Abstract
It has been shown previously that the fusion glycoprotein of human respiratory syncytial virus (RSV-F) interacts with cellular heparan sulfate. Synthetic overlapping peptides derived from the F-protein sequence of RSV subtype A (strain A2) were tested for their ability to bind heparin using heparin-agarose affinity chromatography (HAAC). This evaluation identified 15 peptides representing eight linear heparin-binding domains (HBDs) located within F1 and F2 and spanning the protease cleavage activation site. All peptides bound to Vero and A549 cells, and binding was inhibited by soluble heparins and diminished by either enzymatic treatment to remove cell surface glycosaminoglycans or by treatment with sodium chlorate to decrease cellular sulfation. RSV-F HBD peptides were less likely to bind to glycosaminoglycan-deficient CHO-745 cells than parental CHO-K1 cells that express these molecules. Three RSV-F HBD peptides (F16, F26, and F55) inhibited virus infectivity; two of these peptides (F16 and F55) inhibited binding of virus to Vero cells, while the third (F26) did not. These studies provided evidence that two of the linear HBDs mapped by peptides F16 and F55 may mediate one of the first steps in the attachment of virus to cells while the third, F26, inhibited infectivity at a postattachment step, suggesting that interactions with cell surface glycosaminoglycans may play a role in infectivity of some RSV strains.
Human respiratory syncytial virus (RSV), which belongs to the Pneumovirus genus of the Paramyxoviridae family, is the major cause of lower respiratory tract disease in infants and young children worldwide and is responsible for a large proportion of lower respiratory tract infections in the elderly. RSV has a genome approximately 15,222 nucleotides long that carries 10 major mRNAs that produce 11 viral proteins. Three of these are associated with the virion envelope and include the attachment (G) glycoprotein, the fusion (F) glycoprotein, and a small hydrophobic protein of unknown function (for a review, see reference 12).
Like other paramyxoviruses, RSV is thought to initiate infection by attaching to the host cell using the envelope glycoproteins. It is postulated that following binding, a series of changes in the conformation of F-protein trimers allows fusion of the viral envelope with the host cell membrane, permitting virus entry (39). Although specific cellular receptors for either the attachment or the fusion glycoprotein of RSV have not yet been identified, there is abundant evidence that RSV binds to cellular glycosaminoglycans (GAGs) (5, 16, 19, 20, 26, 27, 35, 38, 45, 47, 57, 58, 59). While it appears that these interactions facilitate virus entry, it does not rule out the possibility that alternative pathways also exist (26, 58).
GAGs are long, unbranched polysaccharide chains consisting of repeating disaccharide units of an amino sugar (either N-acetylglucosamine or N-acetylgalactosamine) and a hexuronic acid (either glucuronic acid or its epimer, l-iduronic acid) that are covalently attached via an O-glycosidic linkage between xylose in the sugar chain and serine in the host cell membrane protein to form proteoglycans. During GAG synthesis, the chain of sugar residues may be further modified by N- and/or O-sulfation, de-N-acetylation, and by varying the chain length. While GAGs are found on the surface of almost all animal cells, the number and position of sulfate groups, N-acetyl groups, chain length, as well as the total number of saccharide chains per protein molecule contribute to the observed heterogeneity in GAG expression, which in turn determines the specificity of interactions with a wide variety of binding proteins (43). Postsynthetic modifications of GAGs also appear to be tissue specific and developmentally regulated (1, 4, 6, 31, 54).
Many viruses depend on cell surface GAGs to initiate infection in vitro (7, 9, 10, 14, 32, 34, 37, 41, 52, 55, 56, 62, 63). Previous work demonstrated that RSV also interacts with cell surface GAGs, as attachment and infectivity were diminished in the presence of soluble GAGs such as heparan sulfate and chondroitin sulfate B (20, 26, 45) or decreased by enzymatic removal of heparin-like GAGs following treatment with GAG lyases and by pretreatment of cells with chlorate to decrease cellular sulfation (26). In a series of experiments using CHO cell lines deficient in individual enzymes required for postsynthetic GAG modification and in virus inhibition studies using chemically modified heparin chains of varying lengths, Hallak et al. showed that RSV infection was dependent on the specific expression of cellular GAGs containing l-iduronic acid as well as N-sulfated residues and had a minimum structural requirement for 10 saccharide units (26).
The RSV-G protein has been recognized as the putative attachment protein, based on experiments using antiserum raised against purified G glycoprotein that diminished binding of RSV virus to HEp-2 cells while antiserum raised against RSV-F did not (42). Previous work in our laboratory showed that RSV-G bound immobilized heparan sulfate with high affinity, while others have demonstrated that soluble RSV-G bound to cell surface GAGs (20, 45). Subsequently, we used overlapping synthetic peptides to identify a linear heparin-binding region within the G ectodomains of subtype A and B viruses, represented by the sequences 184AICKRIPNKKPGKKT198 and 183KSICKTIPSNKPKKK197, respectively, that bound immobilized heparin as well as cell surface GAGs and that inhibited infection of Vero cell monolayers with the homologous subtype (20). RSV-G heparin-binding linear peptides were characterized by the motif XBBX, identified in other mammalian heparin-binding proteins, where B represents a positively charged basic residue and X represents any other uncharged amino acid (for a review, see reference 8). This suggests that RSV-G binds to cells via charge-charge interactions between basic residues in the heparin-binding domain and negatively charged sulfates present on cell surface GAGs. However, these interactions between the G heparin-binding domain and cellular GAGs do not appear to be essential for virus infectivity, because viruses engineered as G-HBD deletion mutants replicated in tissue culture and in the respiratory tract of infected mice, and viruses lacking RSV-G entirely were infectious in vitro but were attenuated in vivo (36, 58). Coincidentally, the G-HBD mimics the fractalkine-binding site that mediates attachment to CX3CR1 and, like fractalkine, RSV-G binding to CX3CR1 triggers a cell-signaling cascade (61). It is not known if heparin binding is a prerequisite that promotes high-affinity interactions between RSV-G and CX3CR1.
Interestingly, RSV viruses lacking RSV-G were still sensitive to inhibition by soluble heparin (19, 57, 58). Also, RSV-F derived from either a subtype A virus, A2, or a subtype B virus, cp52, bound immobilized heparin (19). Taken together, these data indicated that the fusion glycoprotein could function as the attachment protein in the absence of RSV-G and that at least some of the RSV-F cell interactions that facilitated virus attachment and infectivity were with cellular GAGs (19, 57, 58).
RSV-F is initially translated into a 70-kDa inactive precursor, F0, which is cotranslationally modified by the addition of N-linked carbohydrates (11, 21). F0 is then endoproteolytically cleaved by a cellular trypsin-like protease, furin, at one or both of the two known highly basic cleavage sites into two disulfide-linked subunits, consisting of F1 and F2 of varying molecular weights depending upon the cleavage site(s) used (23, 24, 65). Upon cleavage, the highly conserved hydrophobic fusion peptide containing the FLG sequence at or near the amino terminus of F1 is thought to facilitate virus fusion by insertion into the target cell membrane (for a review, see reference 29). In addition to the basic regions found at each furin cleavage site, 106RARR109 and 131KKRKRR136, four other potential heparin-binding domains have been discussed but not experimentally defined (57). In the present study, we have used overlapping synthetic RSV-F peptides to map linear domains within the fusion protein that are important for heparin-binding and characterized these interactions on cells permissive for RSV infection. We provide evidence that eight regions in RSV-F bind heparin and cellular GAGs and three regions facilitate infection at either an attachment or postattachment step.
MATERIALS AND METHODS
Cells and viruses.
Vero, HEp-2, and A549 cells were grown in Eagle's medium containing Earle's salts (EMEM) (Mediatech, Inc., Herndon, VA) with 10% fetal bovine serum (FBS) (Cambrex, Walkersville, MD), 4 mM l-glutamine (Mediatech, Inc.), amphotericin B at 250 μg/ml (Quality Biological, Gaithersburg, MD), and 10,000 U/ml penicillin-streptomycin (Quality Biological). Chinese hamster ovary (CHO) K1 and 745 cells were grown in Hams F12 medium (Mediatech, Inc.) with 10% FBS. Human respiratory syncytial virus (RSV) strains A2 and 18537 and RSV/B1/cp52 (cp52) virus stocks were obtained from R. Chanock, B. Murphy, and S. Whitehead, LID, National Institute of Allergy and Infecious Disease, National Institutes of Health, and prepared in either HEp-2 (for strains A2 and 18537) or Vero cell (for cp52) monolayers as previously described, and aliquots were stored at −70°C (19). Virus was sucrose purified as previously described (47). Infectivity of virus stocks was determined on Vero cell monolayers and calculated using the method of Reed and Muench; titers are reported as 50% tissue culture infectious doses (33).
Synthesis of RSV-F overlapping peptides.
Amino acid sequences for F peptides were deduced from F-gene nucleotide sequences for RSV subtype A strains with four amino acid mutations in the consensus that distinguish it from the published sequence for RSV-A2-F (RSH1CE; accession number M11486.1) (11). Seventy-seven peptides representing amino acids 26 to 574 of RSV-F were synthesized with biotin-serine-glycine-serine-glycine (SGSG) at their amino terminus and contained 17 amino acids of RSV-F sequence with a 10-amino-acid overlap and 7-amino-acid offset. Two additional RSV-F peptides, F78 and F79, were synthesized to represent the carboxy terminus of F2 (121N→R136) and amino terminus of F1 (137F→V152) following cleavage of F0, respectively. Peptide F79 was biotinylated at the carboxy terminus, and as such it contains a free amine. Peptides were synthesized by 9-fluorenylmethoxy carbonyl (Fmoc) solid-phase chemistry (PharmAust Chemistry Ltd., Australia; formerly Mimitopes, Raleigh, NC) with an average purity of 80 to 90%. Peptide RT2.2 (RT; 459ENREILKEPVHGVYY474), derived from the human immunodeficiency virus type 1 (HIV-1) consensus sequence of the pol gene from the Los Alamos database, was used as a negative control peptide. Peptide VN (346AKKQRFRHRNRKG358) was derived from vitronectin and represented a peptide with a known mammalian consensus heparin-binding motif, XBBXBX (boldface letters represent basic residues) in heparin-agarose affinity chromatography (HAAC) and cell-binding assays (13). Peptide F24 (187VLTSKVLDLKNYIDKQL203) represents an RSV-F cell-binding peptide that does not bind heparin. Biotinylated peptides representing the RSV-G heparin-binding domain, Ga19 and Ga20, have been previously described (20). Biotinylation efficiency of synthetic peptides was compared prior to use.
Polyanion-binding assay.
Individual peptides were screened in a polyanion-binding assay as previously described with the following modifications (7). CARBO-BIND plates (Corning Inc., Corning, NY) coated with or without heparin were blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h at room temperature. Biotinylated peptides were added to coated and uncoated wells starting at an initial concentration of 20 μg/ml (50 μl per well) in PBS and using serial twofold dilutions, followed by addition of streptavidin-horseradish peroxidase (1:500) (Kirkegaard & Perry Laboratories [KPL], Gaithersburg, MD) and the colorimetric substrate 2,2′-azino-di-(3-ethylbenzthizoline)-6-sulfonate (ABTS) (KPL). Absorbances were read at 405 nm, and specific binding was determined by subtracting the absorbance obtained for each peptide in uncoated wells from the corresponding absorbance in heparin-coated wells. Values that were at least two times greater than the corrected value obtained for peptide RT2.2 were considered heparin binding. Heparin-binding activity was confirmed using heparin-agarose affinity chromatography, described below.
HAAC.
Peptides reactive in the polyanion-binding assay were evaluated by HAAC as described previously (20), with some modifications. Heparin-agarose beads or cross-linked 4% Sepharose CL4B beads (Sigma, St. Louis, MO) were equilibrated with 1 ml PBS at room temperature for 30 min on an orbital rocker. Each peptide was tested at a concentration of 12.5 μg/ml in PBS with the exception of peptides F36, F39, and F75, which were tested at 50 μg/ml. Peptides (200 μl) were incubated with 100 μl of heparin-coated or control agarose beads at room temperature for 30 min and then washed five times with 1 ml PBS. Peptides were eluted using 200 μl of PBS containing 2 mg/ml low-molecular-weight heparin (LMWH) (porcine intestinal mucosa; Mr = 6,000; Sigma) or, where noted, bovine lung heparin (BLH) (Calbiochem, La Jolla, CA) and specific elution revealed by the peptide detection assay described below. Final washes were tested in parallel with the corresponding eluates to ensure that the majority of unbound peptide was removed prior to specific elution.
Peptide detection assay.
Each final wash and eluate from heparin-coated and control agarose beads were subjected to serial threefold dilutions in PBS starting with 50 μl of undiluted material in 96-well polystyrene plates (Immunlon 1B), and biotinylated peptides were detected as previously described (20). Peptides were considered heparin binding if all of the following criteria were met: (i) the optical density at 405 nm (OD405) of each peptide eluted from heparin-agarose beads was at least two times the absorbance of the corresponding final wash for that peptide at the same dilution, (ii) the absorbance (OD405) of each peptide eluted from heparin-agarose beads was at least two times greater than the signal obtained for the same peptide eluted from control agarose beads, and (iii) the absorbance (OD405) of each peptide eluted from heparin-agarose beads was at least two times greater than the value obtained for peptide RT2.2 after specific elution with heparin.
Detection of peptide binding to the cell surface.
Biotinylated peptides were tested for their ability to bind to live cells as previously described (see the legend to Fig. 2 for details as well as reference 20). RSV-F peptides with minimum mean absorbance values greater than 0.1 were considered to be cell binding, as this cutoff was more than three standard deviations above the mean value obtained for the negative control peptide, RT2.2, which had mean absorbance values of <0.01 in this assay. Cells were visualized microscopically to be certain that monolayers were intact following addition of ABTS and prior to reading. Binding to Vero, HEp-2, and A549 cells was evaluated in separate experiments.
FIG. 2.
Peptide binding to Vero cells. Biotinylated peptides (10 μg/ml) were incubated for 1 h at 4°C on Vero cells, washed with PBS, and fixed with 4% paraformaldehyde. Cell-binding peptides were detected with streptavidin-horseradish peroxidase/ABTS. Absorbances were read at 405 nm, and OD values greater than 0.1 were considered positive. Error bars indicate the standard deviations of the mean OD readings.
Detection of peptide binding to the cell surface using flow cytometry.
Confluent Vero cells were removed from culture flasks using 50 mM EDTA and washed three times with fluorescent-activated cell sorter (FACS) buffer (PBS, without calcium or magnesium, with 1% FBS). Cells (5 × 105 per sample) were incubated with 10 μg/ml (in 100 μl) of each RSV-F peptide, VN peptide, peptide RT2.2, heparan sulfate monoclonal antibody HepSS-1, F5810E4 (Seikagaku, Tokyo, Japan), or an immunoglobulin M (IgM) isotype control monoclonal antibody (Coulter, Miami, FL) for 1 h at 4°C in the dark, followed by three washes with FACS buffer. Streptavidin-fluorescein isothiocyanate (FITC) (KPL) at 1:100 or goat anti-mouse (IgG, IgA, and IgM)-FITC (KPL) at 1:20 added to peptide-cell mixtures or antibody-cell mixtures, respectively, and incubated for 1 h at 4°C in the dark was used to detect binding of biotinylated peptides or heparin sulfate-specific monoclonal antibodies to cells, respectively. After three washes, samples were analyzed on a Becton-Dickinson LSR flow cytometer using CellQuest software.
Characterization of peptide-cell surface GAG interactions. (i) GAG lyase treatment of cells.
Peptide binding was analyzed by flow cytometry following treatment of Vero cells with lyases that cleave specific GAGs as a first step in evaluating the dependence on cell surface GAG expression. Vero cells removed from flasks using 50 mM EDTA were washed three times in PBS containing 1% BSA. Cells (5 × 105 per sample) were untreated or were treated with 4 U chondroitinase C (Sigma) or a mixture of 4 U heparin lyase I (Sigma) (that preferentially cleaves highly sulfated glycosaminoglycans at l-iduronic acid-2-O-sulfate) and 4 U heparin lyase III (Seikagaku) (that preferentially cleaves unsulfated regions within glycosaminoglycans at either l-iduronic or glucuronic acid) in PBS containing 0.5 mM MgCl2, 0.5 mM CaCl2, and 0.5% BSA, incubated for 3 h at 37°C, and then washed in FACS buffer. Peptide binding was determined by flow cytometry as described above. Data are reported for experiments that demonstrated a significant decrease in mean fluorescence for binding of heparan sulfate-specific monoclonal antibodies and for VN peptide on lyase-treated cells relative to mean fluorescence on untreated cells.
(ii) Detection of peptide binding to CHO cells.
To further characterize the dependence of peptide binding with cell surface GAG molecules, reactivity of each peptide with parental CHO-K1 and GAG-deficient CHO-745 cells was compared in parallel using flow cytometry as described above, except peptides were tested at 50 μg/ml.
(iii) Sodium chlorate treatment of cells.
Reactivity of peptides on sodium chlorate-treated cells was examined to determine if binding was dependent on cellular sulfation. Vero cells were grown in sulfate-deficient medium consisting of Joklick's minimal essential medium (Cambrex) with 10% dialyzed FBS, 4 mM l-glutamine, 250 μg/ml amphotericin B, 200 mg/liter CaCl2, and 50 mM sodium chlorate (Sigma) for 24 h at 36°C. Untreated controls cells were grown in EMEM containing 10% FBS. Cell binding was determined by flow cytometry as described above. Data are reported for experiments that showed a significant decrease in mean fluorescence intensity of binding for heparan sulfate-specific monoclonal antibodies and for the VN peptide on chlorate-treated cells relative to the mean fluorescence intensity on cells cultured in EMEM in the absence of chlorate.
(iv) Competitive binding studies.
To further evaluate the specificity of the interaction between peptides and sulfated versus that of unsulfated cellular glycosaminoglycans, peptide binding to cell surface molecules was assessed in the presence of soluble competitors. Dose curves were generated for each peptide to determine peptide concentrations that were equivalent to 50% of saturation on live cell monolayers as described previously (20). Binding of RSV-F peptides at the indicated concentration was compared in the presence and absence of 2 mg/ml of LMWH (Sigma), BLH (Sigma), hyaluronic acid (Sigma), K5 polysaccharide (K5) (kindly provided by Willie Vann, CBER, FDA), sucrose octasulfate (SOS) (Sigma), or aurintricarboxylic acid (ATA) (Sigma). Percent inhibition was determined by comparing peptide binding in the absence of competitor to binding in wells containing the soluble competitor.
(v) Peptide inhibition of virus binding to cells.
Saturating and 50% saturation concentrations were determined for peptides and sucrose gradient-purified A2 virus, respectively, on Vero cell monolayers. Binding of sucrose gradient-purified A2 virus was compared in the presence and absence of saturating amounts of peptide. Briefly, peptide-virus mixtures were incubated on Vero cell monolayers for 1 h at 4°C followed by three washes with PBS. Cells were fixed with 4% paraformaldehyde, air dried, and blocked with 5% BSA in PBS-Tween. Virus was detected using a pool of RSV-F monoclonal antibodies diluted 1:1,000, and plates were incubated overnight at room temperature and washed three times with PBS-Tween prior to addition of goat anti-mouse-IgG conjugated to horseradish peroxidase (1:1,000) for 3 h at 37°C. Plates were washed three times with PBS-Tween before adding ABTS substrate and reading absorbances at 405 nm. Percent inhibition of virus binding was determined by comparing the absorbance in the presence of the RSV-F peptide to binding of virus in the presence of an irrelevant peptide or binding in peptide-free wells. The RSV-F monoclonal antibody pool used to detect virus binding did not bind any of the F peptides used in these studies (data not shown).
(vi) Infectivity inhibition.
Peptides were assayed for their ability to inhibit RSV A2, 18537, and cp52 infectivity as previously described (20) with the following modifications. Peptides were diluted to a concentration of 50 μg/ml, and 50-μl aliquots of each were added in triplicate to Vero cells in a 96-well plate. After 45 min of incubation at 4°C, virus (diluted to yield 20 to 40 plaques/well in 50 μl) was added to peptide-treated and untreated control wells. Virus-peptide mixtures were allowed to adsorb for 2 h at room temperature, after which monolayers were washed and overlaid with 1% carboxymethyl cellulose (ICN, Aurora, OH) in Liebovitz 15 media (Mediatech, Inc.) containing 5% FBS and antibiotics. Three days postinoculation for assays using A2 and 18537 and six days postinoculation of cp52, assays were fixed with 80% methanol and subjected to RSV-specific immunostain, as previously described (19). Plates were scanned and images of each well magnified in Microsoft Paint to facilitate plaque counting. Percent inhibition of virus infectivity was determined by comparing the numbers of plaques in peptide-treated wells to the numbers in untreated control wells. RSV-F-specific monoclonal antibodies with known neutralizing activity and a control monoclonal antibody that does not neutralize virus were diluted to 1:320 in serum-free EMEM and tested in parallel. Individual peptides were also evaluated in parallel on Vero monolayers for cellular toxicity using a metabolic inhibition test as previously described (15). Virus inhibition data are the means of at least two assays and are reported only for peptide concentrations that were nontoxic to cells.
Statistical analysis.
Peptide binding and virus inhibition in the presence and absence of peptides were compared using Student's t test, and results are expressed as the means ±1 standard deviation or as the means ±1 standard error of the mean as noted in the figure legends. Results having P values of <0.05 were considered significant.
RESULTS
RSV-F synthetic peptides bind in HAAC.
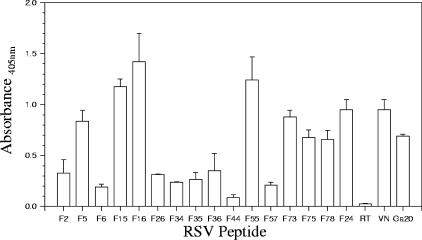
Synthetic peptides derived from the F-protein amino acid sequence were initially screened in a polyanion-binding assay to identify regions with heparin-binding activity (data not shown). Of the 79 RSV-F peptides tested, 18 bound in the polyanion-binding assay and were subsequently analyzed by HAAC to confirm reactivity (Fig. 1). Table 1 shows the sequence and relative positions of 15 RSV-F peptides (F2, F5, F6, F15, F16, F26, F34, F35, F36, F44, F55, F57, F73, F75, and F78) that bound immobilized heparin and were specifically eluted by soluble LMWH or BLH. Control peptides representing the putative vitronectin heparin-binding domain (VN) and the RSV-Ga HBD, peptide Ga20, also bound heparin agarose and were specifically eluted. Peptides RT2.2 and F24, which do not bind heparin, served as negative controls in this and in subsequent experiments. RSV-F peptides, VN, and Ga20 did not bind to cross-linked CL4B agarose lacking heparin, indicating that the binding observed in HAAC was heparin specific. Three RSV-F peptides (F12, F39, and F43) that reacted in the polyanion-binding assay were not heparin binding in HAAC tests, and these peptides were excluded from further analysis (Fig. 1).
FIG. 1.
HAAC of RSV-F biotinylated peptides. One hundred microliters of heparin-agarose (closed bars) or cross-linked 4% Sepharose CL4B beads (open bars) was equilibrated with 1 ml PBS prior to mixing with individual RSV-F or control peptides at a concentration of 12.5 μg/ml in PBS, with the exception of peptides F36, F39, and F75, which were tested at concentrations of 50 μg/ml. Samples were incubated at room temperature for 30 min, followed by five washes with 1 ml PBS. An aliquot of final wash from each reaction was collected and tested in order to ensure the majority of unbound peptide was removed. Peptides were eluted with 200 μl of PBS containing 2 mg/ml bovine lung heparin. Fifty microliters of final wash or eluted peptide was adsorbed to microtiter plates, and the presence of biotinylated peptides was confirmed by the peptide detection assay. Specific elution by soluble heparin was indicated if the absorbance (OD450) of the eluted peptides from the heparin-agarose was two or more times that seen in the final wash at the same dilution and two or more times that seen in the material eluted from Sepharose-CL4B beads for the same peptide. Data are representative of the means of one of at least two experiments,with the error bars indicating the standard deviations of the means.
TABLE 1.
Amino acid sequence of RSV-F heparin-binding and control peptides
| Peptidea | Region | Amino acid sequence |
|---|---|---|
| F2 | F2 | 33YQSTCSAVSKGYLSALR49 |
| F5 | F2 | 54TSVITIELSNIKKIKCN70 |
| F6 | F2 | 61LSNIKKIKCNGTDAKVK77 |
| F78 | F2 | 120NNAKKTNVTLSKKRKRR136 |
| F15 | F2-1 | 124KTNVTLSKKRKRRFLGF140 |
| F16 | F2-1 | 131KKRKRRFLGFLLGVGSA147 |
| F26 | F1 | 201KQLLPIVNKQSCSISNI217 |
| F34 | F1 | 257LLSLINDMPITNDQKKL273 |
| F35 | F1 | 264MPITNDQKKLMSNNVQI280 |
| F36 | F1 | 271KKLMSNNVQIVRQQSYS287 |
| F44 | F1 | 327KEGSNICLTRTDRGWYC343 |
| F55 | F1 | 404SSVITSLGAIVSCYGKT420 |
| F57 | F1 | 418GKTKCTASNKNRGIIKT434 |
| F73 | F1 | 530IIIVIIVILLSLIAVGL546 |
| F75 | F1 | 544VGLLLYCKARSTPVTLS560 |
| Ga19b | G | 184AICKRIPNKKPGKKT198 |
| Ga20b | G | 179NPTCWAICKRIPNKK193 |
| RT2.2b | HIV-1 pol | 459ENREILKEPVHGVYY474 |
| VN-HBDc | HBD consensus | 346AKKQRFRHRNRKG358 |
Peptides were biotinylated at the amino terminus through an SGSG linker via Fmoc chemistry. F denotes peptides representing the RSV fusion protein, Ga denotes peptides from the RSV subtype A attachment protein, RT2.2 is a peptide derived from the HIV pol gene, and VN denotes a consensus sequence for a heparin-binding domain derived from the vitronectin sequence. Peptides are depicted as single-letter codes.
Ga and RT2.2 peptides were derived from consensus sequences for RSV subtype A G or HIV pol gene sequences obtained the Los Alamos database.
From Feldman et al. (20).
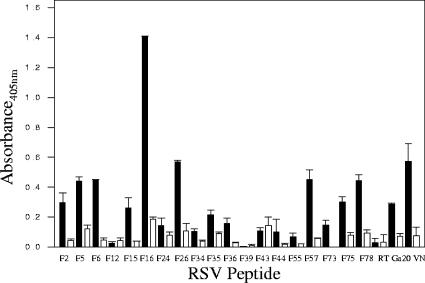
RSV-F peptide binding to Vero cells after GAG lyase treatment.
The ability of peptides to bind to RSV-permissive cells was examined using monolayers of live Vero, HEp-2, and A549 cells. As expected, VN and Ga20 peptides bound to each cell line tested, while RT2.2 did not bind. Each of the 15 RSV-F peptides identified as heparin binding by HAAC bound to Vero cells at a concentration of 10 μg/ml (Fig. 2). Similar reactivity was seen on A549 cells, while HEp-2 cells bound 12 of the 15 RSV-F peptides, including F2, F5, F15, F16, F26, F35, F36, F55, F57, F73, F75, and F78 (data not shown). Peptide binding on Vero cells was further evaluated using flow cytometry, and this method confirmed that each of the 15 RSV-F peptides bound to Vero cells, although binding of peptide F44 in this assay was noticeably diminished when mean fluorescence was compared to that seen with the other F heparin-binding peptides and to binding seen with VN (data not shown).
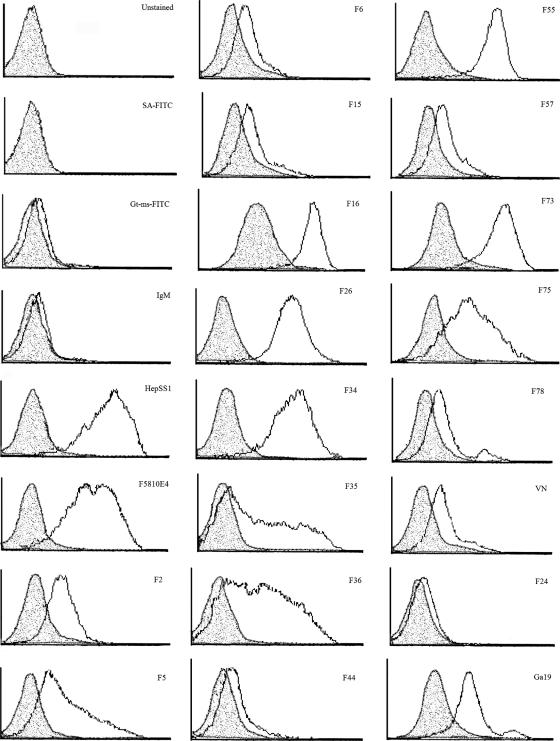
Flow cytometry was then used to assess peptide binding following GAG lyase treatment to reduce or remove GAGs from the cell surface. Binding of VN, Ga19, and each of the 15 RSV-F peptides was decreased following treatment with a mixture of heparin lyase I and III compared to binding on untreated cells (Fig. 3), while binding after chondroitinase C treatment was essentially unchanged (data not shown). Interestingly, binding of RSV-F peptides F16, F73, F75, and Ga19 was decreased but not abolished by GAG lyase treatment, suggesting that these peptides may bind to GAGs that are lyase resistant or to non-heparin molecules. Binding of heparan sulfate-specific monoclonal antibodies HepSS-1 and F5810E4 was abrogated following heparin lyase I and III treatment, indicating adequate digestion of cellular GAGs and suggesting that residual peptide binding following lyase treatment was not likely to be due to an inefficient digestion.
FIG. 3.
Peptide binding to Vero cells treated with GAG lyases. Vero cells (5 × 105 per sample) were treated in suspension for 2.5 h at 37°C without enzymes or in the presence of 4 U chondroitinase C (Sigma) or a mixture of 4 U heparin lyase I (Sigma) and 4 U heparin lyase III (Seikagaku, Japan) in GAG lyase buffer. Cells were then incubated with 10 μg/ml biotinylated peptide or HepSS1 or F5810E4 anti-heparin sulfate antibody for 1 h at 4°C, followed by three washes with FACS buffer. Goat anti-mouse (IgG, IgA, and IgM)-FITC at 1:20 or streptavidin-FITC at 1:100 was added to peptide cell mixtures for 1 h at 4°C in the dark to detect binding of anti-heparin sulfate monoclonal antibodies or biotinylated peptides, respectively, and were used in the absence of antibodies and peptides to measure background fluorescence due to nonspecific binding of the FITC conjugate. Samples were analyzed for staining on a Becton-Dickinson LSR flow cytometer. Unshaded histogram, binding to untreated cells; shaded histogram, binding after heparin lyase I plus heparin lyase III treatment.
RSV-F peptide binding on cells lacking GAGs.
To confirm that the RSV-F peptides identified by HAAC bind to cellular glycosaminoglycans, reactivity on parental CHO-K1 cells was compared to binding on mutant CHO-745 cells that lack xylose transferase and are reportedly GAG negative (17, 18). Cell binding of individual RSV-F peptides was examined by flow cytometry (Table 2). Many of the RSV-F peptides (7 of 15) showed an 80 to 98% reduction in mean fluorescence intensity on CHO-745 cells compared to binding on CHO-K1 cells, while four (F16, F57, F73, and F75) had only a modest 43 to 77% reduction.
TABLE 2.
Peptide binding to CHO-K1 (Gag-positive) and CHO-745 (Gag-deficient) cells
| Peptide or antibodya | % Positive for binding to:
|
% Reduction in binding | |
|---|---|---|---|
| CHO-K1 | CHO-745 | ||
| Peptide | |||
| F2 | 95 | 5 | 95 |
| F5 | 84 | 2 | 98 |
| F6 | 6 | 1 | NDc |
| F15 | 5 | 1 | ND |
| F16 | 99 | 27 | 73 |
| F26 | 98 | 4 | 96 |
| F34 | 98 | 2 | 98 |
| F35 | 55 | 1 | 98 |
| F36 | 87 | 3 | 97 |
| F44 | 5 | 2 | ND |
| F55 | 98 | 4 | 96 |
| F57 | 13 | 3 | 77 |
| F73 | 98 | 56 | 43 |
| F75 | 60 | 27 | 55 |
| F78 | 6 | 1 | ND |
| VN | 11 | 1 | 91 |
| Ga19 | 96 | 9 | 91 |
| Antibody | |||
| IgM control | 1 | 0 | ND |
| HepSS-1b | 97 | 1 | 99 |
| F5810E4b | 97 | 1 | 99 |
Biotinylated peptides (50 μg/ml) or antibodies were incubated with cells for 1 h at 4°C, followed by three washes with FACS buffer; peptide or antibody binding was evaluated by flow cytometry.
Heparan sulfate-specific monoclonal antibodies (Seikagaku).
ND, binding to CHO-K1 cells was <10%, so reduction in binding was not determined.
RSV-F peptide binding on cells following sodium chlorate treatment.
Peptide binding to Vero cells grown in sulfate-deficient medium in the presence of sodium chlorate was examined by flow cytometry and compared in parallel to binding on untreated Vero cells in order to identify peptides with sulfate-dependent interactions (Table 3). All peptides showed a greater than 70% decrease in binding with the exception of peptides F16 and F73, which demonstrated 35% and 49% decreases, respectively.
TABLE 3.
Peptide binding to Vero cells following chlorate treatmenta
| Peptide or antibody | % Positive with or without NaClO3 treatment
|
% Reduction in binding | |
|---|---|---|---|
| Without | With | ||
| Peptide | |||
| F2 | 29 | 3 | 90 |
| F5 | 93 | 3 | 97 |
| F6 | 26 | 2 | 92 |
| F15 | 31 | 3 | 90 |
| F16 | 98 | 64 | 35 |
| F26 | 95 | 28 | 71 |
| F34 | 93 | 13 | 86 |
| F35 | 44 | 2 | 95 |
| F36 | 65 | 2 | 97 |
| F44 | 15 | 4 | 73 |
| F55 | 98 | 24 | 76 |
| F57 | 47 | 4 | 91 |
| F73 | 98 | 50 | 49 |
| F75 | 73 | 21 | 71 |
| F78 | 36 | 4 | 89 |
| F24 | 43 | 4 | 91 |
| VN | 52 | 15 | 71 |
| Ga19 | 96 | 21 | 78 |
| Antibody | |||
| IgM negative control | 17 | 4 | 76 |
| HepSS-1b | 92 | 57 | 38 |
| F5810E4b | 92 | 77 | 16 |
Vero cells were grown for 24 h in sulfate-deficient media containing 50 mM sodium chlorate and then used to examine sulfate-dependent binding. Peptides (10 μg/ml) or antibodies were incubated on cells for 1 h at 4°C, followed by three washes with FACS buffer, and binding was determined by flow cytometry.
Heparan sulfate-specific monoclonal antibodies (Seikagaku).
RSV-F peptide binding to cells in the presence of soluble competitors.
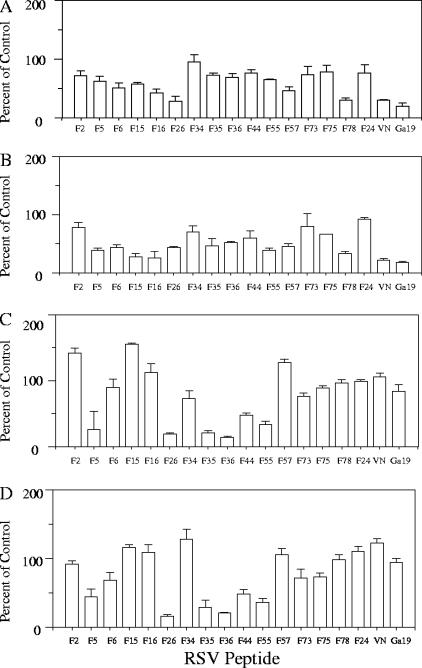
RSV-F peptide binding in the presence of soluble competitors was evaluated in order to characterize the specificity of each peptide interaction with cellular glycosaminoglycans and/or sulfated molecules. Peptide concentrations equal to 50% saturation were determined for each RSV-F and control peptide (data not shown). Binding was then examined in the presence or absence of LMWH, BLH, hyaluronic acid, and SOS (Fig. 4). Twelve of the 15 RSV-F peptides were inhibited by at least one of the soluble competitors. LMWH (Mr, ∼6,000 Da) reduced binding by approximately 50% for F6, F16, F26, F57, and F78. BLH inhibited binding of F5, F6, F15, F16, F26, F35, F36, F55, F57, and F78 by approximately 50%. Unsulfated, negatively charged hyaluronic acid inhibited binding of peptides that mapped to four regions, including a region in F2, represented by F5, the N-heptad repeat region of F1, represented by F26, and two domains in the stalk, represented by peptides F35, F36, F44, and F55. In the presence of SOS, F5, F26, F35, F36, F44, and F55 binding was diminished, while ATA decreased binding of only F16 and F35 by 50% or more and F55 by ∼45% (data not shown). An uncharged bacterial polysaccharide, K5, which lacks sulfate, carboxylic side chains, and iduronic acid, decreased binding of a single peptide, F73, which is also uncharged and hydrophobic (data not shown). Soluble competitors did not inhibit binding of F2, F34, or F75. Control peptides VN and Ga19 showed at least 50% inhibition in the presence of either LMWH or BLH, while F24, a non-heparin-binding peptide with cell-binding activity derived from the F ectodomain, showed no inhibition irrespective of competitor, as expected.
FIG. 4.
Inhibition of heparin-binding peptides by soluble GAGs. Binding of individual peptides in the presence of soluble heparin or other competitors was examined on Vero cell monolayers. Biotinylated peptide (at 50% saturation binding) and competitor were mixed for 30 min in a 96-well plate and then transferred to Vero monolayers, allowed to bind for 2 h at 4°C, and then washed and fixed with 4% paraformaldehyde. Cell-binding biotinylated peptides were detected using avidin-conjugated horseradish peroxidase. Data are representative of one of at least three experiments, with error bars indicating the standard deviations of the means. (A) Low-molecular-size heparin; (B) bovine lung heparin; (C) hyaluronic acid; (D) sucrose octasulfate.
Identification of RSV-F peptides that block virus binding.
The ability of each of the 15 RSV-F peptides to inhibit virus attachment was examined in competitive binding assays on Vero cell monolayers. Binding of sucrose gradient-purified A2 was significantly decreased in the presence of RSV-F peptides F15, F16, and F55, while the remaining 12 F peptides, peptide Ga20, and VN had little or no effect on A2 virus attachment (Fig. 5). The reciprocal experiment to examine the ability of purified virus to inhibit peptide binding revealed no inhibition (data not shown). Similarly, RSV-F peptides did not inhibit binding of purified F protein to Vero cells, and F55 was the only RSV-F peptide able to block binding of cp52 virus (data not shown).
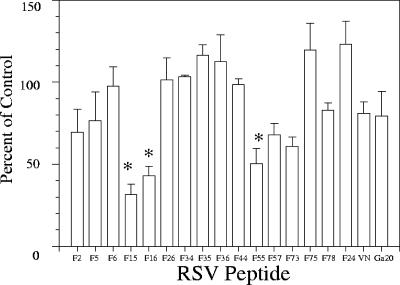
FIG. 5.
RSV-F heparin-binding peptides inhibit A2 binding to Vero cells. Binding of purified A2 in the presence of RSV-F HBD peptides was examined on Vero monolayers. Peptide (at saturation binding) was mixed with purified A2 used at 50% of saturation and then incubated on Vero cells for 1 h prior to washing and fixing with 4% paraformaldehyde. Monolayers were air dried and then blocked with 5% BSA-PBS-Tween and incubated overnight with an RSV-F monoclonal antibody pool, and virus binding was detected by enzyme-linked immunosorbent assay using goat anti-mouse horseradish peroxidase. Data compare binding of virus in peptide-containing wells to binding in the absence of peptide and are representative of one of at least two experiments, with error bars indicating the standard deviations of the means of duplicate wells. *, P < 0.05.
Inhibition of RSV infectivity by RSV-F heparin-binding peptides.
The ability of each peptide to inhibit infectivity of RSV subtype A (strain A2) and subtype B (18537 and cp52) was examined next. Twelve of the 15 RSV-F peptides as well as peptides VN and RT2.2 did not inhibit infectivity of any of the RSV strains tested (Fig. 6). In contrast, RSV-F peptides F16, F26, and F55 had inhibitory activity. Peptide F16 significantly inhibited A2 and 18537 infectivity by 69% and 55%, respectively. However, F16 had only minimal inhibitory activity against cp52 (28% reduction). RSV-F peptide F55 inhibited each of the three viruses tested by 73%, 64%, and 40% for strains A2, 18537, and cp52, respectively. Interestingly, RSV-F peptide F26 significantly decreased A2 infectivity by 33% compared to the negative control peptide RT2.2 (P < 0.05). However, F26 inhibition of 18537 was not significant, and it had no effect on cp52 infectivity.
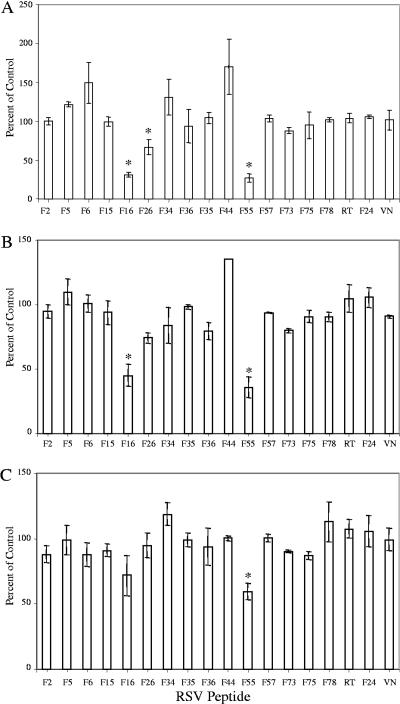
FIG. 6.
Inhibition of RSV infectivity by RSV-F HBD peptides. Vero cells were preincubated with 50 μl of peptide (50 μg/ml). Peptides were adsorbed for 45 min at 4°C, followed by the addition of virus (A2, panel A; 18537, panel B; cp52, panel C) for 2 h at room temperature. Cells were then washed to remove unbound virus and overlaid with 1% carboxymethyl cellulose in L15 medium. Three days (A2 and 18537) or 6 days (cp52) postinoculation, assays were fixed with 80% methanol and subjected to RSV-specific immunostain. Plaques were counted, and the percent inhibition of virus infectivity of treated wells was determined versus untreated control wells. The data represent the means of at least two separate experiments, with the error bars indicating the standard error of the mean. *, P < 0.05 compared to RT.
DISCUSSION
Many viruses, including dengue virus (9), Japanese encephalitis virus (41), West Nile virus (41), herpes simplex virus (63), foot-and-mouth disease virus (34), Sindbis virus (7, 37), coronavirus (14), and HIV (52), attach to cells via cell surface glycosaminoglycans. RSV also binds to cellular GAGs, and this interaction appears to be mediated by both the F and G envelope glycoproteins. In this study, we identified several linear domains within the F glycoprotein that bind heparin and determined that interactions between cellular GAGs and some of the F heparin-binding domains may play a role in virus infectivity at either an attachment or postattachment step.
Fifteen RSV-F peptides that bound to and were specifically eluted from immobilized heparin identified eight linear heparin-binding domains distributed across the fusion protein. These domains were located within F1 (represented by 201K→I217 and the consensus sequences 257L→S287, 327K→C343, and 404S→T434) and F2 (represented by 33Y→R49 and the consensus sequence 54T→K77) subunits and included a segment between the two known furin cleavage sites (represented by the consensus sequence 120N→A147) as well as a region spanning the transmembrane and cytoplasmic domains (represented by the consensus sequence 530I→S550) (Fig. 7). Thus, we were able to confirm previous predictions for RSV-F HBDs with one exception: we were not able to demonstrate heparin-binding activity for the 106RARR109 sequence that characterizes the alternative furin cleavage site, as F11, which contained this sequence, did not bind in the polyanion screening assay and F12, which reacted in the polyanion assay, did not bind in HAAC.
FIG. 7.
RSV-F glycoprotein consensus amino acid sequence for subtype A viruses. Seventy-nine overlapping peptides 17 amino acids long were synthesized with a 7-amino-acid offset. Amino acids contributing to linear heparin binding domains are in boldface type. The corresponding peptides are represented by dashed lines under the primary sequence, and peptides with infectivity-inhibiting activity are in boldface type.
All RSV-F HBD peptides bound to Vero and A549 cells, and most (12 of 15 peptides) bound to HEp-2 cells. Although there appears to be a high degree of conservation of binding sites on primate cells, differences probably reflect heterogeneity in GAG expression among the cell lines tested (3, 48).
Several lines of evidence suggested that binding of these RSV-F HBD peptides was, for the most part, mediated by interactions with cell surface GAGs. First, enzymatic treatment of cells with a combination of heparin lyases removed exposed GAGs and decreased or abolished binding of each of the RSV-F HBD peptides, while this treatment did not abrogate binding of the control peptide. RSV-F HBD peptide binding was not altered by treatment with chondroitinase C, suggesting that the cellular GAGs mediating these interactions are heparin-like. Residual binding after lyase treatment seen with peptides F16, F73, F75, and Ga19 suggested that these peptides bound GAGs that were lyase resistant or that they also bound to non-heparin molecules.
Second, when the reactivity of peptides with parental CHO cells was compared with that seen on GAG-negative CHO-745 cells, there was a significant decrease in binding observed for many RSV-F HBD peptides. Peptides F6, F15, F44, and F78 did not bind well to CHO-K1 cells, and consequently the reduction in binding on GAG-negative cells could not be calculated. Interestingly, F16, F73, and F75 bound to 27%, 56%, and 27%, respectively, of CHO-745 cells, providing additional evidence that these peptides may also bind non-heparin molecules.
Third, when sulfation was inhibited by chlorate, most RSV-F HBD peptides showed a reduction in binding compared to that seen on untreated cells, signifying that many of these interactions were sulfate dependent. The exceptions to this were F16 and F73, which bound to 65% and 51% of cells after chlorate treatment, respectively, indicating that their reactivity was not dependent on O-sulfation, the form most sensitive to sulfate deprivation and chlorate treatment. Binding of F16 could be mediated via N-sulfated residues, which are more resistant to chlorate treatment and which would be consistent with the reports by Hallak et al. that describe the relative importance of N-linked sulfates in RSV binding and infection, or F16 could bind via non-heparin moieties (26). Alternatively, the period of sulfate depletion used in these experiments may have been too short to affect highly sulfated GAGs already expressed on the surface of Vero cells, as treatment diminished but did not completely abrogate binding of the heparan sulfate monoclonal antibodies used as controls. It is not surprising that chlorate treatment did not diminish binding of F73, as this atypical heparin-binding peptide is uncharged.
Fourth, binding of most RSV-F HBD peptides could be blocked by soluble heparins, including LMWH and BLH, consistent with previous observations that indicated that RSV infection is facilitated by interactions with l-iduronic acid and N-sulfate-containing GAGs (25). However, only 6 of the 15 peptides tested (F5, F26, F35, F36, F44, and F55) were also specifically inhibited by both SOS and hyaluronic acid, an unsulfated GAG, suggesting that while some RSV-F binding could be attributed to sulfate-dependent charge-charge interactions, binding may also involve sulfate-independent interactions with GAGS as well. Interestingly, peptides F15, F16, and F78, which have the highest net positive charge of all the peptides, were not inhibited in the presence of SOS, while ATA inhibited F16 binding. Taken together with the results of binding after GAG-lyase or chlorate treatment, the data suggested that F16 binding was consistent with previous work that indicated RSV infection had a structural requirement for disaccharides at least 10 U long and involved both sulfate-dependent as well as sulfate- and GAG-independent interactions (26). Uncharged, hydrophobic F73 binding was blocked only in the presence of the uncharged unsulfated competitor, K5, indicating the unique specificity, and perhaps distinct function, of GAG-peptide interactions within the transmembrane (TM) region (data not shown). Three RSV-F HBD peptides, F2, F34, and F75, were not inhibited by any of the soluble competitors tested; these data may reflect the relatively higher affinity of these peptides for cellular GAGs than for the competitor. While it seems unlikely that the TM or cytoplasmic tail regions mapped by F73 and F75 would be utilized for virus attachment and entry, it is possible that TM-GAG or cytoplasmic tail-GAG interactions could influence budding and release of mature virions from the cell.
Interestingly, three peptides, F16, F26, and F55, significantly inhibited infectivity of subtype A and/or B strains. Two of these peptides, F16 and F55, also blocked virus attachment. This was a surprise, because it is generally believed that RSV-G serves as the primary attachment protein. If RSV-G mediated the first step in virus entry, one would not expect RSV-F peptides to block virus attachment at all. However, F16 and F55 blocked both attachment and infectivity, suggesting that these interactions may represent one of the initial steps between the virus and host cell, at least in vitro. This does not rule out the possibility that F16 and F55 bind to RSV-G or inhibit virus attachment via interference with RSV-G-GAG interactions by competing for the same binding site on the cell, and we are currently investigating this possibility. Binding of cp52 virus, which lacks RSV-G, was blocked by F15 and F55, lending additional support to the idea that RSV-F-GAG interactions mediate an early event in cellular attachment. Ga20, a heparin-binding peptide with known virus inhibitory activity, did not block virus attachment, signifying that RSV-G-GAG interactions may represent a critical step in entry that occurs subsequent to virus binding.
While F16 blocked virus infectivity, overlapping peptides F15 and F78 did not. There are several possible explanations for this. The spatial arrangement of basic residues in F16 may be very different from that found in peptides F15 and F78, and as a result F15 and F78 cannot bind appropriately oriented sulfate groups necessary to block infection. F16 may have the advantage of not only blocking virus-GAG interactions but may also contain sufficient hydrophobic finger sequence to inhibit membrane insertion events and interactions with Rho-A that are thought to be essential for virus-membrane fusion (50, 51). Along these lines, it is also possible that extracellular proteases removed the basic residues from the carboxy terminus of RSV-F2, as is the case for fusion proteins of virulent strains of Sendai and Newcastle disease virus (28, 49, 60). Therefore, attachment and infectivity would not be mediated by interactions between GAGs and the region represented by peptides F15 and F78, because this portion of the protein may be missing in the mature, fully processed virion, and as a result F15 and F78 would not be effective inhibitors. In addition, F15 and F78 interactions appear to be dependent on sulfated GAGs which may not be necessary for virus entry, while F16 binding appears to involve other types of molecules which may eventually prove to be critical for virus infectivity. Lastly, we also observed differences in the proportion of cells that bind RSV-F peptides that inhibit infection versus those that do not. The failure of F15 and F78 to saturate binding sites on a high percentage of cells as revealed by flow cytometry may explain the lack of infectivity inhibition even when attachment was diminished, as was the case for cp52 in the presence of F15. The regions mapped by both F16 and F55 are strictly conserved across human subtype A and B virus strains, possibly indicating preservation of an important structural feature of the fusion glycoprotein needed for receptor binding.
In contrast, F26, which also inhibited infectivity of RSV-A2, did not diminish virus binding to cells, indicating that its effect probably occurred at a postattachment step. F26 maps to a site immediately adjacent to the F1 N-terminal heptad repeat region, and our data are in agreement with proposed models for fusion proteins that place the N-terminal heptad repeat in the internal core of an F trimer that does not itself interact with the target membrane (46, 64). The close proximity of the F26 heparin-binding domain to the N-heptad repeat region suggests a role for F-heparin interactions in the formation or function of the six-helix bundle that can occur subsequent to attachment. The subtype B strains tested were not inhibited by F26, possibly due to four mutations that differentiate A versus B strains in this region.
There has been little effort to distinguish the relative heparin-binding contributions of RSV-F and -G glycoproteins. It is known that RSV-F can bind to cellular heparan sulfate and that RSV-F alone is capable of mediating attachment and infectivity (19, 36, 57, 58). We describe several linear domains within RSV-F that may mediate these interactions. However, there are several important limitations to our findings that must be noted. For example, we cannot comment on conformational RSV-F epitopes that may also bind GAGs or other cell surface molecules. The experiments reported herein used cell lines known to be permissive for RSV, so our findings may reflect virus adaptations acquired during tissue culture passage. These studies should be repeated on primary human lung epithelial cells in order to see if RSV-F HBD peptides retain the ability to bind cells and inhibit infectivity, as this may be more relevant to understanding RSV human infection. Also, we did not evaluate the reactivity of scrambled RSV-F peptides in each assay. In the absence of this control, we may have overestimated the regions of RSV-F that bind cellular GAGs. In addition, we have not compared binding affinities of the peptides on heparin or on cells in a rigorous way, so we cannot say if differences observed in the ability to block virus attachment or infectivity correlate with differences in affinity of binding. Finally, it would appear that the RSV-G HBD is not essential for virus infection; likewise, the significance of RSV-F HBDs should be assessed using RSV-G deletion mutant viruses engineered to express fusion proteins that lack one or more of the heparin-binding domains, or that mutate the residues responsible for heparin binding within a specific domain, to see if these changes abrogate infection or fusion.
Previous work showed that RSV infects cells devoid of GAGs, indicating that these molecules are not essential for virus entry and calling into question the relevance of experiments that examine virus-GAG binding, because these interactions may have little biological significance (57). However, in one study, heparin also inhibited the initial tissue culture passage of primary isolates, indicating that RSV-GAG interactions may also be important during natural infection (59). It appears that the mechanisms RSV uses for cell binding, entry, and fusion are likely to be complex and involve multiple types of interactions, including, but perhaps not limited to, interactions with the fracktalkine receptor (CX3CR1) (61), RhoA (50), annexin II (44), ICAM (2), toll receptors (53), the receptor for tumor necrosis factor (40), or the receptors for surfactant protein A or D by virus particles coated with these molecules (22, 30) as well as with GAGs. Our data provide evidence that, in vitro, some regions of RSV-F bind to cells via sulfated molecules, that a portion of these interactions appear to be with cell-surface GAGs, and that these interactions play a role in RSV attachment as well as in subsequent steps that lead to infection.
Acknowledgments
We thank Mike Brennan, Andrew Byrnes, and Marcela Parra for their critical reviews of the manuscript. We also thank Brian Murphy and Steve Whitehead (NIAID, NIH) for kindly providing RSVB1/cp-52 virus and Jeffrey Esko for providing CHO cell lines.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Bandtlow, C. E., and D. R. Zimmerman. 2000. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol. Rev. 80:1267-1290. [DOI] [PubMed] [Google Scholar]
- 2.Behera, A. K., H. Matsuse, M. Kumar, X. Kong, R. F. Lockely, and S. S. Mohapatra. 2001. Blocking intercellular adhesion molecule-1 on human epithelial cells decreases respiratory syncytial virus infection. Biochem. Biophys. Res. Commun. 280:188-195. [DOI] [PubMed] [Google Scholar]
- 3.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 79:11588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernfield, M., and R. D. Sanderson. 1990. Syndecan, a developmentally regulated cell surface proteoglycan that binds extracellular matrix and growth factors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 327:171-186. [DOI] [PubMed] [Google Scholar]
- 5.Bourgeois, C., J. B. Bour, K. Lidholt, C. Gauthray, and P. Pothier. 1998. Heparin-like structure on respiratory syncytial virus are involved in its infectivity in vitro. J. Virol. 72:7221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brauker, J. H., M. S. Trautman, and M. Bernfield. 1991. Syndecan, a cell surface proteoglycan, exhibits a molecular polymorphism during lung development. Dev. Biol. 147:285-292. [DOI] [PubMed] [Google Scholar]
- 7.Byrnes, A. P., and D. E. Griffin. 1998. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 72:7349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardin, A. D., and H. J. Weintraub. 1989. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9:21-32. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 10.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. L., Y. T. Huang, and G. W. Wertz. 1984. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 81:7683-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1351. In B. N. Fields, D. M. Knipe, P. M. Howley, et al (ed.), Fields virology, vol. 1, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 13.de Boer, H. C., K. T. Preissner, B. N. Bouma, and P. G. de Groot. 1992. Binding of vitronectin-thrombin-antithrombin III complex to human endothelial cells is mediated by the heparin binding site of vitronectin. J. Biol. Chem. 267:2264-2268. [PubMed] [Google Scholar]
- 14.de Haan, C. A., Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. Rottier. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 79:14451-14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denizot, F., and R. Lang. 1986. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89:271-277. [DOI] [PubMed] [Google Scholar]
- 16.Escribano-Romero, E., J. Rawling, B. Garcia-Barreno, and J. A. Melero. 2004. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J. Virol. 78:3524-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esko, J. D., A. Elgavish, T. Prasthofer, W. H. Taylor, and J. L. Weinke. 1986. Sulfate transport-deficient mutants of Chinese hamster ovary cells. Sulfation of glycosaminoglycans dependent on cysteine. J. Biol. Chem. 261:15725-15733. [PubMed] [Google Scholar]
- 18.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82:3197-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernie, B. F., G. Dapolito, P. J. Cote, Jr., and J. L. Gerin. 1985. Kinetics of synthesis of respiratory syncytial virus glycoproteins. J. Gen. Virol. 66:1983-1990. [DOI] [PubMed] [Google Scholar]
- 22.Ghildyal, R., C. Hartley, A. Varrasso, J. Meanger, D. R. Voelker, E. M. Anders, and J. Mills. 1999. Surfactant protein A binds to the fusion glycoprotein of respiratory syncytial virus and neutralizes virion infectivity. J. Infect. Dis. 180:2009-2013. [DOI] [PubMed] [Google Scholar]
- 23.Gotoh, B., Y. Ohnishi, N. M. Inocencio, E. Esaki, K. Nakayama, P. J. Barr, G. Thomas, and Y. Nagai. 1992. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J. Virol. 66:6391-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruber, C., and S. Levine. 1983. Respiratory syncytial virus polypeptides. III. The envelope-associated proteins. J. Gen. Virol. 64:825-832. [DOI] [PubMed] [Google Scholar]
- 25.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 26.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris, J., and D. Werling. 2003. Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response. Cell. Microbiol. 5:671-680. [DOI] [PubMed] [Google Scholar]
- 28.Heminaway, B. R., Y. Yang, Y. Tanaka, M. Panin, Y. T. Huang, and M. S. Galinski. 1995. Role of basic residues in the proteolytic activation of Sendai virus fusion glycoprotein. Virus Res. 36:15-35. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 30.Hickling, T. P., R. Malhotra, H. Bright, W. McDowell, E. D. Blair, and R. B. Sim. 2000. Lung surfactant protein A provides a route of entry for respiratory syncytial virus into host cells. Viral Immunol. 13:125-135. [DOI] [PubMed] [Google Scholar]
- 31.Horwitz, A. L., and R. C. Crystal. 1975. Content and synthesis of glycosaminoglycans in the developing lung. J. Clin. Investig. 56:1312-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao, J. C., C. S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72:8374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsiung, G. D. 1994. Virus assay, neutralization test, and antiviral assay, p. 46. In V. F. Chan (ed.), Hsiung's diagnostic virology. Yale University Press, New Haven, CT.
- 34.Jackson, T., F. M. Ellard, R. A. Ghazaleh, S. M. Brookes, W. E. Blakemore, A. H. Corteyn, D. I. Staurt, J. W. Newman, and A. M. King. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 36.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 39.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langedijk, J. P., B. L. de Groot, H. J. Berendsen, and J. T. van Oirschot. 1998. Structural homology of the central conserved region of the attachment protein G or respiratory syncytial virus with the fourth subdomain of 55-kDa tumor necrosis factor receptor. Virology 243:293-302. [DOI] [PubMed] [Google Scholar]
- 41.Lee, E., R. A. Hall, and M. Lobigs. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J. Virol. 78:8271-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration the glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl, U., M. Kusche-Gulberg, and L. Kjellen. 1998. Regulated diversity of heparan sulfate. J. Biol. Chem. 273:24979-24982. [DOI] [PubMed] [Google Scholar]
- 44.Malhotra, R., M. Ward, H. Bright, R. Priest, M. R. Foster, M. Hurle, E. Blair, and M. Bird. 2003. Isolation and characterisation of potential respiratory syncytial virus receptor(s) on epithelial cells. Microbes Infect. 5:123-133. [DOI] [PubMed] [Google Scholar]
- 45.Martinez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 46.Matthews, J. M., T. F. Young, S. P. Tucker, and J. P. Mackay. 2000. The core of the respiratory syncytial virus fusion protein is a trimeric coiled coil. J. Virol. 74:5911-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mbiguino, A., and J. Menezes. 1991. Purification of human respiratory syncytial virus: superiority of sucrose gradient over percoll, renografin and metrizamide gradients. J. Virol. Methods 31:161-170. [DOI] [PubMed] [Google Scholar]
- 48.Mourao, P. A., and G. M. Machado-Santelli. 1978. Sulfated glycosaminoglycans of cells grown in culture: dermatan sulfate disappearance in successive fibroblast subcultures. Cell Diff. 7:367-374. [DOI] [PubMed] [Google Scholar]
- 49.Nagai, Y., H. D. Klenk, and R. Rott. 1976. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology 72:494-508. [DOI] [PubMed] [Google Scholar]
- 50.Pastey, M., J. E. Crowe, Jr., and B. S. Graham. 1999. RhoA interacts with the fusion glycoprotein of respiratory syncytial virus and facilitates virus-induced syncytium formation. J. Virol. 73:7262-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pastey, M., T. Gower, P. Spearman, J. Crowe, and B. Graham. 2000. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat. Med. 6:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel, M., M. Yanagishita, G. Roderiquez, D. C. Bou-Habib, T. Oravecz, V. C. Hascall, and M. A. Norcross. 1993. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res. Hum. Retrovir. 9:167-174. [DOI] [PubMed] [Google Scholar]
- 53.Rudd, B. D., E. Burstein, C. S. Duckett, X. Li, and N. W. Lukacs. 2005. Differential role for TLR3 in respiratory syncytial virus-induced chemokine expression. J. Virol. 79:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanderson, R. D., and M. Bernfield. 1988. Molecular polymorphism of a cell surface proteoglycan: distinct structures on simple and stratified epithelia. Proc. Natl. Acad. Sci. USA 85:9562-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shafti-Keramat, S., A. Handisurya, E. Kriehuber, G. Meneguzzi, K. Slupetzky, and R. Kirnbauer. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 77:13125-13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent of cellular glycosaminoglycans for attachment than complete virus. Virology 294:296-304. [DOI] [PubMed] [Google Scholar]
- 58.Teng, M. N., and P. L. Collins. 2002. The central conserved cystine noose of the attachment G protein of human respiratory syncytial virus is not required for efficient viral infection in vitro or in vivo. J. Virol. 76:6164-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 60.Toyoda, T., T. Sakaguchi, K. Imai, N. M. Inocencio, B. Gotoh, M. Hamaguchi, and Y. Nagai. 1987. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology 158:242-247. [DOI] [PubMed] [Google Scholar]
- 61.Tripp, R. A., L. P. Jones, L. M. Haynes, H. Zheng, P. M. Murphy, and L. J. Anderson. 2001. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat. Immunol. 2:732-738. [DOI] [PubMed] [Google Scholar]
- 62.Voigt, A., D. Sawitzky, H. Zeichhardt, and K. O. Habermehl. 1995. Cellular receptor structures for pseudorabies virus are blocked by antithrombin III. Med. Microbiol. Immunol. 184:97-103. [DOI] [PubMed] [Google Scholar]
- 63.WuDunn, D., and P. G. Spear. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 63:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]