Abstract
The motifs involved in the various functions of the human immunodeficiency virus type 1 (HIV-1) gp41 cytoplasmic tail (CT), particularly those related to the intracellular trafficking and assembly of envelope glycoproteins (Env) onto core particles, have generally been assessed with a restricted panel of T-cell laboratory-adapted virus strains. Here, we investigated gp41 CT sequences derived from individuals infected with HIV-1 viruses of various subtypes. We identified four patients harboring HIV variants with a natural polymorphism in the membrane-proximal tyrosine-based signal Y712SPL or the Y802W803 diaromatic motif, which are two major determinants of Env intracellular trafficking. Confocal microscopy showed that the intracellular distribution of Env with a mutation in the tyrosine or diaromatic motif differed from that of Env with no mutation in these motifs. Surprisingly, the gp41 CTs of the primary viruses also had differential effects on the intracellular distribution of Env, independently of mutations in the tyrosine or diaromatic motifs, suggesting the involvement of additional determinants. Furthermore, analyses of virus replication kinetics indicated that the effects of mutations in the tyrosine or diaromatic motifs on viral replication depended on the gp41 CT context. These effects were at least partly due to differences in the efficiency of Env incorporation into virions. Thus, polymorphisms in primary HIV-1 gp41 CTs at the quasispecies or subtype level can influence the intracellular distribution of Env, its incorporation into virions, and viral replication capacity.
The envelope glycoproteins (Env) of human immunodeficiency virus type 1 (HIV-1) are primarily responsible for the specific binding of virions to target cells and for the fusion of viral and cellular membranes in the entry process (27). Env is synthesized as a 160-kDa precursor, which is then processed during its trafficking through the secretory pathway to yield a surface subunit, gp120, noncovalently attached to a transmembrane subunit, gp41. The gp41 subunit consists of an ectodomain exposed on the virion surface, a hydrophobic transmembrane anchor, and a cytoplasmic tail (CT). During HIV-1 assembly, Env is incorporated at the surface of the viral particle as a trimeric complex consisting of three gp120 and gp41 subunits (51, 53). HIV-1 particles initially interact with target cells via gp120 and the CD4 receptor (45). Binding to CD4 leads to a change in the conformation of gp120, revealing a binding site for one of the viral coreceptors, typically the chemokine receptor CXCR4 or CCR5 (15, 20, 28). Subsequent coreceptor engagement triggers changes in the structure of the gp41 ectodomain, leading to the fusion of the viral and target cell membranes (28).
Lentiviruses, including HIV-1, are unusual in having transmembrane glycoproteins with much longer CTs (∼150 amino acids) than most other retroviruses (20 to 50 amino acids) (27). Early mutational studies, in which deletions or truncations were introduced into the CT of HIV-1 gp41, showed that this region played an important role in regulating Env protein functions and viral replication (18, 24, 31, 54). The effects of the gp41 CT on viral replication are cell type dependent. In many transformed T-cell lines, primary peripheral blood mononuclear cells (PBMC), and macrophages, viral replication is dependent on the integrity of the gp41 CT, whereas other T-cell lines (e.g., MT4) permit the replication of HIV-1 with an almost entirely truncated gp41 tail (1, 35). In general, viral replication kinetics and Env incorporation seem to be directly correlated.
Many studies have tried to identify the precise motifs involved in the various functions assigned to the gp41 CT. Two groups of motifs have been identified. The first group consists of three structurally conserved amphipathic α-helical domains, designated lentivirus lytic peptides 1, 2, and 3 (LLP-1, LLP-2, and LLP-3) (Fig. 1) (10, 19, 30). LLP domains have been implicated in various functions, including Env cell surface expression, Env fusogenicity, and incorporation of Env into virus particles (8, 29, 36, 42). Several studies have suggested that Env is incorporated into virions via interactions between LLP and the matrix region of Gag, but there is still controversy concerning the involvement of either LLP-1 or LLP-2/LLP-3 domains in this process (12, 26, 29, 36, 42). The second group of motifs regulates the intracellular trafficking of Env. At steady state, Env is located principally in the trans-Golgi network (TGN) (5). This intracellular distribution results from dynamic cycling of Env between the cell surface, the endosomal compartment, and the TGN. Newly synthesized Env proteins undergo endocytosis after their arrival at the cell surface (44). Env internalization is mediated by the interaction of Y712SPL, a membrane-proximal tyrosine-based signal in the gp41 CT, with the adaptor protein (AP) complexes of the cellular sorting machinery, involving the clathrin adaptor AP-2 in particular (4, 6, 37). The gp41 CT also interacts with the TGN- and endosome-based clathrin-associated adaptor AP-1. The dileucine motif at the C terminus of gp41 is involved in this interaction with AP-1, which has been shown to play a role in determining the subcellular distribution of Env (4, 52). Y802W803, a diaromatic motif involved in the retrograde transport of Env to the TGN, was recently identified in the gp41 CT (5). This motif seems to interact with TIP47, a protein required for the retrograde transport of mannose 6-phosphate receptors to the TGN from late endosomes. The concentration of Env in the TGN at steady state results partly from this retrograde transport of Env from the cell surface to the TGN (5).
FIG. 1.
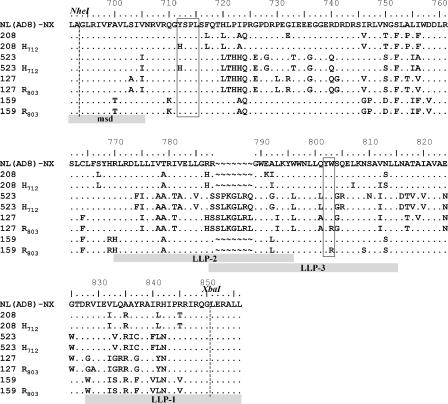
Cytoplasmic domain sequences of Env clones from patients 208, 523, 127, and 159 used in this study. Sequence alignments for gp41 CTs are shown for NL(AD8)-NX and for a viral variant with a mutation in the tyrosine or diaromatic motif or with no mutation in these determinants from each patient. The Env sequences upstream from the NheI restriction site and downstream from the XbaI restriction site are derived from NL(AD8). Amino acid identity (.), insertion/deletion (∼), and substitution are indicated. Highlighted domains include the C-terminal region of the membrane-spanning domain (msd) and the amphipathic alpha-helical domains LLP-1, LLP-2, and LLP-3. Amino acid numbers correspond to the HXB2 sequence.
Modifications of the motifs regulating the intracellular distribution of Env have been shown to affect viral assembly and replication in various ways. Mutations of the tyrosine in the membrane-proximal tyrosine-based signal Y712SPL disturb the polarized release of HIV-1 in epithelial cells (32, 33). This motif is similarly required for the polarized release of HIV-1 from a localized, cap-like region of the plasma membrane overlaying the uropod in lymphocytes (16). The tyrosine-based signal (Y712SPL) and the diaromatic motif (Y802W803) are also important for optimal Env incorporation and viral replication (5, 9, 14, 49). The effects of mutations in the Y712SPL signal on Env incorporation and viral replication are cell dependent, consistent with previous results for an almost fully truncated gp41 CT (1, 14, 35, 49).
These data demonstrate the existence of a complex system regulating the intracellular distribution of Env that probably plays an important role in HIV-1 assembly and replication. However, the motifs involved in the various functions of the gp41 CT, and particularly those involved in Env intracellular trafficking, have generally been assessed with a restricted panel of T-cell laboratory-adapted (TCLA) virus strains, such as HIV-1 HXB2 or NL4-3 molecular clones. Adaptation to continuous growth in established T-cell lines is accompanied by genetic changes affecting the viral Env proteins. For example, these changes expand tropism for established T-cell lines and increase sensitivity to antibody neutralization over that of primary viruses subjected to the selective pressure of the immune system in vivo (34, 50).
Here, we focused on gp41 CT sequences from viruses obtained from patients. We carried out an analysis, at the quasispecies level, of Env sequences obtained from individuals infected with HIV-1 viruses of various subtypes and identified four patients harboring HIV variants with mutations in the membrane-proximal tyrosine-based signal Y712SPL or the Y802W803 diaromatic motif. Based on this polymorphism, we hypothesized that these gp41 CTs from primary HIV-1 might affect the intracellular distribution of Env, with possible consequences for viral assembly and replication capacity. We tested this hypothesis by using virus-based Env expressors and replication-competent viral clones carrying gp41 CT sequences derived from the viral variants with a mutation in the tyrosine or diaromatic motif and from a coexisting viral variant with no mutation in these determinants. Confocal microscopy showed that the intracellular distribution of Env proteins containing a mutation in the tyrosine or diaromatic motif differed from that of the corresponding Env proteins with no mutation in these motifs. This observation suggests that the tyrosine and diaromatic motifs are involved in a primary-Env context. Surprisingly, we also observed that the gp41 CTs of the various primary viruses studied here had differential effects on the intracellular distribution of Env, independently of mutations in the tyrosine or diaromatic motifs, suggesting the involvement of additional determinants within the CT. Furthermore, analyses of virus replication kinetics in T-cell lines or PBMC and macrophages indicated that the effects of mutations in the tyrosine and diaromatic motifs on viral replication depended on the gp41 CT context. These effects were at least partly due to the efficiency of Env incorporation into virions. Thus, polymorphisms in primary HIV-1 gp41 CTs at the quasispecies or subtype level may influence the intracellular distribution of Env, its incorporation into virions, and viral replication capacity.
MATERIALS AND METHODS
Patient-derived envelope selection and characterization of tropism.
Viral env genes were isolated from four patients (patients 208, 523, 127, and 159) selected from a group of 30 HIV-1-positive patients followed at the Department of Infectious Diseases of the Croix-Rousse Hospital, Lyon, France. All patients gave written, informed consent. The procedure used to amplify full-length env genes from the PBMC DNA of patients 127 and 159 has been described elsewhere (2, 48). Full-length env genes were amplified in similar ways from patients 208 and 523. All PCR products were inserted into the EcoRI site of pCR2.1 (Topo TA cloning kit; Invitrogen). The nucleotide sequences of full-length primary env genes (three or four clones for each patient) were analyzed using sequence analysis (Applied Biosystems), and the corresponding deduced amino acid sequences were aligned using Clustal W (46), with manual corrections. One Env sequence carrying a mutated Y712/H-SPL membrane-proximal tyrosine-based signal was identified among the Env clones isolated from patients 208 and 523, and one Env sequence with a mutated Y802-W803/R diaromatic motif was identified among the Env clones isolated from patients 127 and 159. Env sequences 127 and 159, with a mutated Y802-W803/R diaromatic motif, have already been deposited in the GenBank database under accession numbers AY231152 and AF041128, respectively (2, 48). The alignment of env gene sequences with reference sequences and the construction of neighbor-joining trees (13) showed that patient 208 was infected with a subtype B virus, whereas patient 523 was infected with a subtype G virus. Patients 127 and 159 were previously shown to be infected with a circulating recombinant form (CRF), CRF02_AG, and a subtype B virus, respectively (2, 48). The chemokine receptor usage pattern of the selected Env proteins was determined, using a recombinant phenotypic assay, as previously described (47). All Env proteins mediated viral entry, using CCR5 as a coreceptor.
Env expression vectors and infectious proviral clones.
Full-length primary Env from patients 208, 523, 127, and 159 was produced from the pCI expression vector (Promega) containing the various env genes inserted into the EcoRI site downstream from the cytomegalovirus promoter. Viral-based Env expressors carrying gp41 CT sequences derived from primary Env were constructed in a pNL(AD8) background (21), which is an R5 derivative of pNL4-3 containing pAD8-1 sequences in the gp120 and gp41 ectodomains. We used pNL4-3 as starting material for plasmid construction. The NL4-3 Env expressor was constructed by deleting a fragment encompassing the gag and pol genes between the PstI (nucleotide [nt] 1415 of pNL4-3) and EcoRI (nt 5743) sites. The NL(AD8) expressor was obtained by replacing a sequence containing the env gene in the NL4-3 expressor, between the EcoRI (nt 1415) and XhoI (nt 4559) sites, with the corresponding sequence from pNL(AD8). Primary env CT sequences were cloned by constructing the Env expressor variant NL(AD8)-NX (NX-WT), in which the NheI and XbaI restriction sites were inserted into the sequences corresponding to the membrane-spanning domain of gp41 and at the end of the env gene, respectively. Mutagenesis was carried out in pSK-AD8, which carries a fragment of the NL(AD8) expressor encompassing the gp41 sequence inserted between the BsaBI and XhoI sites of pBluescript SKII (Stratagene). Sequences were modified using a QuikChange site-directed mutagenesis kit (Stratagene) and the following primers: Nhe41(+) (5′ATAGTAGGAGGGCTAGCAGGTTTAAGAATAG3′), Nhe41(−) (5′CTATTCTTAAACCTGCTAGCCCTCCTACTAT3′), Xba41(+) (5′ATAAGACAGGGTCTAGAAAGGGCTTTGCTATAAG3′), and Xba41(−) (5′CTTATAGCAAAGCCCTTTCTAGACCCTGTCTTAT3′). Following mutagenesis, the BsaBI/XhoI fragment was excised and inserted into the NX-WT expressor, previously digested with the same enzymes. The DNA sequences encoding the CTs of primary Env proteins were amplified by PCR, using primers similar to those described for the site-directed mutagenesis of the AD8 expressor. We modified only single nucleotides in primer sequences, to ensure a perfect match with the various env genes. The PCR products were inserted into pCR2.1 (Topo TA cloning kit; Invitrogen), removed by digestion with NheI/XbaI, and inserted into the corresponding sites of the NX-WT expressor, giving rise to the NX-208, NX-208H712, NX-523, NX-523H712, NX-127, NH-127R803, NX-159, and NX-159R803 Env expressors. Env expressor NL(AD8)-NX, carrying mutated Y712SPL membrane-proximal tyrosine-based signals (NXA712 and NXH712) or mutated Y802W803 diaromatic motifs (NXS802-L803 and NXR803), was also generated by site-directed mutagenesis with the appropriate primers, as previously described. The sequences were amplified and the mutations verified by DNA sequencing.
Replication-competent pNL(AD8)-NX proviral constructs carrying sequences encoding gp41 CTs from primary Env proteins were obtained by replacing the BsaBI/XhoI fragment of pNL(AD8) with the corresponding fragment excised from the NL(AD8)-NX expressors.
Cell culture.
293T, HeLa, and their derivative P4P cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (100 IU/ml penicillin and 100 μg/ml streptomycin). MT4.R5 T cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics. PBMC were obtained from the buffy coats of HIV-1-seronegative donors by Ficoll-Hypaque density gradient centrifugation. Monocytes were purified from PBMC by adhesion to plastic, as described by Perez-Bercoff et al. (41). We obtained monocyte-derived macrophages (MDM) by allowing harvested monocytes to differentiate into macrophages for 7 days in 12-well plates (8 × 105 cells/well) containing RPMI 1640 medium supplemented with 10% human AB serum and antibiotics. Nonadherent peripheral blood mononuclear cells were treated with phytohemagglutinin-P (5 μg/ml; Difco Laboratories) in RPMI 1640 medium supplemented with interleukin 2 (10 ng/ml; Roche), 20% fetal calf serum, and antibiotics. After 3 days, cells were washed free of phytohemagglutinin-P and maintained in RPMI 1640 medium supplemented with interleukin 2, 10% fetal calf serum, and antibiotics.
Subcellular distribution of Env glycoproteins.
HeLa cells were spread on glass coverslips in six-well plates (2 × 105 cells/well) 24 h before transfection. Cells were transfected with 1 μg of Env expressor (cotransfection with 0.1 μg pCI-rev for pCi Env expressors) or proviral construct DNA, using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The transfected cells were incubated for 40 h, washed twice with phosphate-buffered saline (PBS), and treated with 50 μg of cycloheximide/ml for 3 h. Cells were then fixed by incubation in 4% paraformaldehyde in PBS for 20 min. After fixation, they were quenched and permeabilized by incubation for 30 min in 0.05% saponin-0.2% bovine serum albumin in PBS. Cells were incubated for 1 h at room temperature with primary antibodies: an anti-CD71 mouse monoclonal antibody (MAb) (transferrin receptor; Serotec), anti-TGN-46 sheep polyclonal antibodies (Serotec), an anti-CD63 mouse MAb (Serotec), a mouse MAb recognizing p17 (matrix) but not Pr55Gag (Advanced Biotechnologies), and anti-Env human MAb 2G12 (provided by H. Katinger). The corresponding fluorescent Alexa 488- and 594-conjugated secondary antibodies were used (Molecular Probes). Coverslips were washed and mounted on microscope slides with Immuno-Fluore (MP Biomedicals). The transfection conditions used yielded about 5% positive cells with moderate Env staining. Images of representative cells were acquired on each slide with an Olympus FluoView 500 confocal microscope equipped with argon (488 nm) and HeNe (546 nm) lasers, a 60× PlanApo oil immersion objective, and Fluoview 4.3 software.
The colocalization of Env and TGN-46 proteins was quantified for cells expressing Env NX-208, NX-208H712, NX-523, NX-523H712, NX-127, NH-127R803, NX-159, and NX-159R803. Cells were imaged in each channel, along the z axis, using Fluoview 4.3 software, and 10 in-focus sections (0.2 μm each) from each z stack were analyzed for colocalization by using ImarisColoc (Imaris, Bitplane). The threshold values for each channel used to quantify colocalization were determined using the automatic threshold function of the software. Colocalization was defined as the overlap of two channels in three dimensions and was calculated automatically by the program. Pearson channel correlation coefficients (R) in a studied volume (1 indicating perfect colocalization, 0 indicating no correlation) were calculated for 10 cells for each sample and are given as means ± standard deviations.
Electron microscopy.
HeLa cells (106) were spread in T25 flasks 24 h before transfection with 10 μg of plasmid DNA, using Lipofectamine 2000 (Invitrogen). Cells were washed with PBS 12 to 18 h after transfection, and fresh medium was added. Cells were harvested 40 h after transfection, pelleted by low-speed centrifugation, and fixed by incubation for 16 h in 4% paraformaldehyde, 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2). The pellet was washed in PBS and postfixed by incubation for 1 h with 2% osmium tetroxide (Electron Microscopy Science). It was dehydrated in a graded series of ethanol solutions, cleared in propylene oxide, and embedded in Epon resin (Sigma), which was allowed to polymerize for 48 h at 60°C. Ultrathin sections were cut, placed on electron microscopy grids, and stained with 5% uranyl acetate and 5% lead citrate before observation with a Jeol 1230 transmission electron microscope.
Production of viral stocks.
Viruses for the infection of T cells and MDM were obtained by transfecting 293T cells. We spread 3.5 × 106 cells in T75 flasks 24 h before transfection with 30 μg of plasmid DNA by the calcium phosphate method according to the kit manufacturer's instructions (Invitrogen). Cells were washed with PBS 12 to 18 h after transfection, and fresh medium was added. Viral supernatants were harvested 48 h after transfection, centrifuged at low speed, filtered (0.45-μm pores), and then frozen at −80°C. All virus stocks were sampled for detection of the p24 capsid protein by an Innotest HIV antigen enzyme-linked immunosorbent assay (ELISA) kit (Ingen) before freezing. The 50% tissue culture infective dose (TCID50) value of each HIV-1 stock was determined on P4P cells (CD4+ CXCR4+ CCR5+ adherent HeLa cells) as previously described (3). Similar TCID50 values per ng of p24 were obtained for the various viral stocks produced in 293T cells.
Viruses for Env incorporation assays were generated by transfecting HeLa and MT4.R5 cells or infecting PBMC. For HeLa cell transfections, 106 cells were spread in T25 flasks 24 h before transfection with 10 μg of plasmid DNA, using Lipofectamine 2000 (Invitrogen). Cells were washed 12 to 18 h after transfection, and fresh medium was added. Viral supernatants were harvested 48 h after transfection, centrifuged at low speed, filtered, and immediately used for virus purification. For MT4.R5 cell transfections, a suspension of 2 × 106 cells was transfected with 10 μg of plasmid DNA, using Lipofectamine 2000 (Invitrogen). Viral supernatants were harvested 72 h after transfection, centrifuged at low speed, filtered, and immediately used for virus purification. For PBMC infections, 2 × 106 cells were pelleted and resuspended in 1 ml of RPMI medium containing 100, 500, or 1,000 ng p24 equivalent of virus, depending on viral growth. Cells were incubated for 2 h at 37°C and then thoroughly washed and resuspended in 5 ml of RPMI 1640 medium supplemented with interleukin 2, 20% fetal calf serum, and antibiotics. Cells were incubated for 3 days at 37°C and then washed, resuspended in 10 ml of media, and incubated at 37°C for a further 3 days. Viral supernatants were harvested, centrifuged at low speed, filtered, and immediately used for virus purification. All virus stocks were sampled for quantification of the p24 capsid protein before virus purification.
Lysates of HeLa cells were prepared at the time of virus collection. Pelleted cells were washed with PBS, repelleted by centrifugation, and resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer containing dithiothreitol. A sample was removed for p24 analysis, and cell lysates were then frozen at −80°C for subsequent Western blot analysis, as described below.
Viral replication kinetics.
The concentration of the viral stocks produced as described above from 293T cells was normalized on the basis of p24 capsid protein concentration. For the infection of MT4.R5 cells, 106 cells were pelleted and resuspended in 1 ml of complete RPMI 1640 medium containing 5 ng p24 equivalent of virus. Cells were incubated for 2 h at 37°C, washed, resuspended in 4 ml of medium, and returned to the incubator at 37°C. Samples of culture supernatants were taken every 2 days for p24 capsid protein determination. After the removal of the sample, cell suspensions were split 1:4 and returned to the incubator at 37°C. For PBMC infection, 106 cells were pelleted by centrifugation and resuspended in 1 ml of RPMI medium containing 10 ng p24 equivalent of virus. The cells were incubated at 37°C for 2 h, washed, resuspended in 2 ml of RPMI 1640 medium supplemented with interleukin 2, 20% fetal calf serum, and antibiotics, and returned to the incubator at 37°C. The culture medium was sampled every 2 days for p24 capsid protein determination and was then completely replaced with fresh medium. For MDM infection, 8 × 105 cells/well were incubated for 2 h at 37°C in 1 ml of RPMI medium containing 20 ng p24 equivalent of virus per well, in 12-well plates. Cells were then washed, and 1 ml of RPMI 1640 medium supplemented with 10% human AB serum and antibiotics was added before cells were returned to incubation at 37°C. The culture medium was sampled every 2 days for p24 capsid protein determination and was then completely replaced with fresh medium.
Env incorporation assays.
Viruses produced, as described above, from HeLa cells, MT4.R5 cells, and PBMC were used to assess Env incorporation into virions. Culture supernatants containing the viruses were overlaid on a 20% sucrose cushion in a Beckman SW28 tube, and particles were pelleted by centrifugation for 2 h at 50,000 × g and 4°C. Viral pellets were resuspended in a small volume of TNE buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5 mM EDTA) supplemented with 1% Triton X-100. An aliquot was removed for p24 capsid protein determination by ELISA, and resuspended pellets were frozen at −80°C for quantitative gp120 ELISA and Western blot analysis, as described below.
For quantitative gp120 ELISA, Immunolon II plates (Dynex) were coated by incubation overnight at 4°C with a 5-μg/ml solution of anti-gp120 sheep polyclonal antibody D7324 (Aalto) in Tris-buffered saline (TBS; 100 mM Tris-HCl [pH 7.5], 150 mM NaCl). The plates were washed with 0.5% Tween 20 in TBS (TBS-T). Nonspecific binding sites were saturated by incubation for 1 h at room temperature with 2% newborn calf serum in TBS. Dilutions of the virus lysate samples in TBS supplemented with 10% fetal calf serum and 1% Nonidet P-40 were added, and the plates were incubated for 2 h at 37°C. The samples used contained equal amounts of p24, as shown by ELISA. The plates were washed, and human MAb 2G12 diluted (1 μg/ml) in TBS supplemented with 0.5% Tween 20, 20% newborn calf serum, and 10% fetal calf serum (TBS.T.N.F) was added and allowed to bind for 1 h at 37°C. The plates were then washed, and a goat anti-human immunoglobulin (Biosource) conjugated with peroxidase and diluted 1:2,000 in TBS.T.N.F was added. The plates were incubated for 30 min at room temperature and washed, and a mixture of H2O2 and o-phenylene-diamine was added. The plates were left for 30 min at room temperature in the dark, and color development was then stopped by adding 50 μl of 2 N H2SO4. Absorbance at 490 nm (A490) was determined. Dilutions of purified gp120IIIB (Advanced BioScience Laboratories) were used to construct a standard curve. Similar ELISAs were also carried out with a pool of HIV-1-positive human sera, for the detection of gp120.
We assessed Env incorporation by using Western blotting only for viruses produced in HeLa cells. Based on the p24 ELISA data, we prepared equal amounts of p24 per sample in SDS-polyacrylamide gel electrophoresis reducing sample buffer. Samples were boiled for 3 minutes and subjected to electrophoresis in an SDS-10% polyacrylamide gel. The protein bands were then electroblotted onto a nitrocellulose membrane. The membrane was blocked by incubation with 5% nonfat milk powder in TBS supplemented with 0.01% Tween 20, washed, incubated with primary antibody, washed, incubated with a horseradish peroxidase-conjugated secondary antibody, washed, and developed with enhanced chemiluminescent substrate (Amersham-Pharmacia). Images were captured by a Chemi-Smart 3000 gel documentation system, using Chemi-Capt software for image acquisition (Vilber Lourmat). The membrane was initially probed for gp120, using goat polyclonal anti-gp120 antibodies (Biogenesis). After blot development, the antibodies were removed from the blot by incubation with Restore Western blot stripping buffer (Pierce), and the membrane was washed with 0.01% Tween 20 in TBS. The Western blotting procedure was then repeated, using a mouse MAb directed against p24 capsid protein (13B5D10; bioMerieux) or gp41 (Abcam).
Nucleotide sequence accession numbers.
Env sequences 208, 208H712, 523, and 523H712 have been submitted to GenBank and assigned accession numbers EF033657 to EF033660.
RESULTS
The intracellular distribution of primary HIV-1 Env glycoproteins.
It has been shown that the Env proteins of TCLA viruses, such as HXB2, are mostly present in the TGN at steady state, due to determinants located within the gp41 CT (5). Based on the polymorphism observed in the gp41 CTs of various primary Env proteins, we hypothesized that the variant forms of Env might display different intracellular distributions. We investigated the virus populations of four patients harboring variants characterized by polymorphisms in the tyrosine Y712SPL motif (patients 208 and 523) or in the Y802W803 diaromatic motif (patients 127 and 159). Patients 208 and 159 were infected with a subtype B HIV-1 virus, whereas patient 523 was infected with a subtype G virus and patient 127 was infected with a circulating recombinant form, CRF02_AG. Full-length primary env genes from patients 208, 523, 127, and 159 were inserted into the pCI expression vector. The corresponding proteins were transiently produced in HeLa cells, and their intracellular distribution at steady state was compared with that of TGN-46, an endogenous marker of the TGN. Cells were analyzed 40 h after transfection and treated with cycloheximide to eliminate the newly synthesized Env from the early-secretory pathway.
Laser confocal microscopy studies revealed that the primary Env 208, 523, 127, and 159 clones, harboring gp41 CTs containing no mutation in the tyrosine or diaromatic motif, had different intracellular distributions. The primary Env 208 protein was concentrated principally in the perinuclear region and colocalized with TGN-46, as previously reported for other Env proteins from TCLA viruses (Fig. 2) (5). However, other primary Env proteins, such as Env 523, did not colocalize with TGN-46, and labeling for these proteins was mostly associated with vesicular structures dispersed throughout the cytoplasm (Fig. 2). Clones containing a mutation in the tyrosine motif showed a less concentrated intracellular staining, with persistent labeling of peripheral speckles for Env 208 and a similar pattern of vesicular staining for Env 523 (Fig. 2). Env 127 and 159 had intermediate distributions, with Env staining mostly concentrated in the perinuclear region but with only a partial overlap with TGN-46 labeling (data not shown). Thus, the intracellular distribution of Env from primary viruses may differ from the reported pattern for TCLA Env.
FIG. 2.
Localization of primary HIV-1 Env proteins by confocal microscopy. HeLa cells were transfected with the NL4-3 Env expressor or pCI expression vectors encoding full-length Env 208, 208H712, 523, and 523H712. The transfected cells were fixed and processed for immunofluorescence with the 2G12 anti-Env MAb and anti-TGN-46 polyclonal antibodies. Before fixation, all the cells were treated with 50 μg of cycloheximide/ml for 3 h. Colocalization was assessed by confocal microscopy. A series of optical sections at 0.2-μm intervals was recorded. A representative medial section is shown. Scale bar, 20 μm.
The gp41 CTs from primary viruses have differential effects on the intracellular distribution of Env.
We investigated whether the determinants governing the intracellular distribution of primary Env were located within the CT of gp41, by constructing chimeric env genes in which the gp41 CTs of env genes from patients 208, 523, 127, and 159 replaced the corresponding region of the molecular clone NL(AD8). These constructs were produced by transferring the gp41 CT sequences of primary Env clones into the NL(AD8)-NX expressor, as described in Materials and Methods. The proteins were transiently produced in HeLa cells, and their intracellular distribution at steady state was compared with that of TGN-46 by laser confocal microscopy. Cells were analyzed 40 h after transfection and treated with cycloheximide. The ImarisColoc module was used to assess Env and TGN-46 colocalization, as described in Materials and Methods. Pearson channel correlation coefficients (R) for the studied volume were calculated for 10 cells for each sample and are given as means ± standard deviations.
The pattern of Env NX-208 staining matched that of TGN-46 in the perinuclear area (R = 0.70 ± 0.04) (Fig. 3A). In contrast, intracellular staining for Env NX-208H712 was less concentrated, with persistent labeling of peripheral speckles (R = 0.21 ± 0.03) (Fig. 3A). Cell surface staining was observed in the cells producing the largest amounts of Env NX-208H712. Env NX-523 was found in vesicular structures dispersed throughout the cytoplasm (R = 0.24 ± 0.04). Env NX-523H712 displayed a similar pattern of vesicular staining (R = 0.18 ± 0.03) (Fig. 3A). Env NX-127 and NX-159 staining was observed mainly in the perinuclear region, with only a partial overlap with TGN-46 labeling (for Env NX-127, R = 0.51 ± 0.06; for Env NX-159, R = 0.50 ± 0.09) (Fig. 3B). The corresponding clones containing the Y802-W803/R mutation displayed a less concentrated staining around the nucleus (for Env NX-127R803, R = 0.41 ± 0.07; for Env NX-159R803, R = 0.37 ± 0.06). These results are consistent with those obtained with the full-length Env and suggest that the CT of the primary Env protein determines the specific intracellular distribution of the protein.
FIG.3.
Localization of Env NL(AD8) carrying the gp41 CT from primary viruses by confocal microscopy. HeLa cells were transfected with the Env expressors NX-208, NX-208H712, NX-523, and NX-523H712 (A); the Env expressors NX-127, NX127R803, NX-159, and NX-159R803 (B); and the Env expressors NX-WT, NXA712, NXH712, NXS802-L803, and NXR803 (C). The transfected cells were fixed and processed for immunofluorescence with the 2G12 anti-Env MAb and anti-TGN-46 polyclonal antibodies. Before fixation, all the cells were treated with 50 μg of cycloheximide/ml for 3 h. Colocalization was assessed by confocal microscopy as described in the legend to Fig. 2. Scale bar, 20 μm.
FIG. 3—
Continued.
We evaluated the impact of mutations in the tyrosine and diaromatic motifs on the parental NL(AD8) Env, by analyzing the intracellular distribution of Env NL(AD8)-NX containing either mutations characterized in previous studies (mutation Y712/A-SPL in the tyrosine motif or Y802W803/SL in the diaromatic motif) or mutations found in the primary Env proteins used in this study (mutation Y712/H-SPL in the tyrosine motif or Y802-W803/R in the diaromatic motif). Env NX-WT, which has a gp41 CT identical to that of the NL4-3 TCLA virus, was detected mostly in the perinuclear region and colocalized with TGN-46, as previously described for Env from TCLA viruses (Fig. 3C). Mutations in the tyrosine motif (either Y712/A-SPL or Y712/H-SPL) resulted in less-concentrated intracellular staining, with the persistent labeling of peripheral speckles, similar to that observed with Env NX-208H712. Mutations in the diaromatic motif resulted in less-concentrated staining in the perinuclear area, giving a pattern similar to that observed with Env NX-127R803 and NX-159R803.
FIG. 3—
Continued.
Thus, determinants located within the gp41 CTs of primary Env proteins control the distribution of these proteins, directing some of them to vesicular structures dispersed throughout the cytoplasm. Furthermore, the intracellular distributions of primary Env proteins with CTs harboring mutations within the tyrosine or diaromatic motif differed, to various extents, from that of the corresponding Env clones without mutations in these determinants, confirming the involvement of these motifs in primary-Env contexts.
Env proteins carrying the gp41 CT from primary viruses are redistributed to the cell surface when coexpressed with Gag.
We investigated possible differences in the intracellular distributions of Env proteins harboring gp41 CT sequences from primary viruses produced together with Gag, by transferring the gp41 CT sequences from NL(AD8)-NX expressors in the HIV-1 molecular clone pNL(AD8)-NX, as described in Materials and Methods. Molecular pNL(AD8)-NX clones containing mutations in the tyrosine or diaromatic motif were obtained in a similar way. HeLa cells were transiently transfected with the various proviral plasmids, to check for the production of Env proteins. No significant difference in p24 production was found between the various NL(AD8)-NX viruses, indicating the absence of unanticipated defects in the RNA sequence and the synthesis of a functional Rev protein. HeLa cell lysates were analyzed by Western blotting for specific characterization of Env glycoprotein production. After being stained for gp120, the membrane was stripped and reprobed for p24, to rule out differences in transfection efficiency and loading. All Env proteins were correctly produced and processed (data not shown).
The intracellular distribution of Env in transfected HeLa cells was compared with those of TGN-46 and p17 (matrix) viral proteins by laser confocal microscopy. The anti-p17 antibody used specifically recognizes the mature form of matrix but not the unprocessed precursor Pr55Gag (38, 39). Since the majority of Gag processing occurs only after virus particles are formed, the staining obtained with this antibody most likely represents the sites at which virus assembly occurs (23). Cells were analyzed 40 h after transfection and treated with cycloheximide. When produced together with Gag, Env NX-WT displayed a pattern of punctate staining at the cell surface (Fig. 4A). A similar intracellular distribution was observed with the NL(AD8)-NX viruses carrying mutated gp41 CTs (data not shown) or primary gp41 CTs (Fig. 4A). This cell surface staining was accompanied by diffuse cytosolic staining with various intensities, depending on the nature of the CT. Env and Gag showed high degrees of colocalization at the cell surface, for all viruses except the NL(AD8)-NX-159R803 virus (Fig. 4A). For this virus, some of the punctate staining at the cell surface corresponded to Gag proteins only. The colocalization of staining for Env proteins, such as NX-WT or NX-208, and TGN-46 was much weaker than that observed for Env and TGN-46 in the absence of Gag (data not shown). These results suggest that Env proteins, including those carrying primary gp41 CTs with markedly divergent sequences, are found, at least in part, at the cell surface in the presence of Gag. The changes in colocalization between Gag and Env induced by the gp41 CT of Env NH-159R803 were the only changes strong enough to be discerned clearly by confocal microscopy.
FIG.4.
Coexpression of Env NL(AD8) carrying the gp41 CT from primary viruses with Gag: analyses of Env distribution by confocal microscopy and viral budding by electron microscopy. (A) HeLa cells transfected with pNL(AD8)-NX-WT, pNL(AD8)-NX-159, pNL(AD8)-NX-159R803, pNL(AD8)-NX-523, and pNL(AD8)-NX-523H712 were fixed and processed for immunofluorescence with 2G12 anti-Env MAb and anti-p17 MAb. Before fixation, all the cells were treated with 50 μg/ml of cycloheximide for 3 h. Protein distribution was assessed by confocal microscopy as described in the legend to Fig. 2. Scale bar, 20 μm. (B) Transmission electron micrograph of HeLa cells transfected with pNL(AD8)-NX-WT or pNL(AD8)-NX-159. Viral budding and particle release were observed principally at the cell surface, where both immature and mature (see arrow) viruses were present. Scale bar, 0.2 μm.
FIG. 4—
Continued.
HeLa cells transfected with proviral plasmids were also analyzed by electron microscopy. Predominant budding at the cell surface was observed for all NL(AD8)-NX viruses carrying mutated gp41 CTs or primary gp41 CTs, as shown previously by confocal microscopy analyses. The electron micrographs of HeLa cells transfected with the pNL(AD8)-NX-WT or pNL(AD8)-NX-159 proviral plasmid shown in Fig. 4B are representative of those obtained for all NL(AD8)-NX viruses. Thus, the various Env proteins studied had no apparent effect on the site of viral budding in HeLa cells.
The gp41 CTs from primary viruses have differential effects on viral replication kinetics.
We characterized the phenotype of the NL(AD8)-NX viruses carrying primary gp41 CT or mutated gp41 CT in the spread of infection, using a T-cell line (MT4.R5), primary human PBMC, and macrophages for replication kinetics experiments. The amounts of virus obtained by transfecting 293T cells with pNL(AD8)-NX proviral clones containing primary or mutated gp41 CTs were normalized such that all samples contained equal amounts of p24 capsid protein. The TCID50 values per ng of p24 determined for P4P cells (CD4+ CXCR4+ CCR5+ adherent HeLa cells) were similar for the various viral stocks produced in 293T cells (data not shown). The inoculum was adapted for each cell line, based on permissiveness to NL(AD8)-NX viruses. Replication kinetics were compared by determining the levels of p24 capsid protein in cell culture supernatants.
In MT4.R5 cells, the NL(AD8)-NX-WT, 208, 523, and 523H712 viruses replicated with similar kinetics (Fig. 5A). In contrast, the replication kinetics of the NL(AD8)-NX-208H712 virus was strongly affected, with p24 levels lower by a factor of 30.5 than those for the isogenic viral clone with no mutation in the tyrosine motif, 4 days after infection. The NL(AD8)-NX-127 and 159 viruses had similar replication efficiencies (Fig. 5A). The NL(AD8)-NX-127R803 virus showed a slight decrease in replication rate, whereas the NL(AD8)-NX-159R803 virus was more strongly affected, with p24 levels 4 days after infection lower than those for the isogenic viral clone with no mutation in the diaromatic motif by a factor of 12.9. All the control NL(AD8)-NX viruses containing a mutation in the tyrosine or diaromatic motif had similar replication kinetics in MT4.R5 cells, with the exception of the NL(AD8)-NXS802-L803 virus, which replicated at a slightly lower rate (Fig. 5A). The strongest impact on viral replication was observed with the NL(AD8)-NX-208H712 and NL(AD8)-NX-159R803 viruses, which contain gp41 CTs derived from subtype B viruses. We checked that the observed effects were not related to differences in the amino acid sequence of the gp41 CT other than that affecting the tyrosine or diaromatic motif (Fig. 1), by generating the proviral constructs pNL(AD8)-NX-208H712 and pNL(AD8)-NX-159R803, in which the mutation was eliminated and the wild-type tyrosine or diaromatic motif restored. These viruses had replication kinetics similar to those of NL(AD8)-NX-208 and NL(AD8)-NX-159, demonstrating that the effects on viral replication observed with the NL(AD8)-NX-208H712 and NL(AD8)-NX-159R803 viruses were due to modification of the tyrosine or diaromatic motif (data not shown). Thus, the gp41 CTs from some primary Env proteins influence viral replication kinetics in MT4.R5 cells, whereas these cells are similarly permissive for the growth of NL(AD8) viruses harboring mutations in the tyrosine and diaromatic motifs. The latter observation is consistent with previous reports that the parental MT4 cell line is permissive for the growth of TCLA viruses containing mutations in the tyrosine motif or even a C-terminal truncation of the gp41 CT (14, 35).
FIG. 5.
Replication kinetics of NL(AD8)-NX viruses carrying the gp41 CT from primary viruses. Virus stocks, obtained by transfecting 293T cells with the indicated molecular clones, were normalized for p24 capsid protein concentration and used to infect MT4.R5 (A), PBMC (B), or MDM (C) cells as described in Materials and Methods. Variations in p24 concentrations were monitored in the culture supernatant over time. Each experiment was performed at least twice, in duplicate, with similar results in terms of both the hierarchy and the extent of replication. Error bars indicate standard deviations for duplicate infections.
NL(AD8)-NX-WT was the virus that replicated most rapidly in PBMC (Fig. 5B). The replication kinetics of the NL(AD8)-NX-523, 523H712, and 208 viruses were slightly slower, whereas the effect on the replication kinetics of the NL(AD8)-NX-208H712 virus was greater, with p24 levels in the culture supernatant 6 days after infection lower than those for the isogenic viral clone with no mutation in the tyrosine motif by a factor of up to 15.3. The NL(AD8)-NX-127 and 159 viruses had replication kinetics of the same order of magnitude (Fig. 5B). The corresponding isogenic viral clones with mutated diaromatic motifs replicated more slowly. The NL(AD8)-NX-159R803 virus showed the largest effect on replication kinetics, with p24 levels in the culture supernatant falling below 500 pg/ml for 10 days after infection. All NL(AD8)-NX viruses containing a mutation in the tyrosine or diaromatic motif displayed a slight decrease in replication kinetics, with the exception of NL(AD8)-NXS802-L803, which was more strongly affected (Fig. 5B). Thus, although maximal yields differed, the hierarchies of replication rates were similar in PBMC and MT4 cells.
NL(AD8)-NX-WT also replicated more rapidly than any other virus in macrophages (Fig. 5C). Interestingly, the hierarchy of replication kinetics observed for viruses containing primary CTs with a mutation in the tyrosine motif differed from the kinetics observed in MT4.R5 cells and PBMC. NL(AD8)-NX-523H712 replicated slightly faster than NL(AD8)-NX-523. Surprisingly, NL(AD8)-NX-208H712 replicated more rapidly than NL(AD8)-NX-208, with a maximum difference of a factor of 3.5 in p24 levels in the culture supernatant, 10 days after infection. Conversely, the hierarchy of replication kinetics for viruses containing primary gp41 CTs with mutations in the diaromatic motif closely matched that observed in PBMC and MT4.R5 cells (Fig. 5C). The difference in replication kinetics was greatest between the NL(AD8)-NX-159 and 159R803 viruses, with a maximum difference of a factor of 114 in p24 levels in the culture supernatant, 10 days after infection. NL(AD8)-NX viruses containing a mutation in the CT replicated at a markedly lower rate, with the exception of NL(AD8)-NXA712, which displayed replication kinetics similar to those for NL(AD8)-NX-WT (Fig. 5C).
Thus, the impact of mutations in the tyrosine or diaromatic motif differed, according to the primary gp41 CTs studied. In particular, viruses with gp41 CTs derived from the two subtype B HIV-1-infected patients (patients 208 and 159) included in this study were the most strongly affected. Furthermore, the impact of mutations in the tyrosine and diaromatic motifs was also cell dependent, suggesting that the mechanisms controlling the intracellular distribution of Env may differ in the cell types used here.
The gp41 CTs from primary viruses have differential effects on Env incorporation.
We assessed the efficiency of Env incorporation for the various NL(AD8)-NX viruses carrying primary gp41 CTs or mutated gp41 CTs, by setting up a sensitive Env ELISA and assaying lysates of purified virions. Various antibodies (human MAb 2G12 and a pool of human sera from HIV-1-infected patients) for detecting the gp120 captured on the solid phase were tested. The data obtained with the human MAb 2G12 are reported here, as the background was lowest for this antibody. However, similar results were obtained with the pool of human sera, showing that the differences in gp120 incorporation between the various viruses did not depend on variations in the antigenic properties of Env.
Following the transfection of HeLa cells, the NL(AD8)-NX-WT virus incorporated the highest level of gp120 (Fig. 6A). NL(AD8)-NX-208, 208H712, 523, and 523H712 virus lysates contained similar amounts of gp120. Only viruses containing primary gp41 CTs with a mutation in the diaromatic motif incorporated less gp120 than the corresponding virus with no mutation. Mutations in the tyrosine or diaromatic motifs of the gp41 CT of the NL(AD8)-NX virus did not affect Env incorporation. The amounts of gp120, gp41, and p24 found in the various virus lysates were determined by Western blotting, as described in Materials and Methods. The results were consistent with those obtained by ELISA, demonstrating that the difference in gp120 incorporation observed in ELISA reflected Env incorporation rather than excess shedding. Indeed, similar defects in gp120 and gp41 incorporation were observed in NL(AD8)-NX-159R803, in comparisons with the NL(AD8)-NX-159 and NL(AD8)-NX-WT viruses (Fig. 6D).
FIG. 6.
Env incorporation into NL(AD8)-NX viruses carrying the gp41 CT from primary viruses. Env incorporation into virions was assessed, using viruses purified by centrifugation through a 20% sucrose cushion. Viral pellets were lysed in TNE buffer containing 1% Triton X-100, and gp120 was quantified by ELISA. (A) Virions prepared from HeLa cells transfected with pNL(AD8)-NX-208, 208H712, 523, 523H712, 127, 127R803, 159, and 159R803; pNL(AD8)-NX-WT, NXA712, NXH712, NXS802-L803, and NXR803; and pNL4-3 Δenv (11). (B) Virions prepared from MT4.R5 cells transfected with the same proviral vectors. (C) Virions prepared from PBMC infected with viruses NL(AD8)-NX-208, 208H712, 523, 523H712, 127, 127R803, 159, and 159R803 and NL(AD8)-NX-WT, NXA712, NXH712, NXS802-L803, and NXR803. Data shown are the means for two independent experiments. Error bars indicate standard deviations. (D) Western blot analysis of Env incorporation into virions prepared from HeLa cells transfected with pNL(AD8)-NX-WT, 127, 127R803, 159, and 159R803. Membranes were probed with anti-gp120, anti-gp41, or anti-p24 capsid protein antibodies (see Materials and Methods). gp120 and gp41 protein bands were quantified by densitometry analysis using Bio-1D software (Vilber Lourmat). Results are presented as the amounts of gp120 and gp41 incorporated into virions, expressed as percentages of wild-type (WT) levels.
In MT4.R5 cells, the NL(AD8)-NX-WT virus incorporated the highest level of gp120 (Fig. 6B). Interestingly, gp120 incorporation levels were much lower for NL(AD8)-NX-208H712 than for NL(AD8)-NX-208 virus. The levels of gp120 incorporation for NL(AD8)-NX-523 and 523H712 remained similar to that previously observed in HeLa cells. The differences in gp120 incorporation between NL(AD8)-NX-127, 127R803, 159, and 159R803 were also similar to those in HeLa cells. Mutations of the tyrosine or diaromatic motif in the gp41 CTs of NL(AD8)-NX viruses slightly decreased gp120 incorporation. The deficit in gp120 incorporation was most marked with the virus containing the Y802W803/SL mutation. Thus, defects in Env incorporation closely matched the replication kinetics of these viruses in MT4.R5 cells. The NL(AD8)-NX-208H712 and 159R803 viruses, which incorporated the smallest amounts of Env, had the slowest replication kinetics.
The hierarchy of Env incorporation observed in MT4.R5 cells was respected in PBMC (Fig. 6C). The largest differences in gp120 incorporation were those between the NL(AD8)-NX-159 and 159R803 viruses and between the NL(AD8)-NX-208 and 208H712 viruses. The difference in gp120 incorporation in MT4.R5 cells and PBMC between the NL(AD8)-NX-208 and 208H712 viruses contrasts with the similar levels of gp120 incorporation observed in HeLa cells, providing evidence for cell type-dependent perturbation of the assembly process by mutation of the tyrosine motif in this primary-CT context. These results demonstrate that the changes in replication kinetics observed for NL(AD8)-NX-159R803 and NL(AD8)-NX-208H712 viruses were probably due to the impaired incorporation of Env in the virions produced by T cells. The NL(AD8)-NX-523 virus remained an exception, as the Y712/H-SPL mutation had no effect on Env incorporation in the various target cells tested.
DISCUSSION
The numerous functions of the gp41 CT, and particularly those involved in determining Env intracellular trafficking and assembly onto core particles, have generally been assessed with only a small number of virus clones, mostly TCLA virus strains. Here, we studied the gp41 CTs of primary Env proteins from four patients infected with HIV-1 of subtype B or G or a circulating recombinant form, CRF02_AG. In addition to the natural genetic diversity between HIV-1 viruses of various subtypes and CRFs, the Env sequences retained for this study also presented a genetic polymorphism at the quasispecies level, within the tyrosine-based Y712SPL or the diaromatic Y802W803 motif. These two motifs have been identified as two of the main determinants regulating Env intracellular trafficking and distribution. Using these primary Env proteins, we showed that genetic differences between HIV-1 primary gp41 CTs at the quasispecies or subtype level can have a specific influence on the intracellular distribution of Env, its incorporation into virions, and viral replication capacity.
Previous confocal microscopy studies have shown that the HIV-1 Env proteins of TCLA viruses, such as HXB2, are located primarily in the TGN at steady state (4, 5). We show here that primary Env proteins may display different distributions in the cytoplasms of HeLa cells. The most striking intracellular distribution pattern was that of Env 523. This Env protein was located in vesicular structures dispersed throughout the cytoplasm. Using a pNL(AD8)-based Env expressor harboring the primary gp41 CT, we showed that the Env 523 CT contained the necessary determinants for a vesicular distribution of Env. Antibody uptake assays, in which living cells were incubated for 1 h at 37°C with anti-Env antibody 2G12 before being fixed and processed, showed that Env-NX-523 was transiently expressed at the cell surfaces of some cells (data not shown). However, confocal microscopy analysis of the subcellular distribution of Env at steady state showed that each preparation of transfected cells contained a mixture of cells with different levels of Env expression. Thus, Env 523 may reach the cell surface in cells producing large amounts of this protein, whereas most of the Env 523 protein may follow a different route toward an intracellular compartment. We tried to characterize the intracellular compartment containing Env NX-523 further, using antibodies directed against the early endosomal marker CD71 (the transferrin receptor) or the late-endosome/multivesicular body marker CD63. Env NX-523 displayed some colocalization with CD71-labeled vesicles, but Env staining persisted in vesicles not labeled with the anti-CD71 antibody (data not shown). No specific colocalization with the late-endosome/multivesicular body marker CD63 was detected (data not shown). Interestingly, different Env subcellular distributions were observed with the proteins from the other three patients studied here. Env 208 behaved like a TCLA Env, with its staining matching that for TGN-46 in the perinuclear area. The tyrosine-based signal was dominant in this context because Env 208H712 showed less-concentrated intracellular staining, with the persistent labeling of peripheral speckles. Other primary Env proteins, such as Env 127 and 159, showed an intermediate intracellular distribution, as they were detected mainly in the perinuclear region, but displayed only partial colocalization with the TGN. The corresponding clone containing the Y802-W803/R mutation showed less-concentrated staining around the nucleus. No specific colocalization with the late-endosome/multivesicular body marker CD63 was detected for the Env NX-208, NX-208H712, NX-127, NX-127R803, NX-159, and NX-159R803 clones (data not shown). Overall, these results suggest that the tyrosine-based signal and the diaromatic motif are involved in the intracellular distribution of primary Env proteins but that the CTs of these Env proteins also contain specific determinants modulating their intracellular distribution, potentially resulting in a distribution different from that previously observed with the TCLA Env. The existence of specific determinants modulating the effect of the tyrosine motif in the Env CT has been suggested for other lentiviruses, such as the simian immunodeficiency virus (SIV). SIV has a long Env CT, similar to that of HIV-1. Tyrosine 721 in SIV (SIV-mac239) corresponds to Y712 in HIV-1. Additional determinants downstream from the tyrosine motif in SIV Env appear to be important because the tyrosine motif affects endocytosis only in the context of a truncated CT, with only marginal effects observed in the context of a full-length CT (7).
We also investigated the effect of the Y712/H-SPL mutation in the tyrosine-based signal and the Y802-W803/R mutation in the diaromatic motif in the context of the Env protein from the NL(AD8) clone. Previously reported mutations (4, 5), Y712/A-SPL in the tyrosine-based signal and Y802W803/SL in the diaromatic motif, were also analyzed for comparison. The Y712/H-SPL mutation in the tyrosine-based signal of Env NL(AD8) resulted in less-concentrated perinuclear staining, with the persistent labeling of peripheral speckles. Env NL(AD8), containing the Y712/A-SPL mutation, was distributed similarly, consistent with previous findings (4). Thus, the Y712/H-SPL and Y712/A-SPL mutations affected the intracellular distribution of NL(AD8) Env in similar ways. These results are consistent with the putative role of the tyrosine-based signal, which is similarly impaired by Y712/A-SPL and Y712/H-SPL mutations. The intracellular distribution of Env NL(AD8), containing a mutation in the diaromatic motif, was also consistent with that previously described for TCLA Env proteins with modifications to this motif (5). Indeed, mutations in the diaromatic motif decreased the concentration of Env staining in the perinuclear area, consistent with the diaromatic motif being involved in the retrograde transport of Env to the TGN. Thus, the introduction of mutations into the tyrosine-based motif or the diaromatic motif of our parental Env NL(AD8) modified the intracellular distribution of Env, as previously described for TCLA Env.
Several lines of evidence have been put forward to suggest that Gag and Env interact. Mutations within the matrix region of Gag affect Env incorporation into virions (17, 22). In polarized epithelial cells, Env expression restricts Gag-mediated budding to the basolateral plasma membrane (33). Glutathione S-transferase pull-down assays have also revealed an interaction between Gag and Env in vitro (12). Confocal microscopy has also shown that most of the assembling HIV-1 particles are present at the plasma membrane in HeLa cells, with Gag staining displaying a punctate pattern coinciding with Env staining (25). We therefore investigated whether the intracellular distribution of primary Env was affected by the coexpression of Gag in the same cell. The gp41 CT of the primary Env protein was used to replace the corresponding sequence in the proviral clone pNL(AD8). Confocal microscopy analysis of HeLa cells transfected with pNL(AD8)-NX containing sequences encoding the various gp41 CTs showed that Gag affected the intracellular distribution of Env, with at least some Env being retained at the cell surface. Gag/Env colocalization was observed for all the Env proteins studied, although the intensities of the Env staining colocalized with Gag were lower for some Env proteins, such as NX-159R803, than for others. Thus, despite causing different intracellular distributions, the CTs of gp41 from primary Env proteins contain conserved determinants mediating direct or indirect interactions with Gag proteins. Such interactions might account for the changes in intracellular distribution observed for primary Env proteins, via competition between Gag proteins and the intracellular effectors interacting with their cytoplasmic tails. The influence of Gag expression on Env trafficking may affect the efficiency of Env retention at the site of virus assembly and its incorporation into virions.
Previous studies have shown that mutations in the tyrosine-based and diaromatic motifs affected the replication of TCLA viruses (5, 14, 49). For instance, the Y712/A-SPL mutation in the tyrosine-based signal affects the replication of HIV-1 NL4-3 viruses in the CEM T-cell line and PBMC (14). Similarly, the HIV-1 HXB2 harboring a gp41 CT with the Y802-W803/SL mutations in the diaromatic motif failed to replicate in the Jurkat T-cell line (5). Based on these published data and our observations of differences in intracellular distribution between the primary Env proteins, we compared the replication kinetics of NL(AD8)-NX viruses harboring primary gp41 CTs or mutated gp41 CTs in MT4.R5 T cells, PBMC, and macrophages. In MT4.R5 cells, we found that some primary gp41 CTs had a marked effect on the replication kinetics of HIV-1, even though these cells have been described as relatively tolerant to mutations or truncations within the gp41 CT of TCLA. Conversely, the NL(AD8)-NX-523 and 523H712 viruses replicated at similar rates. In this case, the presence of a mutation in the tyrosine motif had no effect on viral replication kinetics. None of the NL(AD8)-NX viruses harboring mutations in the diaromatic motif failed to replicate. These data suggest that, in some gp41 CT contexts, the diaromatic motif may be less involved in HIV-1 replication than previously reported for HIV-1 HXB2 and Jurkat T cells (5). There must therefore be additional determinants with different effects on viral replication kinetics, depending on the primary gp41 CT considered, paralleling the differences in intracellular distribution between the primary Env proteins. The overall hierarchies of viral replication were similar in PBMC and MT4.R5 cells. This observation suggests that the mechanisms underlying viral assembly are similar in these two cell types. Interestingly, in MDM, viruses containing a primary CT with a mutation in the tyrosine motif displayed replication kinetics different from those observed in MT4.R5 cells and PBMC. Thus, the determinants of the gp41 CT involved in the various steps of viral replication may differ in PBMC and macrophages, reflecting perhaps the occurrence of assembly at different sites in acutely infected T cells and MDM (40, 43).
Different intracellular Env distributions may result in differences in Env incorporation and thus in virus replication kinetics. Indeed, the replication kinetics of NL(AD8)-NX viruses carrying primary gp41 CTs were correlated with levels of Env incorporation into virions. Several mechanisms may account for the differences in Env incorporation mediated by the various primary gp41 CTs. The gp41 CT may promote Env recruitment to the site of virus assembly by interacting with effectors involved in intracellular-trafficking pathways. Thus, differences in the gp41 CT determinants interacting with effectors involved in intracellular-trafficking pathways and/or differences in the expression or distribution of such effectors may be responsible for the specific effects of primary gp41 CTs in a defined cellular environment. This may account for Env incorporation being influenced by both the gp41 CT and the cellular environment. Moreover, the gp41 CT derived from the two primary HIV-1 viruses of subtype B studied here appeared to be more sensitive to mutations in the tyrosine or diaromatic motif. Interestingly, most previous studies on Env intracellular trafficking and distribution were carried out with TCLA HIV-1 of subtype B. In HIV-1 viruses belonging to subtype G or CRF02_AG, additional unidentified determinants also seem to play a major role in Env trafficking and distribution within the cell. The potential importance of intersubtype variability for Env distribution and viral assembly reported here requires confirmation for other isolates of various subtypes. Env incorporation may also be facilitated by host factors, which may interact differently with the various primary gp41 CTs. A direct or indirect interaction between the gp41 CT and the matrix region of Gag is probably required for optimal Env incorporation. Various studies based on site-directed mutagenesis, deletion, or truncation of the gp41 CT have suggested that Env is incorporated into virions via interactions between LLP and the matrix region of Gag (29, 36, 42). However, the impact of gp41 CT modifications on Env intracellular distribution was not assessed in these studies. We found that the matrix was tolerant to variation in the gp41 CT, and particularly in the LLP, because Env NX-523, harboring a gp41 CT from a subtype G virus, was incorporated at least as well as Env harboring gp41 CT sequences from subtype B viruses.
In conclusion, these data suggest that the genetic diversity observed within the gp41 CTs of primary HIV-1 at the quasispecies or subtype level may have different influences on virus assembly and replication capacity. This may be at least partly due to differences in the gp41 CT determinants interacting with effectors involved in intracellular-trafficking pathways. Further studies are required to characterize these gp41 CT determinants specific to primary viruses belonging to a given subtype or CRF.
Acknowledgments
We thank Eric O. Freed (National Cancer Institute at Frederick, Frederick, Md.) and Hermann Katinger (Institute for Applied Microbiology, University of Natural Resources, Vienna, Austria) for providing us with the pNL(AD8) proviral construct and MAb 2G12, respectively. We also thank C. Berlioz-Torrent for helpful discussions. Our data were generated with the help of the Confocal Microscopy Facility of François Rabelais University.
This work was supported by grants from SIDACTION. Our research is supported by the Region Centre (Equipe ESPRI). M.L. was supported by fellowships from the French Ministry of Research and SIDACTION. B.L. is a fellow of the Region Ile-de-France.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Akari, H., T. Fukumori, and A. Adachi. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74:4891-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ataman-Onal, Y., C. Coiffier, A. Giraud, A. Babic-Erceg, F. Biron, and B. Verrier. 1999. Comparison of complete env gene sequences from individuals with symptomatic primary HIV type 1 infection. AIDS Res. Hum. Retrovir. 15:1035-1039. [DOI] [PubMed] [Google Scholar]
- 3.Barin, F., S. Brunet, D. Brand, C. Moog, R. Peyre, F. Damond, P. Charneau, and F. Barre-Sinoussi. 2004. Interclade neutralization and enhancement of human immunodeficiency virus type 1 identified by an assay using HeLa cells expressing both CD4 receptor and CXCR4/CCR5 coreceptors. J. Infect. Dis. 189:322-327. [DOI] [PubMed] [Google Scholar]
- 4.Berlioz-Torrent, C., B. L. Schacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blot, G., K. Janvier, S. Le Panse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for Env incorporation into virions and infectivity. J. Virol. 77:6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, K., A. Pelchen-Matthews, S. Höning, P. J. Vance, L. Creary, B. S. Haggarty, J. Romano, W. Ballensiefen, J. A. Hoxie, and M. Marsh. 2000. The simian immunodeficiency virus envelope glycoprotein contains multiple signals that regulate its cell surface expression and endocytosis. Traffic 1:661-674. [DOI] [PubMed] [Google Scholar]
- 8.Bultmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit surface expression. J. Virol. 75:5263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervantes-Acosta, G., R. Lodge, G. Lemay, and E. A. Cohen. 2001. Influence of human immunodeficiency virus type 1 envelope glycoprotein YXXL endocytosis/polarization signal on viral accessory protein functions. J. Hum. Virol. 4:249-259. [PubMed] [Google Scholar]
- 10.Chen, S. S., S. F. Lee, and C. T. Wang. 2001. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41. J. Virol. 75:9925-9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel, F., M. D. Hoggan, R. L. Willey, K. Strebel, M. A. Martin, and R. Repaske. 1989. Genetic recombination of human immunodeficiency virus. J. Virol. 63:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosson, P. 1996. Direct interaction between the envelope and matrix proteins in HIV-1. EMBO J. 15:5783-5788. [PMC free article] [PubMed] [Google Scholar]
- 13.Dacheux, L., A. Moreau, Y. Ataman-Onal, F. Bion, B. Verrier, and F. Barin. 2004. Evolutionary dynamics of the glycan shield of human immunodeficiency virus envelope during natural infection and implications for exposure of the 2G12 epitope. J. Virol. 78:12625-12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day, J. R., C. Münk, and J. C. Guatelli. 2004. The membrane-proximal tyrosine-based sorting signal of human immunodeficiency virus type 1 gp41 is required for optimal viral infectivity. J. Virol. 78:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unumatz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Deschambeault, J., J. P. Lalonde, G. Cervantes-Acosta, R. Lodge, E. A. Cohen, and G. Lemay. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J. Virol. 73:5010-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68:1689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenberg, D., and M. Wesson. 1990. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers 29:171-177. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry co-factor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 21.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Gabuzda, D. H., A. Lever, E. Terwilliger, and J. Sodroski. 1992. Effects of deletions in the cytoplasmic domain on biological fuctions of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 66:3306-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hourioux, C., D. Brand, P.-Y. Sizaret, F. Lemiale, S. Lebigot, F. Barin, and P. Roingeard. 2000. Identification of the glycoprotein 41TM cytoplasmic tail domains of human immunodeficiency virus type 1 that interact with Pr55Gag particles. AIDS Res. Hum. Retrovir. 16:1141-1147. [DOI] [PubMed] [Google Scholar]
- 27.Hunter, E., and R. Swanstrom. 1990. Retrovirus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 157:187-253. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptor. J. Biol. Chem. 273:403-409. [DOI] [PubMed] [Google Scholar]
- 29.Kalia, V., S. Sarkar, P. Gupta, and R. C. Montelaro. 2003. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J. Virol. 77:3634-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kliger, Y., and Y. Shai. 1997. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry 36:5157-5169. [DOI] [PubMed] [Google Scholar]
- 31.Lee, S. J., W. Hu, A. G. Fisher, D. J. Looney, V. F. Kao, H. Mitsuya, L. Ratner, and F. Wong-Staal. 1989. Role of the carboxy-terminal portion of the HIV-1 transmembrane protein in viral transmission and cytopathogenicity. AIDS Res. Hum. Retrovir. 5:441-449. [DOI] [PubMed] [Google Scholar]
- 32.Lodge, R., H. Gottlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lodge, R., J. P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 38.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and multivesicular body. J. Virol. 78:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Bercoff, D., A. David, H. Sudry, F. Barre-Sinoussi, and G. Pancino. 2003. Fcgamma receptor-mediated suppression of human immunodeficiency virus type 1 replication in primary human macrophages. J. Virol. 77:4081-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piller, S. C., J. W. Dubay, C. A. Derdeyn, and E. Hunter. 2000. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 from human immunodeficiency virus type 1: effects on glycoprotein incorporation and infectivity. J. Virol. 74:11717-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 44.Rowell, J. F., P. E. Stanhope, and R. F. Siciliano. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J. Immunol. 155:473-488. [PubMed] [Google Scholar]
- 45.Smith, D. H., R. A. Byrn, S. A. Marsters, T. Gregory, J. E. Groopman, and D. J. Capon. 1987. Blocking of HIV infectivity by a soluble, secreted form of the CD4 antigen. Science 238:1704-1707. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vachot, L., Y. Ataman-Önal, C. Terrat, P.-Y. Durand, B. Ponceau, F. Biron, and B. Verrier. 2004. Retrospective study to time the introduction of HIV-1 type 1 non-B subtypes in Lyon, France, using env genes obtained from primary infection samples. AIDS Res. Hum. Retrovir. 20:687-691. [DOI] [PubMed] [Google Scholar]
- 49.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wrin, T., T. P. Loh, J. C. Vennari, H., Schuitemaker, and J. H. Nunberg. 1995. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J. Virol. 69:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 52.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Höning, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, X., S. Kurteva, X. Ren, S. Lee, and J. Sodroski. 2005. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J. Virol. 79:12132-12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, X., X. Yuan, M. F. McLane, T. H. Lee, and M. Essex. 1993. Mutations in the cytoplasmic domain of human immunodeficiency type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J. Virol. 67:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]