Abstract
The most promising vaccine strategies for the induction of cytotoxic-T-lymphocyte responses have been heterologous prime/boost regimens employing a plasmid DNA prime and a live recombinant-vector boost. The priming immunogen in these regimens must elicit antigen-specific memory CD8+ T lymphocytes that will expand following the boosting immunization. Because plasmid DNA immunogens are expensive and their immunogenicity has proven disappointing in human clinical trials, we have been exploring novel priming immunogens that might be used in heterologous immunization regimens. Here we show that priming with a prototype recombinant Mycobacterium smegmatis strain expressing human immunodeficiency virus type 1 (HIV-1) gp120-elicited CD4+ T lymphocytes with a functional profile of helper cells as well as a CD8+ T-lymphocyte population. These CD8+ T lymphocytes rapidly differentiated to memory cells, defined on the basis of their cytokine profile and expression of CD62L and CD27. Moreover, these recombinant-mycobacterium-induced T lymphocytes rapidly expanded following boosting with a recombinant adenovirus expressing HIV-1 Env to gp120-specific CD8+ T lymphocytes. This work demonstrates a remarkable skewing of recombinant-mycobacterium-induced T lymphocytes to durable antigen-specific memory CD8+ T cells and suggests that such immunogens might be used as priming vectors in prime/boost vaccination regimens for the induction of cellular immune responses.
As diseases like AIDS, tuberculosis, and malaria have emerged as important targets for vaccine development, attention has focused on developing strategies for the vaccine induction of cellular immunity (13). Studies of laboratory animals and early-phase clinical trials with humans have shown that live recombinant vectors and plasmid DNA can generate CD4+ and CD8+ T-lymphocyte responses to a variety of pathogenic microorganisms. Importantly, the most-effective strategies for the elicitation of cellular immune responses are heterologous prime/boost regimens.
The first immunogen employed in a prime/boost vaccination regimen for eliciting cytotoxic-T-lymphocyte (CTL) responses should ideally induce a large population of memory CD8+ T lymphocytes. These memory cells should persist in the host and proliferate rapidly after reexposure to the antigen expressed by the boosting immunogen. In mice, such memory T cells have been shown to express the lymph node-homing molecules CD62L (l-selectin) and CCR7 (12), the tumor necrosis factor (TNF) superfamily member CD27 (7), and the interleukin 7 (IL-7) receptor α-chain (CD127) (9, 11). Further, memory CD8+ T cells mediate relatively little cytotoxic activity and secrete high levels of cytokines, including IL-2, gamma interferon (IFN-γ), and TNF-α (27). Characterizing the phenotype and function of CD8+ T lymphocytes induced by a particular vaccine modality can thus provide important evidence concerning the potential utility of that immunogen for the priming of CTL responses.
The factors that regulate the differentiation of CD8+ T cells into memory cells are not fully defined. Studies have shown that inflammatory events, dendritic cell (DC) maturation, and antigen persistence all affect this differentiation process. CD4+ T-cell help has also been shown to enhance and maintain memory CD8+ T-cell populations (23, 28). Since different vaccine vectors should elicit cellular immune responses that differ not only in magnitude but in their functional capabilities, we initiated this study to characterize the T-lymphocyte populations generated with distinct vaccine modalities. Specifically, we analyzed the CD8+ T cells elicited by a plasmid DNA vector and a prototypic mycobacterium expressing the human immunodeficiency virus type 1 (HIV-1) gp120 protein. We show that a recombinant mycobacterial vector induces a cellular immune response that is biased toward memory cells and that can expand dramatically on reexposure to an HIV-1 envelope antigen.
MATERIALS AND METHODS
Antibodies.
The antibodies used in this study were directly coupled to fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), APC-Cy7, perdinin chlorophyll protein (PerCP)-Cy5.5, Alexa Fluor 700, or PE-Cy7. The following monoclonal antibodies (MAbs) were used: anti-CD62L-APC-Cy7 (MEL-14; eBioscience), anti-CD44-FITC (IM7; BD Biosciences), anti-CD107a-FITC (1D4B; BD Biosciences), anti-CD107b-FITC (ABL-93; BD Biosciences), anti-CD8α-PerCP-Cy5.5 (53-6.7; BD Biosciences), anti-IFN-γ-Alexa Fluor 700 (XMG1.2; BD Biosciences), anti-IL-2-APC (JHS6-5H4; BD Biosciences), anti-CD127-PE-Cy7 (A7R34; eBioscience), anti-CD27-APC (LG.7F9; eBioscience), and anti-CD4-APC-Cy7 (GK1.5; BD Biosciences).
Strains and vectors.
The efficient plasmid transformation mutant of Mycobacterium smegmatis, namely, MC2155, was used in all these studies (21). Expression of the gp120 protein by M. smegmatis was engineered as previously described (4). Briefly, the codon-optimized HIV-1 HXB2 env gene was cloned into the integrative pJH223 mycobacterial shuttle plasmid under the control of the Mycobacterium tuberculosis α-antigen promoter, followed by the M. tuberculosis 19-kDa signal sequence. The plasmid was then transformed into the recombinant M. smegmatis MC2155 strain (rSmeg-gp120). To create M. smegmatis expressing the luciferase gene (Smeg-luc), we cloned the codon-optimized firefly luciferase gene into the multicopy pJH222 mycobacterial shuttle plasmid under the control of the α-antigen promoter and then transformed it into the M. smegmatis MC2155 strain. The recombinant, replication-defective adenovirus (human serotype 5) containing the HIV-1 HXB2 env gene (rAd-gp140) was generously provided by Gary Nabel, Vaccine Research Center, NIAID, NIH. For DNA immunization, the codon-optimized HIV-1 HXB2 env gene was cloned into the VRC vector (DNA-gp120). The empty VRC vector was kindly provided by Gary Nabel.
Mice and immunization.
Six- to 8-week-old female BALB/c mice were purchased from Charles River Laboratories (Wilmington, MA) and maintained under specific-pathogenic-free conditions. Research on mice was approved by the Dana-Farber Cancer Institute Animal Care and Use Committee. Groups of mice were immunized either intraperitoneally with rSmeg-gp120 (5 × 107 CFU) or intramuscularly with DNA-gp120 (50 μg of DNA in a 100-μl total injection volume; 50 μl was delivered into each quadricep muscle). Ten weeks after the first immunization, mice were boosted with the same quantity of the same vector. In some experiments, rSmeg-gp120- and DNA-gp120-immunized mice were inoculated intramuscularly with 106 particles of rAd-gp140, either 20 weeks after the first rSmeg-gp120 and DNA-gp120 immunization or 10 weeks after the second immunization.
Phenotypic T-lymphocyte analyses.
Tetrameric H-2Dd complexes folded with the gp120 p18 epitope peptide (RGPGRAFVTI) (24) were prepared as previously described (22). Blood was collected from individual mice in RPMI 1640 medium containing 40 U of heparin per ml, and peripheral blood mononuclear cells were isolated using Lympholyte-M (Cedarlane). Cells were washed with phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) and stained for 15 min at room temperature (RT) with the PE-conjugated H-2Dd/p18 tetramer. The cells were then stained with anti-CD8α, anti-CD62L, anti-CD127, anti-CD27, and anti-CD44 for an additional 15 min at RT, washed once, and fixed with PBS containing 2% paraformaldehyde. In certain experiments, single-cell suspensions were prepared from spleens of individual animals in PBS-2% FBS and the staining was performed as described above. Samples were collected on an LSR II instrument (BD Biosciences) and analyzed using the FlowJo software (Tree Star).
Splenocyte stimulation and intracellular cytokine staining.
Splenocytes were harvested from individual mice washed with PBS-2% FBS, counted, and stained with the PE-conjugated H-2Dd/p18 tetramer. Cells were then resuspended (4 × 106 cells per tube) in RPMI 1640 medium (Cellgro; Mediatech, Inc., Herndon, VA) supplemented with 10% FBS, 25 mM HEPES, 2 mM l-glutamine, 20 U of penicillin per ml, 20 μg of streptomycin per ml, 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids. For CD8+ T-cell stimulation, cells were incubated with Golgi Plug (2 μl/ml), anti-CD28 (2 μg/ml), anti-CD49d (2 μg/ml), anti-CD107a (10 μl/ml), anti-CD107b (5 μl/ml), and the p18 peptide (2 μg/ml). For CD4+ T-cell stimulation, instead of the p18 peptide, the cells were incubated with 2 μg of the Env peptide pool per ml. The pool consisted of 47 overlapping 15-mer peptides spanning the HIV-1 IIIB gp120 protein (Centralized Facility for AIDS Reagents, Potters Bar, United Kingdom) and was used such that each peptide was present at a concentration of 2 μg/ml. Unstimulated cells were incubated with all the above-named reagents except for the peptides. As a positive control, splenocytes were incubated with phorbol myristate acetate (2 μg/ml), ionomycin (10 μg/ml), and Golgi Plug. The cells were incubated at 37°C for 6 h and then washed with PBS-2% FBS and stained with the PE-conjugated H-2Dd/p18 tetramer for 15 min followed by antibodies specific for cell surface molecules for an additional 15 min. Permeabilization was performed overnight with Cytofix/Cytoperm solution (BD Biosciences). Cells were washed with 1× Perm/Wash buffer (BD Biosciences) and then stained with an anticytokine MAb. After an additional washing step with 1× Perm/Wash buffer, the cells were fixed in 2% formaldehyde-PBS. Samples were collected on an LSR II instrument (BD Biosciences) and analyzed using FlowJo software (Tree Star).
IFN-γ ELISPOT assay.
Ninety-six-well Multiscreen hemagglutinin plates (Millipore, Bedford, MA) were coated by overnight incubation (100 μl/well) at 4°C with rat anti-mouse IFN-γ MAb (R4-6A2; BD PharMingen) at 10 μg/ml in PBS. Plates were washed three times with PBS and blocked for 2 h at 37°C with 200 μl/well of PBS containing 5% FBS. Splenocytes were harvested from individual mice 8 weeks after rAd-gp140 immunization and tested as either total splenocytes or splenocytes depleted of CD8+ T cells. Effector cells were plated in triplicate at 5 × 105/well in a 100-μl final volume with medium alone, 1 μg of the p18 epitope peptide per ml, or 1 μg of the Env peptide pool. The pool consisted of 158 overlapping 15-mer peptides spanning HIV-1 HXB2/BaL Env (Vaccine Research Center, NIAID, NIH) and was used such that each peptide was present at a concentration of 1 μg/ml. After 18 h of incubation at 37°C, the plates were washed nine times with distilled PBS containing 0.25% Tween 20 and then with sterile water and incubated for 2 h at RT with 75 μl of 5 μg/ml biotinylated rat anti-mouse IFN-γ MAb (XMG1.2; BD PharMingen) per well. The plates were washed six times, and 100 μl of streptavidin-alkaline phosphatase (Southern Biotechnology Associates, Birmingham, AL) was added at a 1/500 dilution. After 2 h of incubation, plates were washed five times and developed with Nitro Blue Tetrazolium-5-bromo-4-chloro-3-indolylphosphate chromogen (Pierce, Rockford, IL). Plates were analyzed with an enzyme-linked immunospot assay (ELISPOT) reader (Hitech Instruments, Edgemont, PA). The mean number of spots from triplicate wells was calculated for each responder animal and adjusted to represent the mean number of spots per 106 spleen cells. Data are presented as the mean numbers of spots per 106 spleen cells from four animals per group.
M. smegmatis CFU counting.
Mice were infected intraperitoneally with 5 × 107 CFU of rSmeg-gp120 in 200 μl of PBS-0.02% Tween 20. At different time points postimmunization, the spleen and liver were removed from individual mice, homogenized in saline, and serially diluted. Samples were plated on 7H10 agar plates in the presence or absence of kanamycin (20 μg/ml), and CFU were counted.
Bioimaging of recombinant M. smegmatis expressing the luciferase protein.
Bioimaging of mycobacterium-expressing firefly luciferase was done using the model 110 in vivo imaging system (IVIS 110) distributed by Xenogen Corporation (Alameda, CA). Mice were anesthetized with ketamine-xylazine and injected intraperitoneally with 100 μl of an isotonic salt solution containing 30 mg/ml d-luciferin (Xenogen). Fifteen minutes after luciferin injection, photonic emissions were measured using an IVIS 110 charge-coupled-device camera. Luciferase quantification was done using the Living Image software to identify and measure regions of interest.
Statistical analysis.
Data were expressed as means ± standard errors of the means (SE). Statistical tests were performed using Student's t test, and a P value of <0.05 was considered significant.
RESULTS
Kinetics of vaccine-elicited p18-specific CD8+ T cells.
There is a growing consensus that the optimal vaccine regimen for the induction of cellular immune responses will involve a heterologous prime/boost immunization strategy, and the best available priming immunogen to date has been plasmid DNA. However, because of the disappointing immunogenicity of many plasmid DNA vaccine constructs in humans, the exploration of other vaccine immunogens for priming is warranted. We initiated the present studies to evaluate the use of recombinant mycobacteria as a priming immunogen, using M. smegmatis as a prototype mycobacterial vaccine vector.
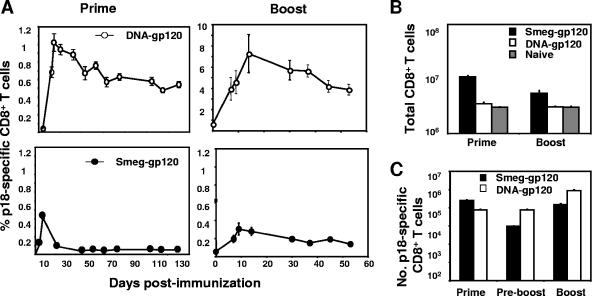
To compare the kinetics of rSmeg-gp120-induced HIV-1-specific CTLs with the kinetics of this cell population elicited by a DNA-gp120 vector, we measured H-2Dd/p18 tetramer-binding CD8+ T cells in the peripheral blood mononuclear cells of mice immunized with these vectors. Mice immunized with rSmeg-gp120 developed lower peak frequencies of p18-specific CD8+ T cells than mice immunized with DNA-gp120, and these cells decreased in frequency over time to low but detectable, stable levels (0.05% ± 0.01% versus 0.02% ± 0.005% in control recombinant-M. smegmatis-immunized mice; P < 0.001) (Fig. 1A). The DNA-gp120 vector induced the relatively slow generation of a p18-specific CD8+ T-cell response; the response peaked on day 14, and no subsequent contraction of the cell population was observed. While the second immunization with the DNA vector resulted in a substantial expansion of p18-specific CD8+ T cells, the magnitude of the boosted response in rSmeg-gp120-immunized mice was quite small, reaching a plateau of 0.2%, but detectable for >18 months (data not shown).
FIG. 1.
Kinetics of HIV-1 gp120 p18-specific CD8+ T-cell responses. Groups of mice were immunized with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg). Ten weeks after being primed, these mice were boosted with the same quantity of the same vector. (A) Peripheral blood was obtained from individual mice at the indicated times, and gp120-specific CD8+ T cells were detected with an H-2Dd/p18 tetramer. Data are presented as the percentages of gated CD8+ T cells that bound the tetramer as measured by flow cytometry. (B and C) Total numbers of CD8+ T lymphocytes (B) and total numbers of p18-specific CD8+ T cells (C) in spleens were determined at the times of the peak immune responses after the first (prime) and second (boost) immunizations and at 10 weeks after the first immunization (preboost). Values are the means of results from five mice per group ± SE.
We next analyzed the CD8+ T-cell responses in the spleens of the mice immunized with rSmeg-gp120 and DNA-gp120. Priming with rSmeg-gp120 was associated with a significant increase in the number of CD8+ T cells found in the spleens (Fig. 1B). Following a second immunization with rSmeg-gp120, the number of splenic CD8+ T cells in the mice remained elevated but was lower than following the first immunization. After mice were primed with rSmeg-gp120, the number of splenic p18-specific CD8+ T cells was three times higher than in DNA-gp120-immunized mice (Fig. 1C). However, 10 weeks following this priming immunization, the total number of splenic p18-specific CD8+ T cells in mice immunized with rSmeg-gp120 had decreased by 96%, while no significant decrease in the number of these virus-specific cells was seen in the spleens of DNA-gp120-immunized mice. At the time of the peak immune response following the second immunization, the p18-specific CD8+ T cells increased in number in the spleens of the DNA-gp120-immunized mice. However, in the rSmeg-gp120-immunized mice, the increase in the p18-specific CD8+ T-cell splenic population was limited and the maximal cell number remained lower than that seen after the priming immunization.
In vivo expression of antigen following DNA-gp120 and rSmeg-gp120 inoculation.
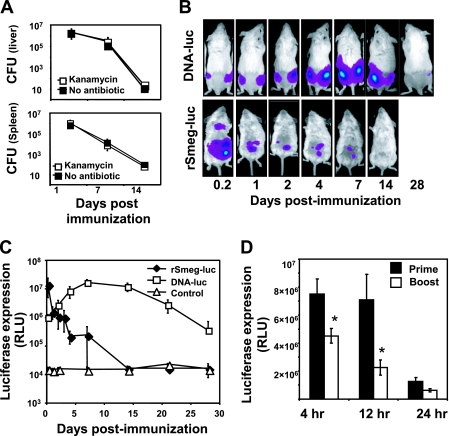
Expression of vaccine antigen by bacterial vectors occurs within the bacteria, and therefore the stability and expression level of the transgene are crucial for inducing efficient immune responses. To assess the potential utility of a recombinant-mycobacterium construct for vaccine priming, we therefore first tested the stability of the rSmeg-gp120 vector by measuring the number of CFU of the bacteria in the spleen and liver. As shown in Fig. 2A, mice controlled rSmeg-gp120 very rapidly and cleared approximately 90% of the inoculated bacteria by 24 h postinoculation. CFU counts were comparable when the bacteria were plated with and without kanamycin, indicating that the gp120 gene insert was stable in M. smegmatis.
FIG. 2.
In vivo expression of the antigen. (A) Mice were immunized with rSmeg-gp120 (5 × 107 CFU), and both the liver and spleen were removed from mice at 1, 7, and 14 days following immunization. Organ homogenates were plated in the presence or absence of kanamycin to determined CFU numbers. Data are the means of results from five mice per group ± SE. (B) Mice were immunized with rSmeg-luc (5 × 107 CFU) or DNA-luc (50 μg), and the levels of luciferase protein expression were measured over time. (C) Mice were either only primed or primed and boosted 6 weeks later, with rSmeg-luc and the levels of luciferase expression being measured over time. Data are the means of results from five mice per group ± SE. *, P < 0.001, compared to the luciferase expression following the priming immunization.
We then evaluated the kinetics of vaccine transgene expression using constructs that encode the luciferase gene (DNA-luc and rSmeg-luc) and IVIS 110 monitoring. Consistent with the kinetics of M. smegmatis clearance from the mice, high expression levels of the luciferase were measured in the rSmeg-luc-immunized mice immediately after the inoculation, and this declined during the following 24 h (Fig. 2B and C). Expression of the luciferase gene by the DNA vector was maximal at 1 week and remained detectable for more than 4 weeks after the immunization.
We then asked whether the minimal increase in immune response observed following a second inoculation of rSmeg-gp120 might be due to low-level antigen expression following the second immunization. Groups of mice were either primed or boosted with rSmeg-luc, and antigen expression was measured in vivo. Figure 2D demonstrates that the second immunization with rSmeg-luc resulted in a significantly lower level of luciferase expression than following the priming immunization (>70% reduction after 12 h). These findings indicate, therefore, that transgene expression by M. smegmatis is stable; however, the magnitude and durability of the expression were far less than observed following DNA immunization. Moreover, antivector immunity may result in low-level transgene expression following homologous boosting.
Expression of maturation-associated molecules on vaccine-elicited tetramer-positive CD8+ T cells.
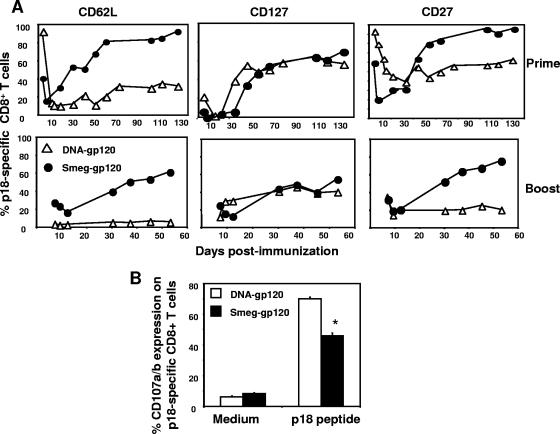
To characterize further the p18-specific CD8+ T cells induced by the two vectors, we evaluated the expression of a number of maturation-associated cell surface molecules on this vaccine-elicited lymphocyte subpopulation. Immediately after the priming immunization, a rapid down-regulation of CD127 and CD62L was seen on the p18-specific CD8+ T cells induced by rSmeg-gp120 and DNA-gp120 (Fig. 3A), reaching the lowest expression levels at the time of the peak immune responses. As the vaccine-induced immune cell populations began to contract, p18-specific CD8+ T cells began to reexpress CD127 and CD62L, with CD62L expression being greatest in mice immunized with rSmeg-gp120.
FIG. 3.
Expression of CD62L, CD127, and CD27 on vaccine-elicited p18-specific CD8+ T cells. Mice were immunized with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg). Ten weeks postimmunization mice were boosted with the same quantity of the same vector. p18-specific CD8+ T cells were detected in the peripheral blood with an H-2Dd/p18 tetramer, and the expression of CD62L, CD127, and CD27 on gated H-2Dd/p18 tetramer-positive CD8+ T cells was determined by flow cytometry. Data are presented as the percentages of tetramer-positive CD8+ T cells that were CD62Lhi, CD127hi, or CD27hi and are the means of results from five mice per group. SE were always less than 10%. (B) Splenocytes were harvested 7 days (rSmeg-gp120) or 14 days (DNA-gp120) after the boost immunization and were cultured for 6 h in the presence of medium alone or p18 peptide (2 μg/ml). Data are presented as the percentages of tetramer-positive CD8+ T cells staining positively for CD107a/b and are the means of results from five mice per group ± SE. *, P < 0.01, compared to the CD107a/b expression in DNA-immunized mice.
In contrast to CD62L and CD127 expression, p18-specific CD8+ T-cell expression of CD27 was tightly linked to the kinetics of the epitope-specific T-cell response in the rSmeg-gp120- but not in the DNA-gp120-immunized mice. CD27 expression was maximally down-regulated at the time of the peak tetramer response on day 7 following priming with rSmeg-gp120 and rose thereafter. In contrast, the down-regulation of CD27 on p18-specific CD8+ T cells in DNA-gp120-immunized mice was much slower and not linked to the kinetics of the p18-specifc CD8+ T cells. Like CD62L, up-regulation of CD27 occurred more rapidly on the tetramer-binding CD8+ T cells in mice immunized with rSmeg-gp120 than in mice receiving the DNA vector.
To determine whether the rapid induction of memory CD8+ T cells is a generalized phenomenon or simply reflects an idiosyncrasy of the gp120 immunization system in H-2Dd mice, H-2kb mice were immunized with an rSmeg-SIINFEKL construct and assessed for the evolution of memory SIINFEKL-specific CD8+ T cells. As in the studies employing the rSmeg-gp120 immunogen, these mice developed tetramer-positive CD8+ T cells expressing memory surface molecules more rapidly than did plasmid DNA-vaccinated animals (data not shown).
Following the boost immunization, the p18-specific CD8+ T cells in mice immunized with rSmeg-gp120 expressed the CD62L and CD27 molecules more rapidly, while their acquisition of CD127 was similar to that seen in the DNA-gp120-immunized mice. Consistent with their more rapid up-regulation of memory-associated molecules, the rSmeg-elicited CD8+ T cells also expressed lower levels of the degranulation-associated molecules CD107a and CD107b than p18-specific CD8+ T cells in mice immunized with the DNA-gp120 vector (P < 0.005). These data therefore demonstrate that immunization of mice with recombinant M. smegmatis induced a particularly rapid differentiation of antigen-specific CD8+ T cells into the memory pool of lymphocytes.
Functional analysis of gp120-specific CD8+ and CD4+ T-cell responses.
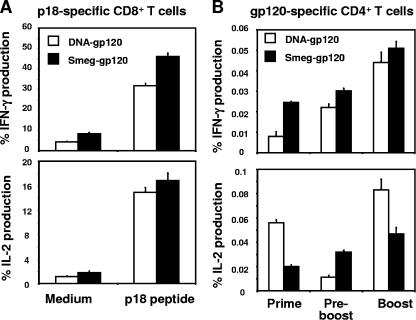
We next sought to investigate the functional potential of the p18-specific CD8+ T cells elicited by rSmeg-gp120 and DNA-gp120. Groups of mice were immunized with rSmeg-gp120 or DNA-gp120, and 10 weeks postimmunization each group was boosted with the same quantity of the same vector. At the time of the peak immune response after boosting, splenocytes were collected from the mice, exposed in vitro to the p18 peptide, stained with the noted monoclonal antibodies, and evaluated by flow cytometry (Fig. 4A). IFN-γ production was detected in a larger percentage of the p18-specific CD8+ T cells in the rSmeg-gp120-immunized mice than in the DNA-gp120-immunized mice (P < 0.05). Levels of production of IL-2 by p18-specific CD8+ T cells were comparable in the mice immunized with rSmeg-gp120 and DNA-gp120.
FIG. 4.
Functional analysis of the vaccine-elicited gp120-specific CD8+ and CD4+ T cells. Mice were immunized with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg) and 10 weeks later were boosted with the same quantity of vector used for priming. (A) Splenocytes were harvested either 7 days (rSmeg-gp120) or 14 days (DNA-gp120) after the boost immunization and were cultured for 6 h in the presence of medium alone or p18 peptide (2 μg/ml). (B) Splenocytes were collected at the time of the peak immune response after the priming or boosting immunization and 10 weeks after the priming and cultured for 6 h in the presence of medium alone or a pool of 47 overlapping peptides spanning the HIV-1 IIIB gp120 protein (2 μg/ml). Data are presented as the percentages of tetramer-positive CD8+ or gp120-responsive CD4+ T cells staining positively for IFN-γ or IL-2. The values are the means of results from five mice per group ± SE.
To determine the ability of these vectors to elicit gp120-specific CD4+ T-cell responses, splenocytes from these mice were exposed in vitro to a pool of peptides spanning the HIV-1 gp120 protein, and then gated CD4+ T cells were assessed for cytokine production. As shown in Fig. 4B, priming of mice with rSmeg-gp120 induced gp120-specific CD4+ T cells that produced higher levels of IFN-γ than were produced by the DNA-gp120-induced CD4+ T cells. Following the boosting immunization, the CD4+ T cells elicited by the two vectors produced comparable levels of IFN-γ. IL-2 production by gp120-specific CD4+ T cells was higher in mice primed with DNA-gp120 than in mice primed with rSmeg-gp120 but did not appear to be durable, since it dramatically decreased by week 10 following the priming immunization to a level that was significantly lower than that seen in rSmeg-gp120-immunized mice. After the boosting immunization, however, IL-2 expression by the gp120-specific CD4+ T cells was higher in DNA-gp120-immunized mice. These findings suggest that gp120-specific CD8+ and CD4+ T cells elicited by immunization with rSmeg-gp120 have a functional profile consistent with that of memory T lymphocytes.
p18-specific CD8+ T cells generated by rSmeg-gp120 efficiently expanded following a heterologous boost immunization with rAd-gp140.
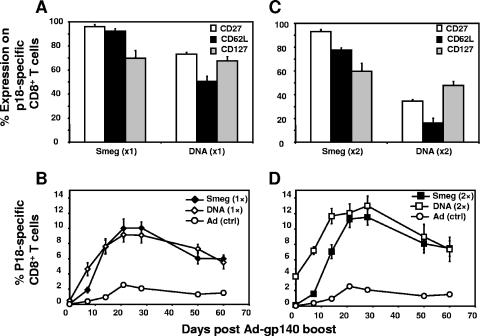
An optimal priming immunogen for a cellular immune response should elicit antigen-specific central memory CD8+ T cells that expand rapidly following reexposure to antigen. Having shown that rSmeg-elicited p18-specific CD8+ T cells differentiate more rapidly into central memory cells than cells generated by DNA immunization, we examined the ability of the p18-specific CD8+ T cells generated by the two vectors to expand following a heterologous boost immunization with rAd-gp140. Groups of mice were injected a single time with rSmeg-gp120 or DNA-gp120, and 20 weeks later the mice were boosted with a suboptimal dose (106 particles) of rAd-gp140. Other groups of mice were injected twice with rSmeg-gp120 or DNA-gp120, 10 weeks apart, and 10 weeks later boosted with rAd-gp140. Figure 5A shows the phenotypic profile of the p18-specific CD8+ T cells in the peripheral blood of each group of immunized mice on the day of the rAd-gp140 boost. In mice primed with rSmeg-gp120, more than 93% of the p18-specific CD8+ T cells expressed CD62L and CD27, surface molecules associated with T-cell memory function, while in DNA-gp120-primed mice, only 50% of the p18-specific CD8+ T cells expressed CD62L and 70% expressed CD27. The differences between the phenotypic profiles of the p18-specific CD8+ T cells elicited by these two vectors were more marked after two immunizations (Fig. 5C). In the rSmeg-gp120-immunized mice, 85% and 92% of these epitope-specific cells expressed CD62L and CD27, respectively, while in the DNA-immunized mice only 15% and 35% of p18-specific CD8+ T cells expressed CD62L and CD27, respectively. No significant differences were observed in the expression of CD127 by p18-specific CD8+ T cells in the rSmeg-gp120- and DNA-gp120-immunized mice. The rSmeg-gp120 and DNA-gp120-immunized mice also differed in the levels of p18-specific CD8+ T cells seen in their peripheral blood. On the day of rAd-gp140 boosting, the rSmeg-gp120- and DNA-gp120-immunized mice demonstrated 0.05% and 0.5% p18-specific CD8+ T cells, respectively, and the second immunization increased these percentages to 0.2% and 4.0%, respectively. Thus, although the rSmeg-gp120-immunized mice had gp120-specifc CD8+ T lymphocytes that were predominantly memory cells, the tetramer-positive cells were a much smaller percentage of the total CD8+ T-cell population in these mice.
FIG. 5.
Heterologous prime-boost immunization using rSmeg-gp120 or DNA-gp120 priming followed by rAd-gp140 boosting. Mice were immunized with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg), and 10 weeks later some mice were similarly immunized a second time. (A and C) Expression of CD62L, CD127, and CD27 on p18-specific CD8+ T cells 20 weeks after the first immunization (×1) (A) and 10 weeks after the second immunization (×2) (C). (B and D) Kinetics of p18-specific CD8+ T cells in rSmeg-gp120 (Smeg)- and DNA-gp120 (DNA)-immunized mice (after the priming [1×] [B] and boosting [2×] [D]) following heterologous immunization with 106 particles of rAd-gp140 [Ad (ctrl)]. Data are the means of results from 5 to 10 mice per group ± SE.
We then inoculated the mice with a suboptimal dose of rAd-gp140 (106 particles) and assessed the kinetics of the generation of p18-specific CD8+ T cells. A suboptimal dose of rAd-gp120 was chosen to facilitate discrimination between the priming efficiency of the plasmid DNA and recombinant-M. smegmatis immunogens. One week after the rAd-gp140 immunization, the DNA-immunized mice (one or two immunizations) had higher p18-specific CD8+ T-cell responses than the rSmeg-gp120-immunized mice (Fig. 5B and D). However, by the second and third weeks postimmunization, rSmeg-gp120-immunized mice had comparable p18-specific CD8+ T-cell responses. The kinetics of the contraction of these immune cell populations in the rSmeg-gp120- and DNA-gp120-immunized mice were comparable. Therefore, although rSmeg-gp120-immunized mice developed only small numbers of antigen-specific CD8+ T cells, these mice generated robust secondary CD8+ T-cell responses following rAd-gp140 boosting that were comparable in magnitude to those generated in the DNA-gp120 prime/rAd-gp140 boost mice.
Functional analysis of the CD8+ and CD4+ T cells elicited by the recombinant-M. smegmatis/DNA-gp120 prime-rAd-gp140 boost immunization.
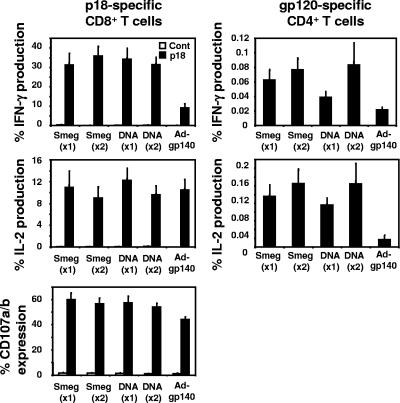
To determine whether priming with DNA-gp120 and rSmeg-gp120 and boosting with rAd-gp140 generated gp120-specific T-cell responses with similar immune potentials, we evaluated the functional properties of the p18-specific CD8+ and CD4+ T cells elicited by this immunization regimen. Following the boost immunization with rAd-gp140, the levels of production of IFN-γ and IL-2 by p18-specific CD8+ T cells were comparable in the rSmeg-gp120- and DNA-gp120-primed mice (Fig. 6). p18-specific CD8+ T-cell production of IFN-γ was greater in these prime/boost vaccine-immunized mice than in mice immunized with rAd-gp140 alone. Levels of expression of CD107a and CD107b on CD8+ p18-specific T cells were also similar in the mice primed with rSmeg-gp120 and in the mice primed with DNA-gp120, with both groups then being boosted with rAd-gp140, and were significantly higher than those seen in mice immunized only with rAd-gp140. gp120-specific CD4+ T cells in mice inoculated with rSmeg-gp120 or DNA-gp120 and then boosted with rAd-gp120 produced comparable levels of IFN-γ and IL-2. Moreover, this cytokine production was higher than that of gp120-specific CD4+ T cells in mice immunized with rAd-gp140 only.
FIG. 6.
Functional analysis of the p18-specific CD8+ T cells and gp120-specific CD4+ T cells elicited by priming with rSmeg-gp120 or DNA-gp120 followed by boosting with rAd-gp140. Mice were immunized either once (×1) or twice (×2) 10 weeks apart with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg). Twenty weeks after the first immunization and 10 weeks after the second immunization, the mice were inoculated with 106 particles of rAd-gp140. Splenocytes were harvested 8 weeks after the immunization with rAd-gp140 and were cultured for 6 h in the presence of medium alone, p18 peptide (2 μg/ml), or the Env peptide pool (2 μg/ml). The intracellular production of IFN-γ and IL-2 by p18-specific CD8+ T cells and gp120-specific CD4+ T cells or CD107a/b expression by p18-specific CD8+ T cells was evaluated. Data are presented as the percentages of tetramer-positive CD8+ T cells staining positively for IFN-γ, IL-2, or CD107a/b and gp120-specific CD4+ T cells staining positively for IFN-γ or IL-2 and are the means of results from five mice per group ± SE.
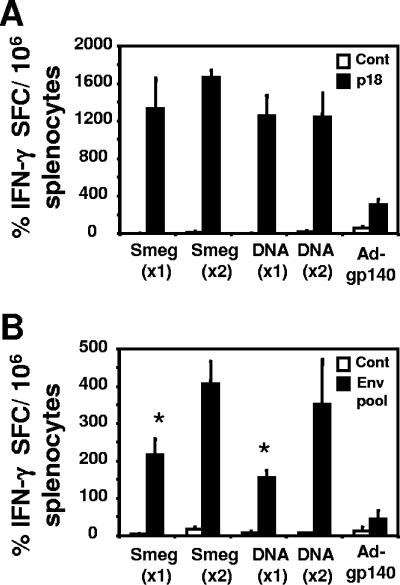
Interestingly, following rAd-gp140 boosting, the mice receiving two priming immunizations with rSmeg-gp120 or DNA-gp120 generated gp120-specific CD4+ T cells that produced more IFN-γ than did mice receiving a single priming immunization (Fig. 7B). These data also support the intracellular cytokine staining data, indicating comparable levels of IFN-γ production by CD8+ T cells in mice primed with rSmeg-gp120 or DNA-gp120 (Fig. 7A). The present findings therefore indicate that after a heterologous boost with rAd-gp140, rSmeg-gp120-primed mice generated vigorous antigen-specific CD8+ and CD4+ T-cells responses that were comparable to those developed in DNA-gp120-primed mice.
FIG. 7.
IFN-γ production by CD8+ and CD4+ T cells following a priming immunization with rSmeg-gp120 or DNA-gp120 and a boosting immunization with rAd-gp140. Mice were immunized either once (×1) or twice (×2), 10 weeks apart, with rSmeg-gp120 (5 × 107 CFU) or DNA-gp120 (50 μg). Twenty weeks after the first immunization or 10 weeks after the second immunization, the mice were inoculated with 106 particles of rAd-gp140. Splenocytes were harvested from individual mice 8 weeks after the immunization with rAd-gp140. IFN-γ production was evaluated by ELISPOT assay using total splenocytes incubated with p18 peptide (1 μg/ml) (A) or by CD8+ T-cell-depleted splenocytes stimulated with a peptide pool consisting of 158 overlapping 15-mer peptides spanning the HIV-1 HXB2/BaL Env protein at a concentration of 1 μg/ml (B). Data are presented as the mean numbers of antigen-specific spots per 106 spleen cells ± SE, with five mice per group. *, P < 0.01 (one immunization versus two immunizations).
DISCUSSION
The present study explored gp120-specific CD8+ T-cell induction following immunization with DNA-gp120 and the prototype mycobacterial vector construct rSmeg-gp120. The natures of these two vaccine modalities are very different. While expression of the gp120 protein by rSmeg-gp120 occurs in the bacteria, the DNA-gp120 immunogen expresses this protein in the host cells. This difference in expression may lead to differences in the gp120 protein itself. The gp120 expressed by the plasmid DNA immunogen has a glycosylation pattern typical of proteins expressed by eukaryotic cells, while the gp120 expressed by rSmeg-gp120 has a very different glycosylation pattern and is acylated due to its fusion to the 19-kDa signal sequence. The kinetics and expression levels of gp120 by these vectors are also likely to be very different. Expression of gp120 by the plasmid DNA vector should occur for a long period of time, while expression by rSmeg-gp120 is likely to be brief. This difference in the kinetics of antigens may impact the development of the memory CD8+ T cells, since these cells have been shown to differentiate into memory cells only after antigen is cleared (10).
An explanation as to why the rSmeg-gp120-immunized mice had a lower-frequency cellular immune response than the DNA-gp120-immunized mice is not readily apparent. We were able to detect transgene expression by rSmeg-luc for more than 24 h after the immunization, a period of time that should be sufficient for maximal programmed CD8+ T-cell expansion and differentiation (10, 26). In fact, it has been shown that ablation of a mouse muscle 10 min after its inoculation with plasmid DNA did not affect the magnitude of the vaccine-elicited CTL response (25), suggesting that a short duration of antigen expression is sufficient for CTL induction. It is also possible that a low-frequency CTL response is generated in mice because this vaccine construct expresses only a small amount of transgene product. Finally, the magnitude of the vaccine-elicited T-cell population in rSmeg-gp120-immunized mice may simply reflect the kinetics of the contraction of these antigen-specific lymphocytes following immunization. At the peak of the immune response, the number of p18-specific CD8+ T cells in the spleen was higher in the rSmeg-gp120- than in the DNA-gp120-immunized mice. However, 96% of these cells died during the contraction phase of the response. A similar contraction has been described following immunization with other vectors, including Listeria monocytogenes and vaccinia virus (2, 6). Whatever the explanation for the generation of a small pool of vaccine-induced, long-lived memory cells, modulation of the kinetics of transgene expression through molecular modification of the vector may increase the number of gp120-specific CD8+ T cells that can be elicited with a recombinant-mycobacterium vaccine.
Surface expression of the CD27 molecule on the recombinant-M. smegmatis-elicited CD8+ T cells decreased rapidly during the first week after priming but increased quickly thereafter. Since CD27 expression has been reported to predict the size of the CD8+ effector T-cell pool (8), the rapid loss of CD27 by the epitope-specific CD8+ T cells in the rSmeg-gp120-immunized mice might be associated with the small number of long-lived antigen-specific cells in these mice. Interestingly, the expression of CD27 is down-regulated in the presence of TNF-α (16, 18), a cytokine that is induced by M. smegmatis immediately after infection (19). The findings in the present study are consistent with the suggestion that the inflammatory properties of the vector influence the kinetics of the elicited antigen-specific CD8+ T cells (2).
The rapid acquisition of memory-associated surface molecules on the vaccine-elicited CD8+ T cells and the decreased expression of the CD107a and CD107b molecules on these cells after stimulation suggest an accelerated differentiation of memory CD8+ T cells in the rSmeg-gp120-immunized mice. The rapid clearance of the vaccine antigen following immunization with recombinant M. smegmatis may explain this rapid induction of memory CD8+ T cells. Since effector cells survive for only a short time after antigen is cleared, the percentages of vaccine-elicited memory CD8+ T cells may be particularly high in rSmeg-gp120-immunized mice. Also, it was recently shown that vaccination of mice with peptide-pulsed, mature DCs resulted in an accelerated differentiation of memory CD8+ T cells (1). Several studies have reported that M. smegmatis up-regulates the expression of major histocompatibility complex class I (MHC I) and costimulatory molecules on DCs, leading to their maturation (5, 15). These findings suggest that M. smegmatis might induce the maturation of DCs and, as a consequence, the rapid generation of memory CD8+ T cells.
CD4+ T-cell help has also been shown to enhance and maintain the memory CD8+ T cells generated after acute infection (23). Indeed, immunization with rSmeg-gp120 induced CD4+ T cells that produced IFN-γ and IL-2. Since CD4+ T cells are required for the maintenance of memory CD8+ T cells, the potent CD4+ T-cell responses generated by rSmeg-gp120 might contribute to the bias toward the differentiation of memory CD8+ T cells in these mice. The ability of this M. smegmatis vector to generate CD4+ T-cell immunity is consistent with the immune response generated in response to other mycobacteria and is important for protective immunity (14, 17). In fact, CD4+ T cells have been shown to be required for the development of cytotoxic CD8+ T cells during M. tuberculosis infection (20). These data suggest that mycobacteria skew the immune response toward CD4+ T-cell immunity, a skewing that is important for maintaining memory CD8+ T cells.
As with all vectors that have been proposed for use as platforms for human immunogens, M. smegmatis has certain disadvantages as a vaccine vector. Live recombinant M. smegmatis may have toxicities, since this bacterium induces a strong inflammatory response. However, the observation that M. smegmatis is rapidly cleared, even from SCID mice, suggests that it may still be safe for use in humans (3). Another potential limitation of the M. smegmatis vector is that it induces a small number of antigen-specific CD8+ T cells in comparison to those elicited by other vectors. This might be a consequence of the in vivo expression of only small amounts of antigen by M. smegmatis or reflect the fact that the localization of bacterium-encoded antigen in the infected cell prevents efficient MHC class I presentation. These potential drawbacks might be circumvented by selecting mutant mycobacteria for use as vectors that are particularly efficient in directing transgene products into MHC class I processing pathways.
An ideal priming vector for a cellular immune response should elicit a large population of CD8+ T lymphocytes that differentiate rapidly into memory cells. This will potentiate a vigorous secondary immune response following boosting. The observation that small numbers of M. smegmatis-elicited p18-specific CD8+ T cells can expand into large populations of functionally competent CTLs following heterologous boosting suggests that recombinant M. smegmatis may be useful as a priming vector in prime/boost vaccine regimens.
Acknowledgments
This research was conducted as part of the Collaboration for AIDS Vaccine Discovery with support from the Bill & Melinda Gates Foundation.
We are grateful to Darci Gorgone, Michelle Lifton, Dan Barouch, and Itai Roni Eyal for technical assistance and scientific discussions. The HIV-1 IIIB gp120-overlapping peptides were provided by the EU Program EVA/MRC Centralized Facility for AIDS Reagents, National Institute for Biological Standards and Control, United Kingdom.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Badovinac, V. P., K. A. Messingham, A. Jabbari, J. S. Haring, and J. T. Harty. 2005. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat. Med. 11:748-756. [DOI] [PubMed] [Google Scholar]
- 2.Badovinac, V. P., B. B. Porter, and J. T. Harty. 2004. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 5:809-817. [DOI] [PubMed] [Google Scholar]
- 3.Bange, F. C., F. M. Collins, and W. R. Jacobs, Jr. 1999. Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuber. Lung Dis. 79:171-180. [DOI] [PubMed] [Google Scholar]
- 4.Cayabyab, M. J., A. H. Hovav, T. Hsu, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, G. J. Fennelly, B. F. Haynes, W. R. Jacobs, Jr., and N. L. Letvin. 2006. Generation of CD8+ T-cell responses by a recombinant nonpathogenic Mycobacterium smegmatis vaccine vector expressing human immunodeficiency virus type 1 Env. J. Virol. 80:1645-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheadle, E. J., D. O'Donnell, P. J. Selby, and A. M. Jackson. 2005. Closely related mycobacterial strains demonstrate contrasting levels of efficacy as antitumor vaccines and are processed for major histocompatibility complex class I presentation by multiple routes in dendritic cells. Infect. Immun. 73:784-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington, L. E., R. van der Most, J. L. Whitton, and R. Ahmed. 2002. Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J. Virol. 76:3329-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriks, J., L. A. Gravestein, K. Tesselaar, R. A. van Lier, T. N. Schumacher, and J. Borst. 2000. CD27 is required for generation and long-term maintenance of T cell immunity. Nat. Immunol. 1:433-440. [DOI] [PubMed] [Google Scholar]
- 8.Hendriks, J., Y. Xiao, and J. Borst. 2003. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J. Exp. Med. 198:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huster, K. M., V. Busch, M. Schiemann, K. Linkemann, K. M. Kerksiek, H. Wagner, and D. H. Busch. 2004. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA 101:5610-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaech, S. M., and R. Ahmed. 2001. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaech, S. M., J. T. Tan, E. J. Wherry, B. T. Konieczny, C. D. Surh, and R. Ahmed. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191-1198. [DOI] [PubMed] [Google Scholar]
- 12.Lauvau, G., S. Vijh, P. Kong, T. Horng, K. Kerksiek, N. Serbina, R. A. Tuma, and E. G. Pamer. 2001. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science 294:1735-1739. [DOI] [PubMed] [Google Scholar]
- 13.Letvin, N. L. 2002. Strategies for an HIV vaccine. J. Clin. Investig. 110:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogues, T., M. E. Goodrich, L. Ryan, R. LaCourse, and R. J. North. 2001. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J. Exp. Med. 193:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neyrolles, O., K. Gould, M. P. Gares, S. Brett, R. Janssen, P. O'Gaora, J. L. Herrmann, M. C. Prevost, E. Perret, J. E. Thole, and D. Young. 2001. Lipoprotein access to MHC class I presentation during infection of murine macrophages with live mycobacteria. J. Immunol. 166:447-457. [DOI] [PubMed] [Google Scholar]
- 16.Orengo, A. M., C. Cantoni, F. Neglia, R. Biassoni, and S. Ferrini. 1997. Reciprocal expression of CD70 and of its receptor, CD27, in human long term-activated T and natural killer (NK) cells: inverse regulation by cytokines and role in induction of cytotoxicity. Clin. Exp. Immunol. 107:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrofsky, M., and L. E. Bermudez. 2005. CD4+ T cells but not CD8+ or γδ+ lymphocytes are required for host protection against Mycobacterium avium infection and dissemination through the intestinal route. Infect. Immun. 73:2621-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranheim, E. A., M. J. Cantwell, and T. J. Kipps. 1995. Expression of CD27 and its ligand, CD70, on chronic lymphocytic leukemia B cells. Blood 85:3556-3565. [PubMed] [Google Scholar]
- 19.Roach, D. R., A. G. Bean, C. Demangel, M. P. France, H. Briscoe, and W. J. Britton. 2002. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J. Immunol. 168:4620-4627. [DOI] [PubMed] [Google Scholar]
- 20.Serbina, N. V., V. Lazarevic, and J. L. Flynn. 2001. CD4(+) T cells are required for the development of cytotoxic CD8(+) T cells during Mycobacterium tuberculosis infection. J. Immunol. 167:6991-7000. [DOI] [PubMed] [Google Scholar]
- 21.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 22.Staats, H. F., C. P. Bradney, W. M. Gwinn, S. S. Jackson, G. D. Sempowski, H. X. Liao, N. L. Letvin, and B. F. Haynes. 2001. Cytokine requirements for induction of systemic and mucosal CTL after nasal immunization. J. Immunol. 167:5386-5394. [DOI] [PubMed] [Google Scholar]
- 23.Sun, J. C., M. A. Williams, and M. J. Bevan. 2004. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 5:927-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi, H., Y. Nakagawa, C. D. Pendleton, R. A. Houghten, K. Yokomuro, R. N. Germain, and J. A. Berzofsky. 1992. Induction of broadly cross-reactive cytotoxic T cells recognizing an HIV-1 envelope determinant. Science 255:333-336. [DOI] [PubMed] [Google Scholar]
- 25.Torres, C. A., A. Iwasaki, B. H. Barber, and H. L. Robinson. 1997. Differential dependence on target site tissue for gene gun and intramuscular DNA immunizations. J. Immunol. 158:4529-4532. [PubMed] [Google Scholar]
- 26.van Stipdonk, M. J., E. E. Lemmens, and S. P. Schoenberger. 2001. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat. Immunol. 2:423-429. [DOI] [PubMed] [Google Scholar]
- 27.Wherry, E. J., and R. Ahmed. 2004. Memory CD8 T-cell differentiation during viral infection. J. Virol. 78:5535-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, Q., F. Y. Yue, X. X. Gu, H. Schwartz, C. M. Kovacs, and M. A. Ostrowski. 2006. OX40 ligation of CD4+ T cells enhances virus-specific CD8+ T cell memory responses independently of IL-2 and CD4+ T regulatory cell inhibition. J. Immunol. 176:2486-2495. [DOI] [PubMed] [Google Scholar]