL-8 by dsRNA-triggered innate antiviral pathways in the context of HCV infection in vitro.
MATERIALS AND METHODS
Cells and viruses.
Human hepatoma Huh7 cells and HEK293 cells were grown in Huh7 medium which contained Dulbecco's minimum essential medium, 10% fetal bovine serum, 1× penicillin, streptomycin, amphotericin B (Fungizone) (except for HEK cells), 10 mM l-glutamine, and 1× nonessential amino acids (all reagents were from Invitrogen, Carlsbad, CA). Huh7 cells were obtained from Apath, LLC. Huh7.5.1 cells were obtained from Francis Chisari (68) and cultured in Huh7 medium. All cell lines were checked for mycoplasma by using a MycoAlert assay (Cambrex Bio Science, Rockland, ME) and found to be mycoplasma-free.
JFH-1 viral stock preparation, cell infection, and titration were performed exactly as described previously (59, 68).
Plasmids.
BB7 replicon plasmid DNA was obtained from Apath, LLC. Generation of JFH-1 RNA and transfection of cells were performed as described previously (59). Various luciferase reporter genes under the control of different forms of CXCL-8 were also used and were obtained from Naofumi Mukaida (44) or generated as described previously (8) (see Fig. 1). Plasmid pQ150 expresses green fluorescent protein (GFP) under the control of the constitutive EF-1α promoter and was provided by Jeffery Vieira. Cytomegalovirus IRF-3 (CMV-IRF-3) (33) was obtained from John Hiscott. Wild-type RIG (RIG-WT), constitutively active (RIG-N), and dominant negative (RIG-C) plasmids were generated as previously described (14, 66).
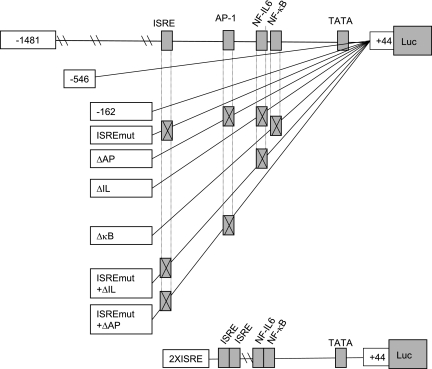
FIG. 1.
Schematic representation of the CXCL-8 promoter. Locations of ISRE, AP-1, NF-IL-6, NF-κB sites are indicated. The names of the reporter constructs are found in the boxes to the left of each schematic. Luc, luciferase.
Transfection.
The day prior to transfection, 3 × 104 cells were plated in black, clear-bottomed, 96-well tissue culture plates. Endotoxin-free plasmid DNA was purified (Endofree kit; QIAGEN, Valencia, CA) and introduced into cells with Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen). For reporter gene studies, unless otherwise indicated, 100 ng of the luciferase gene under the control of the promoter construct of interest was transfected into cells in triplicate or quadruplicate. When expression plasmids were included in the transfection, 50 ng of each was added. Eighteen hours later, stimuli such as recombinant human TNF-α (rhTNF-α) (15 ng/ml; Pierce Biotechnology, Rockford, IL) or virus infection (Sendai virus at a multiplicity of infection [MOI] of 100 hemagglutinin units, or JFH-1 virus at an MOI of 0.01 focus-forming unit) were added to cells. In the case of poly(I) · poly(C) [poly(I:C)] (0.2 μg/well) or JFH-1 RNA (0.25 μg/well), cells were retransfected using Lipofectamine 2000 or transmessenger RNA (QIAGEN) reagents. Six h [for poly(I:C)] or 24 h later (for JFH-1 RNA), luciferase activity was measured on cell lysates using a Britelite assay system (Perkin Elmer, Boston, MA). Before protein harvest, cells were visualized under a fluorescent microscope, and the transfection efficiency was determined by comparing the proportion of GFP-positive cells to the total cell number, as described previously (30, 40, 47). The results, in relative light units, shown in Fig. 2, 3, and 4 are corrected for transfection efficiency.
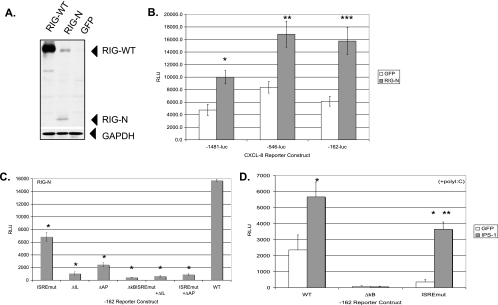
FIG. 2.
Double-stranded RNA signaling pathways activate CXCL-8 transcription. (A) Expression of RIG-WT and constitutively active RIG-N proteins. Huh7 cells were transfected with plasmids expressing RIG-WT, RIG-N, or GFP, and whole-cell protein extracts were harvested 48 h later and analyzed by Western blotting with a polyclonal antiserum to RIG-I. Positions of the proteins are indicated with arrowheads. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cellular protein expression verified equal loading of proteins among the different conditions. (B) RIG-N activates CXCL-8 transcription. Huh7 cells were transfected with RIG-N- or GFP-expressing plasmids along with CXCL-8 reporter genes −1481-luc, −546-luc, or −162-luc, and luciferase readings were measured 24 h posttransfection. RLU, relative light units. Asterisks indicate significance of RIG-N luciferase values versus GFP values (*, P = 0.002; **, P = 0.017; ***, P = 0.003). (C) CXCL-8 promoter mutagenesis. Huh7 cells were transfected with RIG-N or GFP plasmids and the indicated CXCL-8 promoter constructs in the −162 backbone containing various mutations in transcription factor binding sites. Luciferase readings were measured 24 h posttransfection. Asterisks indicate that the luciferase values of mutant promoters compared to that of the wild-type −162 construct were significantly different (P, <0.01). (D) IPS-1 activates CXCL-8 transcription. Huh7 cells were transfected with IPS-1- or GFP-expressing plasmids along with wild-type −162 or mutant −162 promoters containing mutations in the NF-κB and ISRE binding sites, and 20 h posttransfection, cells were transfected with poly(I:C). Luciferase readings were measured 6 h later. The single asterisk indicates that IPS-1 luciferase readings were significantly different from the GFP readings (*, P <0.01). The double asterisk indicates that IPS-1-induced luciferase values from the ISRE mutant promoter were significantly lower than IPS-1 induced luciferase values from the wild-type promoter (**, P = 0.008).
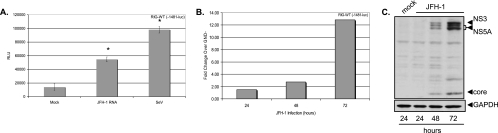
FIG. 3.
HCV RNA and HCV infection activate CXCL-8 transcription. (A) Huh7.5.1 cells were transfected with RIG-WT and the full-length −1481 CXCL-8 luciferase construct, and 24 h later, cells were mock transfected, transfected with JFH-1 RNA, or infected with Sendai virus (SeV) at 100 hemagglutinin inhibitor units. Luciferase readings were measured 24 h later. Asterisks indicate that JFH-1- or Sendai virus-induced CXCL-8 transcription was significantly higher than mock-transfected cells (*, P <0.01). RLU, relative light units. (B) Huh7.5.1 cells were transfected with RIG-WT and the full-length −1481 CXCL-8 luciferase construct, and 24 h later, cells were infected with JFH-1 virus stocks at an MOI of 0.01. Control cells were incubated with an equivalent volume of supernatants from cells transfected with replication-incompetent JFH-1 RNA containing the GND mutation in the viral polymerase. Luciferase readings were taken at the indicated times. Data are expressed as the increase (n-fold) in luciferase values over that of the GND control. (C) Western blot detection of HCV NS3, NS5A, and core proteins in Huh7.5.1 cells infected with JFH-1 at an MOI of 0.01. Blots were also probed with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to verify equal protein loading.
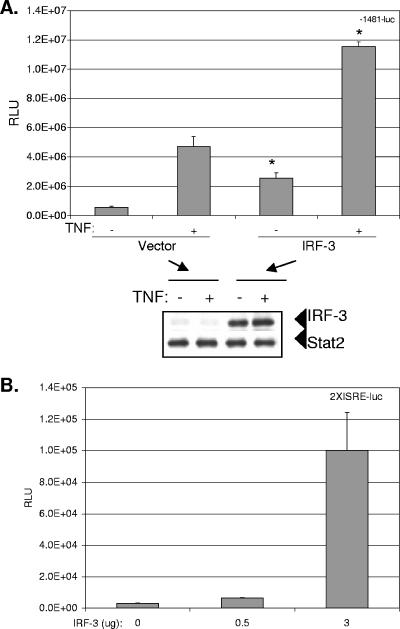
FIG. 4.
IRF-3 transactivates the CXCL-8 promoter. (A) Huh7 cells were plated in 12-well plates and transfected with 0.5 μg of CMV-IRF-3 or vector plasmids along with 0.5 μg of the full-length −1481-luc CXCL-8 promoter-luciferase plasmid. Twenty-four hours later, cells were treated or not treated with 15 ng/ml of rhTNF-α for 6 h before luciferase activity was measured on cell lysates. The Western blot below the graphs depicts the expression of IRF-3 detected with polyclonal antiserum. Stat2 was also detected with polyclonal antiserum and served as a control for equal loading. Asterisks indicate significant stimulation of CXCL-8 transcription by IRF-3 compared to that of the vector control (*, P <0.01). RLU, relative light units. (B) Increasing amounts of IRF-3 were cotransfected with 0.5 μg of the synthetic CXCL-8 promoter, 2XISRE-luc, into Huh7 cells, and cells and lysates were processed as described above.
Western blot analysis.
Protein lysates were quantitated (BCA Protein Assay; Pierce), and equal amounts of total protein (10 to 20 μg) were separated on 4 to 20% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) gels. IRF-3 and Stat2 were detected using polyclonal antiserum (Santa Cruz Biotechnology). HCV proteins were detected using random, deidentified HCV-infected patient serum, as described previously (49). Prior to use, infected serum was inactivated by adding Triton X-100 to a final concentration of 1%. NS5A protein was also detected using a polyclonal antibody to NS5A (Chiron, Emeryville, CA). RIG proteins were detected using a polyclonal antibody (25).
RNA quantitation.
CXCL-8 mRNA was quantitated by real-time reverse transcriptase (RT)-PCR, as recently described (19). Dilutions of precisely quantitated CXCL-8 cDNA in the PCMGSNEO plasmid (64) (kindly provided by Naofumi Mukaida), ranging from 0 to 107 copies per tube, were run in triplicate to generate a standard curve, which served as a reference to calculate the CXCL-8 copy number based on the cycle threshold. RNA copy numbers were normalized to 10 ng of total cellular RNA.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer's specifications (Upstate USA, Charlottesville, VA). Briefly, 2 × 106 cells were plated on 10-cm dishes and then treated with various inducers, including TNF-α, poly(I:C) or encephalomyocarditis virus infection, for various times. Transcription factors were cross-linked to DNA by adding formaldehyde directly to culture medium to a final concentration of 1% and incubated for 10 min at 37°C. Cells were washed and scraped into phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml aprotinin, and 1 μg/ml pepstatin A. Cells were pelleted and resuspended in SDS lysis buffer (Upstate), and DNA was sheared into lengths of 200 to 1,000 base pairs by sonication at 50 V and 30% amplitude for a total of eight times for 5 seconds each on ice. Lysates were cleared by centrifugation for 10 min at 13,000 rpm at 4°C. Supernatants were diluted ninefold in ChIP dilution buffer (Upstate) and precleared with protein A agarose-salmon sperm DNA (50% slurry; Upstate) for 30 min at 4°C with agitation. One to two microliters of IRF-3 antibody (polyclonal, from Michael David, or monoclonal, from Pharmingen) was added to the precleared supernatants and incubated overnight with rotation at 4°C, followed by the addition of protein A agarose-salmon sperm DNA. ChIP samples were washed, DNA protein complexes eluted, cross-links reversed, and DNA extracted. CXCL-8 and IFN-Β promoter-specific PCRs were performed using primer sets CXCL-8F (AAGAAAACTTTCGTCATACTCCG), CXCL-8R (TGGCTTTTTATATCATCACCCTAC), IFN-ΒF (CCTCACAGTTTGTAAATCTTTTTCCC), and IFN-ΒR (ACGAACAGTGTCGCCTACTACCTG), where F and R designate forward and reverse primers.
These primer sets hybridize to regions 1302 to 1473 of the genomic clone of CXCL-8 (NCBI accession number M28130) and to regions 97 to 348 of the IFN-β (NCBI accession number V00534.1) promoter and generate PCR products of 171 (CXCL-8) and 251 (IFN-β) base pairs. Forty-five cycles of PCR were performed as follows: 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds.
Cloning and reporter studies with the CXCL-8 3′UTR.
A 237-base pair region (nucleotides 972 to 1209 of accession number NM000584) from the 1,250-base pair CXCL-8 3′ untranslated region (3′UTR) was amplified by RT-PCR. Briefly, total RNA was extracted by Tri Reagent (Molecular Research Center, Cincinnati, OH) from a THP-1 monocytic cell line (ATCC, Manassas, VA) that was previously stimulated with 10 μg/ml lipopolysaccharide in the presence of cycloheximide (5 μg/ml). cDNA was synthesized and amplified by PCR with the forward primer with a BamHI site (underlined), 5′ GCACCGGATCCGATGTTGTGAGGACATGTG 3′, and the reverse primer with an XbaI site (underlined), 5′ GCCAGTCTAGAACCCTGATTGAAATTTAT 3′. The additional 5′ sequences provided thermal stability to the oligonucleotides and facilitated the restriction digest. The PCR products were purified by phenol-chloroform extraction, followed by ethanol precipitation. The PCR products were cut by BamHI and XbaI sequentially, followed by phenol extraction and ethanol precipitation. The digested PCR products were ligated into an expression vector derived from a gWIZ plasmid (Gene Therapy Systems, Inc., San Diego, CA) that contains an enhanced GFP (EGFP) coding region under the constitutive expression of the CMV/intron A promoter and has a bovine growth hormone 3′UTR. Recombinant colonies were verified by PCR using a forward vector-specific primer and a CXCL-8 3′UTR reverse primer.
Cells (3 × 104 cells per well) in black clear-bottomed 96-well plates were transfected with the GFP plasmids. Transfections were performed as mentioned above. All transfections were performed in quadruplicate. The variance in GFP fluorescence among replicate microwells was <7%. Transfection efficiency in HEK293 was always 70% ± 5%. Data are presented as the mean values ± standard errors of the fluorescence intensity determined by using a ZENYTH 3100 model instrument with the following parameters: excitation filter, 485 nm; emission filter, 535 nm; integration time, 1s; and bottom fluorescence read setting.
Statistics.
Differences between means of triplicate or quadruplicate samples of luciferase or fluorescence readings were compared using Student's t test. A P value of <0.05 was considered significant.
RESULTS
Figure 1 shows a schematic representation of the CXCL-8 promoter constructs used in this study. The figure shows the full-length (−1481) and truncated (−546 and −162) promoters used in this study. The −162 construct was also altered so that the ISRE, AP-1, NF-IL-6, and NF-κB binding sites were mutated to prevent transcription factor binding (8). The 2XISRE construct contains two copies of the ISRE site fused upstream to a minimal CXCL-8 promoter containing the NF-IL-6 and NF-κB binding sites, which was shown previously to be induced by RSV infection (8).
dsRNA sensor proteins activate CXCL-8 transcription.
RIG-I and related CARD-containing cellular proteins have been shown to lead to NF-κB and IRF-3 activation following viral infection or dsRNA stimulation (14, 38, 55, 66). We first demonstrated the functionality of the RIG-WT, RIG-N (constitutively active), and RIG-C (dominant negative) proteins in our experimental system. Wild-type and mutant RIG-expressing plasmids were transfected along with the IFN-β luciferase reporter gene and transfected with poly(I:C), a synthetic source of dsRNA. RIG-WT transfection led to IFN-β transcription only following dsRNA transfection, while RIG-N induced IFN-β transcription independently of dsRNA. In contrast, RIG-C inhibited IFN-β transcription under all conditions (data not shown). We also verified the expression of the RIG-WT and RIG-N proteins (Fig. 2A). We then investigated whether RIG-I modulated CXCL-8 transcription. Huh7 cells were transfected with various CXCL-8 reporter gene constructs in the presence of constitutively active RIG-N or GFP as a negative control. As shown in Fig. 2B, RIG-N transactivated full-length (−1481) and truncated (−546 and −162) CXCL-8 promoter constructs by 2.1-, 2.0-, and 2.6-fold, respectively, relative to that of GFP (all P values were <0.02).
Because dsRNA signaling activates both IRFs and NF-κB and these transcription factors are involved in CXCL-8 induction, we examined the RIG-N activation of CXCL-8 transcription from the −162 constructs containing various mutations in their transcription factor binding sites. As shown in Fig. 2C, RIG-N activated the −162 construct, while the mutation of the ISRE caused a 2.3-fold reduction in luciferase activity (P, <0.01). Mutations of the AP-1, the NF-IL-6, and the NF-κB sites led to 6.5-, 14.8-, and 37.2-fold reductions, respectively, in transcription, while mutations of both the ISRE/NF-IL-6 and the ISRE/AP-1 sites caused 24.3- and 17.6-fold reductions, respectively, in transcription. The data indicate a requirement for NF-κB, NF-IL-6, and AP-1 in RIG-N-mediated induction of CXCL-8 transcription, with a subtle yet consistent regulation by transcription factors binding to the ISRE of the CXCL-8 promoter.
Since IPS-1 is an essential adaptor protein in RIG-I signaling (27, 38, 52, 63), we determined whether it regulates CXCL-8 transcription. Cells were transfected with reporter and expression plasmids and then transfected with poly(I:C) to provide dsRNA. As shown in Fig. 2D, IPS-1 expression activated CXCL-8 transcription from the wild-type −162 construct 2.4-fold compared to that of GFP (P, <0.01), and a mutation of the NF-κB binding site abrogated the response. A mutation of the ISRE binding site in the CXCL-8 promoter revealed that while IPS-1 still stimulated transcription 10.2-fold relative to that of GFP (P < 0.01), the relative levels of transcription were 1.6-fold lower than that of the wild-type CXCL-8 promoter (P = 0.008). In the absence of dsRNA, IPS-1 still activated transcription from the −162 wild-type construct (data not shown). The data indicate that IPS-1 and RIG-I signal to induce the CXCL-8 promoter and that in addition to the well-known role for NF-κB, ISRE-binding proteins also regulate CXCL-8 transcription.
HCV RNA and JFH-1 infection activate CXCL-8 transcription.
Huh7.5.1 cells are defective in dsRNA signaling due to a mutation in RIG-I (55). Therefore, to investigate the effect of transfection of HCV RNA on CXCL-8 transcription, cells were first cotransfected with RIG-WT and the full-length CXCL-8 promoter-luciferase construct and then transfected with JFH-1 viral RNA derived from in vitro transcription. As shown in Fig. 3A, JFH-1 RNA caused a fourfold increase in transcription from the full-length CXCL-8 promoter compared to that of the mock-transfected cells (P, <0.01). As a positive control, cells were infected with Sendai virus, which caused a sevenfold induction of CXCL-8 transcription (P, <0.01). Moreover, as shown in Fig. 3B, when Huh7.5.1 cells were transfected with RIG-WT, followed by JFH-1 virus infection, a progressive increase in CXCL-8 transcription was observed over time. Figure 3C shows the relative levels of HCV NS3, NS5A, and core proteins by Western blotting at 24, 48, and 72 h postinfection, respectively. Collectively, the data indicate that HCV infection activates CXCL-8 transcription via the RIG-I pathway. Note also that transfection of synthetic dsRNA in the form of poly(I:C) also induced CXCL-8 in a RIG-I dependent fashion (data not shown). Thus, the response is not necessarily specific to HCV RNA but may reflect a general cellular response to dsRNA exposure.
IRF-3 stimulates CXCL-8 transcription.
IRF-3 has been shown to regulate the expression of other chemokines such as RANTES (32) and is central to the host's control of HCV infection (15). Since RIG-I signals to IRF-3 (66), we investigated the effects of IRF-3 expression on CXCL-8 transcription and HCV replication. Under basal conditions, IRF-3 induced a highly significant 8.5-fold increase (P, <0.01) in transcription from the full-length −1481 CXCL-8 promoter, while IRF-3 induced a 2.5-fold increase (P, <0.01) in CXCL-8 promoter activity in the presence of TNF-α (Fig. 4A). Shown below Fig. 4 A is a Western blot verifying the specific expression of IRF-3. Moreover, IRF-3 induced a statistically significant dose-dependent increase in activation of the 2XISRE CXCL-8 reporter construct (P, <0.01) (Fig. 4B). IRF-3-5D, a constitutively active mutant of IRF-3 (33), also activated the 2XISRE promoter construct (data not shown). The data indicate that IRF-3 transactivates the CXCL-8 promoter and are consistent with a recent report that during the early course of HCV infection, IRF-3 is activated (34).
IRF-3 binds directly to the CXCL-8 promoter in vivo.
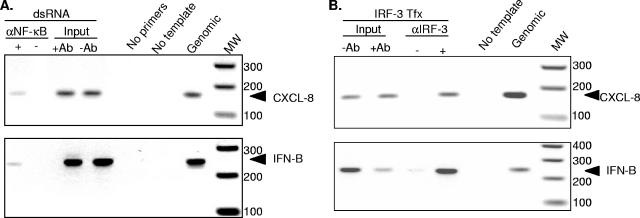
Since IPS-1 and RIG-I signaling to the CXCL-8 promoter requires the ISRE at positions −130 to −162 (Fig. 2C and 2D) and IRF-3 transactivates the CXCL-8 promoter (Fig. 4), we questioned whether IRF-3 binds directly to the CXCL-8 promoter. To investigate this issue, we established ChIP assays which measure binding of transcription factors to DNA in a cellular context. We first demonstrated that NF-κB, a central player in both CXCL-8 (42) and IFN-β (20) transcription, bound to the CXCL-8 and IFN-β promoters in response to double-stranded RNA transfection (Fig. 5A). The interaction was specific since CXCL-8- and IFN-β-specific amplification products were obtained only with extracted genomic DNA, input DNA, and DNA-protein complexes that were immunoprecipitated with anti-NF-κB antiserum. The data indicate that dsRNA treatment results in NF-κB activation and binding to the CXCL-8 and IFN-β promoters. Similarly, when Huh7 cells were transfected with IRF-3, the protein bound specifically to both the CXCL-8 and the IFN-β promoters (Fig. 5B). Cumulatively, the data indicate that dsRNA activates the RIG-I pathway to induce CXCL-8 transcription, which involves the binding of IRF-3 and NF-κB to the CXCL-8 promoter.
FIG. 5.
IRF-3 binds directly to the CXCL-8 promoter in vivo. (A) A confluent 10-cm dish of Huh7 cells was transfected with 20 μg of poly(I:C) for 6 h. Nuclear DNA was isolated from fixed cells and sheared by sonication, and equal amounts were added to immunoprecipitations (IP) with (+) or without (−) antiserum to NF-κB. DNA was extracted from IP, and the CXCL-8 and IFN-β promoters were amplified by PCR. The PCR-positive control was genomic DNA, while PCR-negative controls contained either no primers or no template DNA. Positive controls for the IP included aliquots of the input samples that went into each IP (+antibody [Ab] or −Ab). (B) Huh7 cells were transfected with CMV-IRF3, and 24 h later, nuclear DNA was isolated from fixed cells as described above. Samples were handled as described above except polyclonal antiserum to IRF-3 was used in the immunoprecipitation. Arrows indicate the location of the CXCL-8- and IFN-β-specific PCR products.
RIG-N stabilizes CXCL-8 mRNA in an ARE-dependent manner.
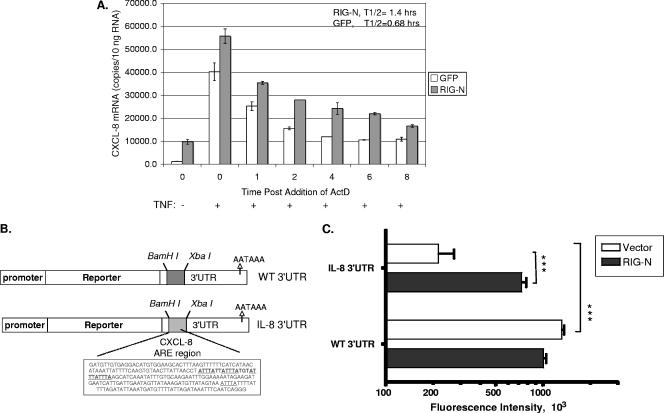
In addition to transcriptional activation, modulation of the half-life of CXCL-8 mRNA plays a major role in CXCL-8 induction in response to various stimuli. For example, TNF-α and virus induction of CXCL-8 involve mRNA stabilization (19, 23, 31, 61) via the binding of regulatory proteins to AREs in the 3′UTR of the mRNA (1, 6, 9, 53). We therefore determined whether RIG-I induction of CXCL-8 also involves stabilization of CXCL-8 mRNA. Huh7 cells were transfected with plasmids expressing RIG-N or GFP and treated with TNF-α to induce CXCL-8 mRNA. Actinomycin D was added to cultures to stop transcription, and CXCL-8 mRNA was quantified by real-time RT-PCR (19). As shown in Fig. 6A, expression of RIG-N led to a 7.6-fold enhancement of basal CXCL-8 mRNA (P, <0.01). Furthermore, RIG-N expression was associated with a 2.1-fold increase in the half-life of CXCL-8 mRNA following TNF-α stimulation. Statistical comparison of the decay rates by a one-phase exponential model (19) showed that this difference is significant (P = 0.03).
FIG. 6.
RIG-N stabilizes CXCL-8 mRNA. (A) Huh7 cells were transfected with RIG-N- or GFP-expressing plasmids, and 24 h later, cells were treated with 10 ng/ml TNF-α for 2 h to induce CXCL-8 mRNA. Actinomycin D was then added to cell cultures to stop transcription, and total cellular RNA was isolated at the indicated times. CXCL-8 mRNA was quantitated by real-time RT-PCR, as described in Materials and Methods. (B) Schematic representation of the CXCL-8 ARE reporter genes. The EGFP protein was placed under the control of a constitutively active CMV promoter. The control of mRNA stability was mediated by sequences in the 3′UTR. The dashed box indicates a 200-nucleotide stuffer sequence which lacks AREs in the vector construct or 237 nucleotides from the CXCL-8 3′UTR that contains AREs. Underlined are the ARE pentamers (AUUUA). (C) Cells (3 × 104 cells per well) in black clear-bottomed 96-well plates were cotransfected with 25 ng of EGFP reporter vectors (CXCL-8 and WT) and 50 ng of modulator vectors (vector and RIG-N). After 48 h, the plates were read by bottom fluorescence. Data are presented as the means ± standard errors of four readings. ***, P values <0.001 using Student's t test.
Since AREs are involved in the posttranscriptional regulation of CXCL-8 mRNA, we performed experiments with an EGFP reporter gene fused to the ARE region of CXCL-8 (Fig. 6B). The CXCL-8 construct contained 237 nucleotides from the CXCL-8 3′UTR that included the ARE (60). A control vector included a sequence of 200 nucleotides which lacked AREs. The CXCL-8 ARE caused a sixfold reduction in EGFP fluorescence compared to that of the wild-type vector (Fig. 6C, compare open bars). Fluorescence levels correlated with mRNA levels as detected by quantitative PCR (data not shown), indicating that the GFP reporter system reflects mRNA changes. Cotransfection experiments with RIG-N demonstrated that RIG-N was able to increase EGFP fluorescence by threefold in the presence of the CXCL-8 ARE compared to that of the wild-type vector that did not contain the ARE (Fig. 6C, solid bars). Overall, these results indicate that RIG-N stabilized CXCL-8 mRNA via ARE-dependent pathways.
DISCUSSION
The inflammatory response to virus infection, which is usually beneficial to the host, is often deregulated in the context of chronic viral infections like those caused by HCV. Indeed, inductions of inflammatory cytokines and chemokines such as CXCL-6, CXCL-8, and TNF-α have been reported in patients with chronic hepatitis C (11, 13, 18, 39, 51). In this case, the inflammatory response may do more harm than good because deregulation of inflammatory cytokines and chemokines creates an environment within the liver that is harsh and leads to hepatocyte turnover and regeneration (7, 17, 35, 37, 57). It has been suggested that chronic inflammation is mechanistically involved in the establishment of cancer (41), in particular hepatocellular carcinoma (5), so a deregulated inflammatory response may also have severe pathological sequelae.
In the current report, we demonstrate that innate antiviral signaling pathways that sense dsRNA during virus infection also trigger inflammatory chemokine expression. RIG-I and IPS-1 induced CXCL-8 transcription, and this involved, as expected, NF-κB. A role for the ISRE site in the CXCL-8 promoter was defined, and we demonstrated for the first time that IRF-3, which is downstream of RIG-I and IPS-1, also activated CXCL-8 transcription by directly binding to the CXCL-8 promoter. Moreover, RIG-N stabilized CXCL-8 mRNA. We propose that the regulation of CXCL-8 production by dsRNA signaling pathways represents a link between innate antiviral and inflammatory pathways. Based on previous studies (24, 50), HCV NS5A and core proteins could also play a role in the CXCL-8 induction observed in the present study. In this case, the mechanisms involved could be independent of dsRNA signaling responses and involve other transcription factors such as NF-κB and AP-1, reflecting the classical mode of CXCL-8 induction.
CXCL-8 induction requires transcriptional activation of the CXCL-8 promoter. Formation of the CXCL-8 “enhanceosome” involves the coordinated assembly of multiple transcription factors (22). Under basal conditions, the CXCL-8 promoter is repressed by at least three mechanisms, including the binding of NF-κB repressing factor to a region that overlaps the NF-κB binding site, the deacetylation of histone proteins by histone deacetylase I (HDAC1), and the binding of octamer-1 (OCT-1) to a complementary site of the CAAT/enhancer-binding protein (C/EBP) binding sites (22). During transcriptional activation, NF-κB enters the nucleus and displaces NRF, OCT-1 is replaced by C/EBP, and CREB-binding protein (CBP)/p300 is recruited, which causes histone acetylation and remodeling of chromatin. In the current report, we demonstrate that NF-κB, AP-1, NF-IL-6, and IRF-3 participate in CXCL-8 transcriptional induction in response to HCV infection. Our data indicate that similar to RSV, HCV induces multiple transcription factors to bind to the CXCL-8 promoter. In other systems, NF-κB binding is essential to CXCL-8 transcriptional induction (42). AP-1, C/EBP, and NF-IL-6 are not required for transcriptional activation in some cells (22, 42), so it is thought that these transcription factors provide maximal gene induction. An important, unresolved issue is whether AP-1 and NF-IL-6 involvement in CXCL-8 induction requires the activation of AP-1 and NF-IL-6 signaling pathways or whether these transcription factors are noninducible and simply serve to amplify the effect of inducible IRF-3 and NF-κB. For example, it has been demonstrated that while the CXCL-8 AP-1 site is not TNF-α inducible, a mutation of the site significantly reduced TNF-α induction of the native CXCL-8 promoter (58).
RIG-I signaling is central for triggering the host response to HCV infection (34). It has been demonstrated that the HCV NS3 protease blocks RIG-I-dependent signaling of IRF-3 and NF-κB activation by its targeted proteolysis of IPS-1 (10, 34, 38). However, the host response may be transiently induced during early points of HCV infection prior to control by NS3/4A (34). Our data now indicate that the RIG-I pathway signaling through IPS-1 can also drive the expression of CXCL-8, thus connecting innate defense signaling to the inflammatory response to virus infection. We found that IRF-3 and NF-κB participate to induce CXCL-8 expression, suggesting that HCV might impose a similar blockade on CXCL-8 induction. We also note that among other transcription factors, NF-κB is essential for CXCL-8 induction and is responsive to many inflammatory stimuli other than dsRNA or virus infection (22). Thus, it is possible that viral triggering and control of CXCL-8 induction will depend on the cumulative and time-dependent cross-talk among signaling networks that is triggered and activated by NF-κB during HCV infection, with the subtle modulation imposed by the dsRNA activation of RIG-I signaling to IRF-3 serving to fine tune the response. Furthermore, while it has been suggested that IPS-1 mediates the bifurcation of the NF-κB and IRF-3 activation pathways, it remains possible that other adaptors of the RIG-I pathway are involved in this process (21).
Although transcriptional activation often plays a central role in gene expression, in certain situations, mRNA stabilization plays a more prominent role (1, 28). For instance, collagen expression in hepatic stellate cells is increased primarily through mRNA stabilization (54). Moreover, we have recently shown that Huh7 and replicon cells constitutively express CXCL-8 mRNA that is regulated posttranscriptionally (19). In the current study, we showed that RIG-N stabilized CXCL-8 mRNA, suggesting that a consequence of dsRNA signaling includes the activation of mRNA stabilizing responses. Note, however, that while ARE-mediated pathways primarily regulate mRNA turnover, they may also influence translation. However, it is likely that the RIG-N effect described in the current report acts at the mRNA level, since the half-life of CXCL-8 was prolonged as demonstrated by the actinomycin D chase experiment. The mRNA stabilization was dependent on AREs in the 3′UTR, since RIG-N increased reporter activity from a vector containing the 3′UTR region that harbors CXCL-8 AREs but not with a 3′UTR that lacked AREs. The CXCL-8 ARE was able to destabilize the reporter activity in the absence of any modulator but also was able to respond to RIG-N-induced stabilization. Thus, this region contains the necessary ARE regulatory and accessory domains for both destabilization and RIG-N-induced stabilization. This region has been shown to contain two functionally different domains in which four AUUUA-containing domains are sufficient for p38 mitogen-activated protein kinase-induced stabilization (60). Thus, it is possible that RIG-N induces the p38 pathway via a yet-unknown process. Since CXCL-8 mRNA stabilization involves the binding of regulatory proteins to the 3′UTR of the mRNA that enhance its mRNA stability (22), it is possible that RIG-N modulates the activity of these proteins. We are currently evaluating these issues. Furthermore, TLR5 and TLR9 signaling results in CXCL-8 induction by increasing the stability of CXCL-8 mRNA (46, 67). The degree to which dsRNA pathways, both TLR3 dependent and independent, regulate CXCL-8 mRNA stability during virus infection requires further study.
In summary, we have shown that dsRNA antiviral pathways cross-talk with inflammatory pathways. The combined action of these pathways is central to the control of acute HCV infection and replication. However, because HCV disrupts antiviral and inflammatory pathways (26, 48), these virus-host interactions likely contribute to HCV persistence and the pathogenesis of HCV-associated liver disease.
Acknowledgments
We thank Francis Chisari, Michael David, John Hiscott, Naofumi Mukaida, and Apath, LLC, for reagents, Benjamin tenOever and Tom Maniatis for technical advice on ChIP assays, and Latifa Al-Haj, Paula McPoland, Jodie Powdrill, and Jeremiah Eng for technical assistance.
S.J.P. is partially supported by NIH grants DK62187 and U19AI66328, A.R.B. by grants AI40218 and AI062885, and M.G. by grants AI060389 and U19AI40035. T.W. is supported by grants from the Japanese Ministry of Health, Labor, and Welfare; the Japan Society for the Promotion of Science; and the Japan Health Science Foundation.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Bakheet, T., M. Frevel, B. R. G. Williams, W. Greer, and K. S. A. Khabar. 2001. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29:246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Bonte, D., C. Francois, S. Castelain, V. Morel, J. Roussel, C. Wychowski, J. Dubuisson, E. Meurs, and G. Duverlie. 2002. Induction of the IL-8 chemokine by the hepatitis C virus NS5A nonstructural protein in an infectious system. Hepatology 36:A495. [Google Scholar]
- 4.Bonte, D., C. Francois, S. Castelain, C. Wychowski, J. Dubuisson, E. F. Meurs, and G. Duverlie. 2004. Positive effect of the hepatitis C virus nonstructural 5A protein on viral multiplication. Arch. Virol. 149:1353-1371. [DOI] [PubMed] [Google Scholar]
- 5.Branda, M., and J. R. Wands. 2006. Signal transduction cascades and hepatitis B and C related hepatocellular carcinoma. Hepatology 43:891-902. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, D. J., and L. Tomanek. 2006. Herpes simplex virus and the chemokines that mediate the inflammation. Curr. Top. Microbiol. Immunol. 303:47-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casola, A., R. P. Garofalo, M. Jamaluddin, S. Vlahopoulos, and A. R. Brasier. 2000. Requirement of a novel upstream response element in respiratory syncytial virus-induced IL-8 gene expression. J. Immunol. 164:5944-5951. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, G., J. Zhong, and F. V. Chisari. 2006. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 103:8499-8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang, E., A. Del Vecchio, S. Smolinski, X. Y. Song, and R. T. Sarisky. 2004. Biomedicines to reduce inflammation but not viral load in chronic HCV—what's the sense? Trends Biotechnol. 22:517-523. [DOI] [PubMed] [Google Scholar]
- 12.Davis, G. L., J. E. Albright, S.F. Cook, and D. M. Rosenberg. 2003. Projecting future complications of chronic hepatitis C in the United States. Liver Transplant. 9:331-338. [DOI] [PubMed] [Google Scholar]
- 13.Falasca, K., C. Ucciferri, M. Dalessandro, P. Zingariello, P. Mancino, C. Petrarca, E. Pizzigallo, P. Conti, and J. Vecchiet. 2006. Cytokine patterns correlate with liver damage in patients with chronic hepatitis B and C. Ann. Clin. Lab. Sci. 36:144-150. [PubMed] [Google Scholar]
- 14.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 16.Girard, S., P. Shalhoub, P. Lescure, A. Sabile, D. E. Misek, S. Hanash, C. Brechot, and L. Beretta. 2002. An altered cellular response to interferon and up-regulation of interleukin-8 induced by the hepatitis C viral protein NS5A uncovered by microarray analysis. Virology 295:272-283. [DOI] [PubMed] [Google Scholar]
- 17.Glass, W. G., H. F. Rosenberg, and P. M. Murphy. 2003. Chemokine regulation of inflammation during acute viral infection. Curr. Opin. Allergy Clin. Immunol. 3:467-473. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Amaro, R., C. Garcia-Monzon, L. Garcia-Buey, R. Moreno-Otero, J. L. Alonso, E. Yague, J. P. Pivel, M. Lopez-Cabrera, E. Fernandez-Ruiz, and F. Sanchez-Madrid. 1994. Induction of tumor necrosis factor alpha production by human hepatocytes in chronic viral hepatitis. J. Exp. Med. 179:841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, J., K. S. Khabar, B. C. Koo, B. R. Williams, and S. J. Polyak. 2006. Stability of CXCL-8 and related AU-Rich mRNAs in the context of hepatitis C virus replication in vitro. J. Infect. Dis. 193:802-811. [DOI] [PubMed] [Google Scholar]
- 20.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010:237-248. [DOI] [PubMed] [Google Scholar]
- 21.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847-855. [PubMed] [Google Scholar]
- 23.Holtmann, H., R. Winzen, P. Holland, S. Eickemeier, E. Hoffmann, D. Wallach, N. L. Malinin, J. A. Cooper, K. Resch, and M. Kracht. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshida, Y., N. Kato, H. Yoshida, Y. Wang, M. Tanaka, T. Goto, M. Otsuka, H. Taniguchi, M. Moriyama, F. Imazeki, O. Yokosuka, T. Kawabe, Y. Shiratori, and M. Omata. 2005. Hepatitis C virus core protein and hepatitis activity are associated through transactivation of interleukin-8. J. Infect. Dis. 192:266-275. [DOI] [PubMed] [Google Scholar]
- 25.Imaizumi, T., S. Aratani, T. Nakajima, M. Carlson, T. Matsumiya, K. Tanji, K. Ookawa, H. Yoshida, S. Tsuchida, T. M. McIntyre, S. M. Prescott, G. A. Zimmerman, and K. Satoh. 2002. Retinoic acid-inducible gene-I is induced in endothelial cells by LPS and regulates expression of COX-2. Biochem. Biophys. Res. Commun. 292:274-279. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, C. L., and M. Gale, Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 27:1-4. [DOI] [PubMed] [Google Scholar]
- 27.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 28.Khabar, K. S. 2005. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J. Interferon Cytokine Res. 25:1-10. [DOI] [PubMed] [Google Scholar]
- 29.Khabar, K. S., F. Al-Zoghaibi, M. N. Al-Ahdal, T. Murayama, M. Dhalla, N. Mukaida, M. Taha, S. T. Al-Sedairy, Y. Siddiqui, G. Kessie, and K. Matsushima. 1997. The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 186:1077-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo, B. C. A., P. McPoland, J. P. Wagoner, O. J. Kane, V. Lohmann, and S. J. Polyak. 2006. Relationships between hepatitis C virus replication and CXCL-8 production in vitro. J. Virol. 80:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leland Booth, J., and J. P. Metcalf. 1999. Type-specific induction of interleukin-8 by adenovirus. Am. J. Respir. Cell Mol. Biol. 21:521-527. [DOI] [PubMed] [Google Scholar]
- 32.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay, C. R. 2001. Chemokines: immunology's high impact factors. Nat. Immunol. 2:95-101. [DOI] [PubMed] [Google Scholar]
- 36.Mamane, Y., C. Heylboreck, P. Genei, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 37.Melchjorsen, J., L. N. Sorensen, and S. R. Paludan. 2003. Expression and function of chemokines during viral infections: from molecular mechanisms to in vivo function. J. Leukoc. Biol. 74:331-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172 [DOI] [PubMed] [Google Scholar]
- 39.Mihm, S., E. Herrmann, U. Sarrazin, M. V. Wagner, B. Kronenberger, S. Zeuzem, and C. Sarrazin. 2004. Association of serum interleukin-8 with virologic response to antiviral therapy in patients with chronic hepatitis C. J. Hepatol. 40:845-852. [DOI] [PubMed] [Google Scholar]
- 40.Miller, K., S. McArdle, M. J. Gale, Jr., D. A. Geller, B. Tenoever, J. Hiscott, D. R. Gretch, and S. J. Polyak. 2004. Effects of the hepatitis C virus core protein on innate cellular defense pathways. J. Interferon Cytokine Res. 24:391-402. [DOI] [PubMed] [Google Scholar]
- 41.Moss, S. F., and M. J. Blaser. 2005. Mechanisms of disease: inflammation and the origins of cancer. Nat. Clin. Pract. Oncol. 2:90-97. [DOI] [PubMed] [Google Scholar]
- 42.Mukaida, N. 2000. Interleukin-8: an expanding universe beyond neutrophil chemotaxis and activation. Int. J. Hematol. 72:391-398. [PubMed] [Google Scholar]
- 43.Mukaida, N., A. Harada, K. Yasumoto, and K. Matsushima. 1992. Properties of ipronflammatory cell type-specific leukocyte chemotactic cytokines, interleukin 8 (IL-8) and monocyte chemotactic and activating factor (MCAF). Microbiol. Immunol. 36:773-789. [DOI] [PubMed] [Google Scholar]
- 44.Mukaida, N., M. Shiroo, and K. Matsushima. 1989. Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J. Immunol. 143:1366-1371. [PubMed] [Google Scholar]
- 45.Murayama, T., N. Mukaida, K. S. Khabar, and K. Matsushima. 1998. Potential involvement of IL-8 in the pathogenesis of human cytomegalovirus infection. J. Leukoc. Biol. 64:62-67. [DOI] [PubMed] [Google Scholar]
- 46.Parilla, N. W., V. S. Hughes, K. M. Lierl, H. R. Wong, and K. Page. 2006. CpG DNA modulates interleukin 1 beta-induced interleukin-8 expression in human bronchial epithelial (16HBE14o-) cells. Respir. Res. 7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plumlee, C. R., C. A. Lazaro, N. Fausto, and S. J. Polyak. 2005. Effect of ethanol on innate antiviral pathways and HCV replication in human liver cells. Virol. J. 2:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polyak, S. J. 2006. Resistance of hepatitis C virus to the host antiviral response. Future Virology. 1:89-98. [Google Scholar]
- 49.Polyak, S. J., D. Paschal, S. McArdle, M. Gale, D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus non-structural 5a protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 50.Polyak, S. J., K. S. A. Khabar, D. M. Paschal, H. J. Ezelle, G. Duverlie, G. N. Barber, D. E. Levy, N. Mukaida, and D. R. Gretch. 2001. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J. Virol. 75:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polyak, S. J., K. S. A. Khabar, M. Rezeiq, and D. R. Gretch. 2001. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J. Virol. 75:6209-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 53.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 54.Stefanovic, B., C. Hellerbrand, M. Holcik, M. Briendl, S. A. Liebhaber, and D. A. Brenner. 1997. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumpter, R., Jr., Y.-M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, D. L., J. Astemborski, R. M. Rai, F. A. Anania, M. Schaeffer, N. Galai, K. Nolt, K. E. Nelson, S. A. Strathdee, L. Johnson, O. Laeyendecker, J. Boitnott, L. E. Wilson, and D. Vlahov. 2000. The natural history of hepatitis C virus infection—-host, viral, and environmental factors. JAMA 284:450-456. [DOI] [PubMed] [Google Scholar]
- 57.Tripp, R. A., C. Oshansky, and R. Alvarez. 2005. Cytokines and respiratory syncytial virus infection. Proc. Am. Thorac. Soc. 2:147-149. [DOI] [PubMed] [Google Scholar]
- 58.Vlahopoulos, S., I. Boldogh, A. Casola, and A. R. Brasier. 1999. Nuclear factor-kappaB-dependent induction of interleukin-8 gene expression by tumor necrosis factor alpha: evidence for an antioxidant sensitive activating pathway distinct from nuclear translocation. Blood 94:1878-1889. [PubMed] [Google Scholar]
- 59.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winzen, R., G. Gowrishankar, F. Bollig, N. Redich, K. Resch, and H. Holtmann. 2004. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol. Cell. Biol. 24:4835-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.World Health Organization and Viral Hepatitis Prevention Board, Antwerp, Belgium. 1999. Global surveillance and control of hepatitis C. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 63.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 64.Yagita, H., T. Nakamura, H. Karasuyama, and K. Okumura. 1989. Monoclonal antibodies specific for murine CD2 reveal its presence on B as well as T cells. Proc. Natl. Acad. Sci. USA 86:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaoka, Y., T. Kudo, H. Lu, A. Casola, A. R. Brasier, and D. Y. Graham. 2004. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology 126:1030-1043. [DOI] [PubMed] [Google Scholar]
- 66.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 67.Yu, Y., H. Zeng, S. Lyons, A. Carlson, D. Merlin, A. S. Neish, and A. T. Gewirtz. 2003. TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 285:G282-G290. [DOI] [PubMed] [Google Scholar]
- 68.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]