Abstract
The 3C protease (3Cpro) from foot-and-mouth disease virus (FMDV), the causative agent of a widespread and economically devastating disease of domestic livestock, is a potential target for antiviral drug design. We have determined the structure of a new crystal form of FMDV 3Cpro, a chymotrypsin-like cysteine protease, which reveals features that are important for catalytic activity. In particular, we show that a surface loop which was disordered in previous structures adopts a β-ribbon structure that is conformationally similar to equivalent regions on other picornaviral 3C proteases and some serine proteases. This β-ribbon folds over the peptide binding cleft and clearly contributes to substrate recognition. Replacement of Cys142 at the tip of the β-ribbon with different amino acids has a significant impact on enzyme activity and shows that higher activity is obtained with more hydrophobic side chains. Comparison of the structure of FMDV 3Cpro with homologous enzyme-peptide complexes suggests that this correlation arises because the side chain of Cys142 contacts the hydrophobic portions of the P2 and P4 residues in the peptide substrate. Collectively, these findings provide compelling evidence for the role of the β-ribbon in catalytic activity and provide valuable insights for the design of FMDV 3Cpro inhibitors.
Foot-and-mouth disease virus (FMDV) causes a highly contagious disease of some of the most common species of domesticated livestock, including cattle, sheep, pigs, and goats (15). This dangerous pathogen is responsible for repeated epidemics around the world that invariably inflict significant economic damage on the agricultural and wider economies of the regions affected (15, 20, 35). Although vaccines that protect against the disease are available, their use, even in the event of an outbreak, is compromised by practical and political difficulties (35). It is therefore desirable to develop alternative methods for disease control, and one promising avenue is to target drugs to the viral enzymes that are necessary for replication.
Like other picornaviruses such as poliovirus (PV), human rhinovirus (HRV), and hepatitis A virus (HAV), FMDV has a single-stranded positive-sense RNA genome that is translated as a single open reading frame. The resulting polyprotein precursor is digested by virally encoded proteases to yield the proteins required for capsid assembly and viral RNA replication. The FMDV 3C protease (3Cpro) (15) performs 10 of the 13 cleavages involved in this dissection and therefore constitutes an attractive target for the design of antiviral agents.
To advance the process of antiviral drug design, we previously determined the crystal structure of FMDV 3Cpro (5, 6). This revealed a close structural similarity to the HAV, HRV, and PV 3Cpro structures (1, 24, 26) and helped to reassert the primacy of the notion that this family of chymotrypsin-like cysteine proteases have a Cys-His-Asp/Glu catalytic triad of residues within their active sites (6). Subsequent studies on HAV 3Cpro have supported this contention (37). One of the most notable differences observed was that a conserved β-ribbon structure that was found in all of the other picornavirus 3Cpro structures solved to date, appeared to be disordered in FMDV 3Cpro. This β-ribbon, which corresponds to a similar feature in some bacterial chymotrypsin-like serine proteases (7, 18), folds over the peptide binding cleft of the enzyme and appears to make significant contributions to substrate specificity, at least for HRV 3Cpro (11, 23). It was therefore somewhat surprising to find it largely disordered in the structure of FMDV 3Cpro; the issue was further complicated because this region of the protein (residues 138 to 150) contained an amino acid substitution (C142S) which was introduced to make the protein soluble (5, 6).
A more practical difficulty with the original crystals of FMDV 3Cpro (space group R3) was that the active sites of both molecules in the asymmetric units were occluded by crystal contacts; this made them unsuitable for crystal soaking experiments which would have allowed structural analysis of enzyme-inhibitor complexes (6). Moreover, this crystal form suffered from variable levels of hemihedral twinning, which was a complicating factor in data processing. To obtain a new crystal form that would be free from both of these problems, we generated a series of mutant FMDV 3Cpro enzymes containing amino acid substitutions that were designed to disrupt contacts between proteins in the R3 crystals and thereby promote alternative packing modes in different crystal forms. The mutation K51Q was found to produce crystals of FMDV 3Cpro in a new space group (P212121) that is not subject to twinning and contains two molecules in the asymmetric unit. The resulting structure of FMDV 3Cpro, determined at 2.2 Å resolution, reveals that in one of these molecules the loop formed by residues 138 to 150 is folded into a β-ribbon structure lying over the peptide binding cleft in a way that closely resembles the equivalent structures observed in the HRV and PV 3Cpro enzymes. We probed the functional significance of this structure by using mutagenesis and showed that replacement of residue 142 at the apical tip of the β-ribbon which lies closest to the active site can have a major impact on the cleavage activity of FMDV 3Cpro. This provides the first experimental demonstration of the importance of the β-ribbon in catalysis for this class of enzyme. Interestingly, in the other molecule of the asymmetric unit, not only is the 138-to-150 loop disordered—as observed previously (6)—but the active-site residues His46 and Asp84 are also severely distorted from the active conformation, apparently because of crystal contacts. This finding is strongly reminiscent of previous observations made with HAV 3Cpro (1, 3, 4) and is indicative of a surprising degree of flexibility in these enzymes that has not generally been observed with chymotrypsin-like serine proteases.
MATERIALS AND METHODS
Preparation and expression of crystal contact mutant enzymes.
Mutant forms of FMDV strain A1061 3Cpro were generated by using the previously described bacterial expression construct as a template (5, 6). Briefly, this expresses the protease with an N-terminal polyhistidine tag, which may be removed by thrombin. Following thrombin cleavage, the protein contains residues 1 to 207 of the 213 amino acids of wild-type 3Cpro and has an additional Gly at the N terminus and an additional His at the C terminus. This construct also incorporates a C163A mutation to abolish proteolytic activity; moreover, it has C95K and C142S mutations that are required for solubility (5, 6). Mutations designed to disrupt protein-protein contacts within the R3 crystals were introduced by using the QuikChange mutagenesis kit (Stratagene) in accordance with the manufacturer's protocols. Mutant proteins were expressed in Escherichia coli BL21(DE3) and purified on TALON resin (BD Biosciences) and by gel filtration chromatography exactly as described previously (5, 6).
Crystallization.
Purified mutant 3Cpro enzymes, concentrated to 10 mg/ml in 50 mM HEPES (pH 7.0)-200 mM NaCl-1 mM EDTA-1 mM β-mercaptoethanol, were used in extensive crystallization trials performed at 4°C and 18°C with commercial screening kits in robotic sitting-drop vapor diffusion experiments. In these experiments, 100 nl of protein was mixed with 100 nl taken from the 100-μl reservoir solution. Optimization of crystallization conditions was performed manually with 2-μl drops over a 200-μl reservoir.
Data collection and structure determination.
X-ray diffraction data were collected from crystals cryocooled to 100 K in a 70:30 volume ratio of reservoir solution (19% polyethylene glycol 4000, 100 mM Tris HCl [pH 8.5], 2 mM sodium acetate) and saturated sucrose, respectively. Diffraction data were processed with the CCP4 program suite (9) and phased by molecular replacement with PHASER (25) by using a superposition of the two models of FMDV 3Cpro from the asymmetric unit of the original R3 crystals (6) as a search model. This resulted in a readily interpretable map revealing two molecules in the asymmetric unit. Model adjustment with O was interleaved with simulated annealing refinement performed with CNS (7a). Final statistics for data collection and refinement are reported in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data collection and refinement parameter | Result |
|---|---|
| Diffraction data | |
| Space group | P212121 |
| a (Å) | 59.39 |
| b (Å) | 71.71 |
| c (Å) | 92.40 |
| Wavelength (Å) | 1.488 |
| Resolution range (Å) | 23.6-2.2 |
| No. of independent reflections | 20,590 |
| Multiplicitya | 3.5 (3.5) |
| Completeness (%) | 99.7 (99.5) |
| I/sI | 11.6 (3.8) |
| Rmerge (%)b | 7.6 (33.9) |
| Model refinement | |
| No. of nonhydrogen atoms/water atoms | 2,818/110 |
| Rmodelc/Rfreed (%) | 22.6/25.5 |
| RMSe deviation from ideal bond lengths (Å) | 0.006 |
| RMS deviation from ideal angles (°) | 1.18 |
| Ramachandran plot, favored/additional (%) | 89.0/10.7 |
| Avg B factor (Å2) | 34.6 |
| Protein Database identification | 2j92 |
Values for the outermost resolution shell (2.32 to 2.20 Å) are in parentheses.
Rmerge = 100 × ΣhΣj|Ihj − Ih|/ΣhΣjIhj, where Ih is the weighted mean intensity of the symmetry-related reflections Ihj.
Rmodel = 100 × Σh|Fobs − Fcalc|/ΣhFobs, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is Rmodel calculated by using a randomly selected 5% sample of reflection data omitted from the refinement.
RMS, root mean square.
Preparation of C142 mutants.
Mutation of the residue at position 142 was performed with a construct that encodes a Δ3B1-3B2-3B3-3C fusion protein; this contains active 3Cpro fused to the upstream 3B peptides found in the FMDV polyprotein (6, 22). This construct also has the C95K and C142S mutations that are required for solubility. Upon expression in E. coli BL21(DE3)/pLysS, 3Cpro cleaves itself away from the 3B peptides and may be purified in a single step on TALON resin via a noncleavable C-terminal polyhistidine tag. Mutations at residue 142 were introduced with the QuikChange mutagenesis kit (Stratagene); mutant 3Cpro proteins were expressed and purified (to greater than 95% homogeneity) exactly as previously described (6). The Cys variant (C142) was prone to aggregation, most likely because of intermolecular disulfide formation (5, 6), which resulted in some preparations showing differing levels of activity (by approximately twofold).
Fluorogenic substrate synthesis.
The internally quenched fluorogenic substrate 4-(4′-dimethylaminophenylazo)benzoic acid-APAKQLLDL[5-(2-aminoethyl)amino-1-naphthalenesulfonic acid (EDANS)]FDLLK was synthesized on an Advanced ChemTech Apex 396 multiple peptide synthesizer by using a standard Fmoc [N-(9-fluorenyl)methoxycarbonyl]-tert-butyl peptide synthesis strategy (2) and 1-hydroxybenzotriazole-2-([1H]benzotriazole-1-yl)1,1,3,3-tetramethyluroniumhexafluorophosphate activation with dimethylformamide as a solvent. The peptide chain was built on Fmoc-Lys-Wang resin. In a typical peptide synthesis, 25 mmol of resin was used. Fmoc removal was achieved with 1 ml of 20% (vol/vol) piperidine in dimethylformamide. All of the couplings including 4-(4′-dimethylaminophenylazo)benzoic acid were performed with a fivefold excess of activated amino acid over the resin free amino groups by Fmoc-amino acid-2-([1H]benzotriazole-1-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate-N-hydroxybenzotriazole-diisopropyl ethylamine (1:1:1:2) activation. Addition of the EDANS-labeled aspartate residue was completed manually by using a twofold excess of activated amino acid over the resin free amino groups Fmoc-Asp(EDANS)-OH-1-hydroxy-7-azabenzotriazole-O-(7-azabenzotriazole-1-yl)- N,N,N′N′-tetramethyluroniumhexafluorophosphate-diisopropyl ethylamine (1:1:1:2). Asp(EDANS) for the synthesis of the fluorogenic peptide was obtained from aspartic acid as described in reference 36. Simultaneous side chain deprotection and cleavage from the resin were performed by adding the deprotection mixture containing trifluoroacetic acid-H2O-tri-isopropylsilane (95:2.5:2.5) for up to 3 h to the dry peptidyl resin. Purification to 98% homogeneity was achieved by reverse-phase high-performance liquid chromatography (HPLC) with an ACE 5 C18 column (250 by 21.2 mm; Hichrom Limited, Berkshire, United Kingdom). The identity of the peptide was confirmed by mass spectrometry.
Quantitative analysis of purified C142 mutants.
For each mutant enzyme, the hydrolysis rate was monitored after the addition of 100 μl of different concentrations of fluorogenic substrate to 100 μl of an enzyme solution (1.6 μM for C142T and C142S and 0.8 μM for C142A, C142L, and C142V). The enzyme concentrations were determined by using an extinction coefficient of 0.41 U of optical density per mg/ml at 280 nm that was calculated from the amino acid sequence. The final enzyme concentrations were 800 and 400 nM, respectively, and the final substrate concentrations were in the range of 2 to 90 mM.
Incubations were performed at 37°C and pH 7.4. All assay solutions were made in a 100 mM sodium phosphate buffer solution containing 1 mM Tris(2-carboxyethyl)-phosphine HCl, 1 mM Na EDTA, and 5% glycerol.
Hydrolysis rates were determined from fluorescence readings (arbitrary fluorescence units per minute). Fluorescence was monitored in 96-well plates by a Cytofluor multiwell plate reader (PerSeptive Biosystems) for up to 20 min. The excitation wavelength range was 340 to 360 nm, and emission was measured at 460 nm. The initial velocity was calculated by using the fluorescence measured at 2-min intervals. The same measurements were carried out three times, and the mean values were transformed (micromolar per minute) and plotted versus the substrate concentration (micromolar). Kinetic parameters were determined by fitting the data obtained to the Michaelis-Menten equation by using the GraFit (21) computer program assuming 100% activity for the enzyme. For all of the enzyme species studied here, it was found that the quenched fluorescent substrate that was used exhibited Km values greater than the highest usable substrate concentration (ca. 100 mM maximum solubility). As a result, the enzymes did not show saturation kinetics but instead had a linear dependence of rate on the concentration of the substrate. In such circumstances, it is impossible to determine individual values for kcat and Km but the slope of the graph yields the specificity constant kcat/Km (12).
RESULTS
Design and crystallization of crystal contact mutants.
Close inspection of the protein-protein contacts in the R3 crystals of FMDV 3Cpro with the program O (19) allowed us to identify five points of contact that were amenable to mutagenesis. We focused on contacts involving amino acid side chains that were distant from the active site and highly exposed to solvent. The selected mutations (K51Q, R68E, R97E, R126E, and L134Q) were engineered, expressed in E. coli, and purified as described in Materials and Methods. All but one of the mutations yielded around 20 mg of purified soluble protein from 4 liters of culture. The L134Q substitution reduced the yield by around 90% and resulted in a protein that exhibited aggregation on a gel filtration column (data not shown); this mutant protein was therefore not used in crystallization experiments.
The remaining four mutant proteins were subjected to extensive sitting-drop vapor diffusion crystallization trials with commercial screening kits (see Materials and Methods). Only in the case of the 3Cpro(K51Q) mutant protein were crystals obtained; optimization yielded single crystals of 3Cpro(K51Q) by mixing equal volumes of 10 mg/ml protein and a reservoir solution composed of 16 to 22% polyethylene glycol 4000, 100 mM Tris HCl (pH 8.5), and 2 mM sodium acetate. These crystals, which diffracted to 2.2 Å at 100 K, belong to space group P212121 with cell dimensions a = 59.4 Å, b = 71.1 Å, and c = 92.4 Å and have two molecules in the asymmetric unit. The structure was solved by molecular replacement and refined to 2.2 Å, yielding an Rfree value of 25.4% (see Materials and Methods). The final model, which has good stereochemistry, contains residues 7 to 207 of molecule A, residues 7 to 206 of molecule B (which is also missing loops 74 to 81 and 141 to 147 inclusive), and 110 water molecules. Data processing and refinement statistics are summarized in Table 1.
Structure of FMDV 3Cpro.
Overall, the structure of the protease in this crystal form is very similar to that obtained in the R3 space group; it adopts the anticipated chymotrypsin-like fold that has been described in detail elsewhere (6) (Fig. 1A and B). Most of the differences involve minor distortions of the exposed portions of the polypeptide backbone due to alterations in crystal packing. Even in the vicinity of the K51Q mutation that was introduced to alter packing from the original R3 space group, the structural changes are relatively modest. This residue lies on a short exposed loop, which appears to be relatively flexible (B = 40 to 62 Å2 compared to an average B factor for the molecule of 35 Å2).
FIG. 1.
Crystal structure of FMDV 3Cpro and comparison with HRV 3Cpro. (A) Structure of molecule A in the new crystal form of FMDV 3Cpro. Secondary structure elements of FMDV 3Cpro are color coded. The strands of the front and back β-sheets of the two β-barrels are green and blue, respectively; helices are pink; and the β-ribbon is orange. β-Strands mentioned in the text are labeled. This and other structural figures were prepared with Pymol (10). (B) Structure of molecule B in the new crystal form of FMDV 3Cpro colored according to the same scheme as in panel A. Note that the B2-C2 β-ribbon present in molecule A is absent in FMDV 3Cpro; the visible residues of this loop are orange. Active-site residues are labeled; note that A163 is Cys in the wild-type protein. (C) Electron difference density maps for residues 138 to 150 of FMDV 3Cpro (molecule A) showing the β-ribbon structure. Shaded blue chicken wire represents the initial Fo-Fc difference map, contoured at 2σ, which was generated with model phases calculated prior to incorporation of the loop residues (depicted as sticks and colored by atom type). (D) Structure of HRV 3Cpro (23) colored according to the scheme used in panel A. (E) Close-up view of the β-sheet in molecule A from which the β-ribbon protrudes. β-Strands mentioned in the text are labeled. Residues involved in the stabilization of this conformation are shown as sticks (see text). (F) Close-up view of the β-sheet in molecule B from which the β-ribbon protrudes. Note the reorganization of Y136 and K137 with respect to molecule A (panel E).
The most striking difference is that the loop formed by residues 138 to 150 within molecule A, which was previously observed to be largely disordered (6), is now found to fold up into a β-ribbon structure that lies over the N-terminal side of the substrate binding cleft (Fig. 1A and C). However, in the other protease molecule present in the asymmetric unit (molecule B), this loop remains largely disordered (Fig. 1B); there was insufficient electron density to allow incorporation of residues 142 to 146 into the model of this molecule. The visible termini of the loop are in configurations slightly different from those observed previously (6).
The 13-residue β-ribbon observed here for FMDV 3Cpro (molecule A) is remarkably similar to the corresponding feature in the HRV (24) (Fig. 1D) and PV (26) 3Cpro enzymes. It is only 1 amino acid longer than the β-ribbons in the HRV and PV 3Cpro enzymes; the extra residue is accommodated at the apical tip of the β-ribbon. In HAV 3Cpro, this feature is extended by 8 residues compared to FMDV 3Cpro. However, for all four picornaviral proteases, the position and orientation of the ribbon with respect to the body of the protease are very similar. The tips, which exhibit the greatest structural variation, are also associated with higher B factors, indicative of a greater degree of flexibility at this extremity; in the case of FMDV 3Cpro, the B factors increase from 28-42 Å2 at the base of the loop to 68-71 Å2 at the tip. The overall structural similarity between the β-ribbons in the different 3C proteases and the observation in the case of HRV 3Cpro that this conformation is stabilized by the presence of substrate analogues (11, 23) strongly suggest that it is likely to be the catalytically relevant structure of this loop. This conclusion is supported by analysis of the impact of mutagenesis of a loop residue on enzyme activity (see below). We suggest that the previous observation that this loop was disordered in FMDV 3Cpro (6) and the present finding that it is also disordered in molecule B are due to the fact that alternative, disordered states of the loop may be trapped in the crystal. It therefore seems evident that this loop is a highly flexible feature on the surface of the protein and that the observed conformation is influenced by contacts between protease molecules within the crystal (though see Discussion).
Comparison of the FMDV 3Cpro structures with and without the β-ribbon reveals that formation of the ribbon is accompanied by switching of the B2 β-strand (Fig. 1E and F). In the presence of the β-ribbon (molecule A), strand B2 pairs up with strand C2 at the bottom of the front four-strand β-sheet of the C-terminal β-barrel (Fig. 1E). This arrangement is identical to that observed in all other picornavirus 3Cpro structures (1, 24, 26). However, when the β-ribbon is not formed, as in molecule B (Fig. 1F), strand B2 peels away from this β-sheet and instead pairs up with the polypeptide linking the N- and C-terminal β-barrels (residues 99 to 114); a mini two-strand β-sheet is thereby formed of residues 137 to 139 (B2′) paired with residues 107 to 109 (A2′), a configuration first seen in the original structure determination (6). This rearrangement also involves an interesting switch in the position of the side chains of Y163 and K137 involving displacements of 6 to 8 Å. The formation of the β-ribbon is in part stabilized by packing of the phenyl moiety of Y136 against a pair of buried hydrophobic residues, M109 and F152, and the formation of an H bond (3.2 Å) between the side chain hydroxyl group of Y136 and the main chain carbonyl oxygen of A107 (Fig. 1E). However, disruption of the β-ribbon displaces Y136, rupturing its H bond with the main chain and leaving the side chain exposed to solvent. Instead, K137, which was previously exposed to solvent, is repositioned to pack against M109 and F152 in a conformation that is stabilized by H bonds to carbonyl main chain oxygen atoms from P149 and L151 (Fig. 1F). Although there are clearly identifiable stabilizing interactions for the two conformations observed for FMDV 3Cpro, the energetic barrier between them must be relatively small.
The formation of the β-ribbon in FMDV 3Cpro is also apparently accompanied by changes in the backbone conformation of loop E1-F1, although these are relatively modest, an unsurprising observation given that this loop has already been found to be relatively flexible (6).
Structure of the active site.
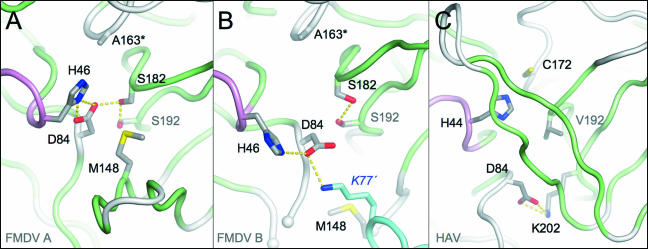
In the active site of molecule A, the conformation of the residues of the catalytic triad—C163 (replaced by Ala in the present structure), H46, and D84—is essentially identical to that observed previously (Fig. 2A) and serves to reinforce the hypothesis that this class of enzymes requires a full triad rather than a Cys-His dyad for proteolytic function (6). Strikingly, in molecule B the triad is severely disrupted and appears to adopt a nonproductive conformation; not only is the side chain of D84 redirected away from its interaction with H46, but H46 itself is also rotated outward, its side chain moving by about 6 Å from the active site (Fig. 2B). This disruption appears to be largely due to contact with a neighboring 3Cpro molecule in the crystal, which inserts the side chain of K77 close to the active site of molecule B and diverts the side chain D84 into making a salt bridge with it. The removal of D84 from the active site evidently creates space, allowing H46 to rotate to a new position. Conceivably, in the wild-type enzyme, H46 would have reduced freedom because of its likely interaction with the catalytic nucleophile C163 (replaced in our structure by Ala). Nonetheless, the displacement of D84 by salt bridging to a lysine residue is remarkably reminiscent of the distortion of the equivalent D84 residue from the active site of HAV 3Cpro due to the formation of an intramolecular salt bridge with K202 (1, 3, 4) (Fig. 2C). This observation originally supported the hypothesis that picornaviral 3C proteases might not require a complete catalytic triad (1, 4). However, this view has been challenged by more recent findings (see Discussion).
FIG. 2.
Comparison of catalytic triads in different FMDV and HAV 3Cpro structures. (A) Close-up view of the active site in molecule A of the present structure. The side chains of key residues are shown as sticks color coded according to atom type. The secondary structure is colored as in Fig. 1A. (B) Equivalent view of the active site of molecule B. Note the interaction between D84 from molecule B and K77′ from a neighboring 3Cpro molecule in the crystal (differentiated by cyan loops and carbon atoms). (C) Active site of HAV 3Cpro showing a similar reorientation of the catalytic Asp (D84) as a result of salt bridge formation with a Lys residue (K202).
Effect on enzyme activity of amino acid substitutions at residue 142 in the β-ribbon.
The FMDV 3Cpro protein used for crystallization trials has two mutations, C95K and C142S, which are required to make the protein soluble. While the C95K substitution is located on the dorsal surface of the protease, well away from the active site, the C142S mutation occurs at the tip of the β-ribbon visualized in molecule A of the present structure. Preliminary HPLC-based peptide cleavage assays indicated that the C142S substitution had no effect on substrate specificity but did reveal a very significant loss of enzyme activity (of about 2 orders of magnitude) (6). In order to probe the molecular basis of this effect and to attempt to produce a soluble enzyme with closer-to-wild-type activity, we engineered alternative substitutions of C142 and assayed the resulting enzymes in peptide cleavage assays with a fluorescently tagged peptide substrate (based on the optimal cleavage sequence that corresponds to the VP1-2A junction [6]); this allowed more accurate quantification of catalytic rates (see Materials and Methods).
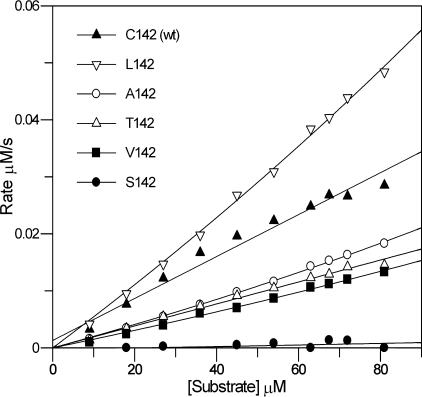
The results of these assays confirm that the C142S mutation drastically reduces the proteolytic activity of the enzyme to <1% of the wild-type activity (Fig. 3; Table 2). Exact comparison of the relative activities of the wild-type and mutant proteins is difficult because of the aggregation behavior of the wild type; this is most likely due to the formation of intermolecular disulfide bonds involving C142 (in spite of the presence of reducing agents), although an intramolecular disulfide between C142 and the active-site nucleophile (C163) cannot be ruled out. Interestingly, replacement of C142 with amino acids such as Thr, Ala, and Val, which are all more apolar than Ser, gave much higher levels of proteolytic activity (41 to 60% of the wild-type activity). The highest activity of all was observed with the most hydrophobic substitution; the C142L mutant enzyme exhibited levels of activity that were comparable to or greater than those observed with wild-type FMDV 3Cpro. This trend strongly suggests that an apolar residue at position 142 is required for optimal activity (Fig. 3; Table 2).
FIG. 3.
Assays of FMDV 3Cpro mutant enzyme activities. See Materials and Methods for experimental details and Table 2 for the kcat/Km values determined for each mutant.
TABLE 2.
Comparative specificity constants for mutant FMDV 3Cpro enzymes with substitutions at residue 142 in the β-ribbon
| Residue 142 | kcat/Km (M−1 s−1)a |
|---|---|
| Cys | 990 ± 20 |
| Ser | <10 |
| Val | 500 ± 70 |
| Thr | 410 ± 70 |
| Ala | 590 ± 2 |
| Leu | 1,510 ± 50 |
kcat/Km values were determined from the linear part of the rate-versus-substrate concentration plot (Fig. 3). Values are the averages from two independent experiments.
DISCUSSION
Protease flexibility and interpretation of crystallographic data.
The results presented in this paper provide valuable new insights into the structure and mechanism of substrate recognition of FMDV 3Cpro. In particular, they serve to clarify the role of the β-ribbon that folds over the substrate binding cleft.
Our findings also provide a useful cautionary note with regard to the interpretation of crystallographic analyses of proteins that have flexible loops. The initial FMDV 3Cpro structure led us to conclude that the β-ribbon feature may be missing from FMDV, apparently in contrast to other picornaviral proteases (6). However, the structure reported here shows that the β-ribbon can indeed be present in FMDV 3Cpro—although the loop is likely to be flexible in solution—and supports the contention that this structural feature is common to all of the picornaviral 3C proteases solved to date (1, 24, 26). Strikingly, it is also found on homologous viral and bacterial proteases (13, 14, 18, 28, 31) (see below).
Moreover, the present study revealed a conformation of FMDV 3Cpro in which the Cys-His-Asp catalytic triad was drastically disrupted, apparently as a result of crystal contacts (Fig. 2). The gross nature of the distortion, which leaves none of the three residues in a position to interact with one another, and comparison with other picornaviral 3Cpro structures with catalytic triads that closely resemble the conformation adopted in serine proteases (6, 24, 26, 37) strongly suggest that this nonproductive conformation is an artifact of crystallization. The distortion observed here for FMDV 3Cpro is reminiscent of the lesser deformation of the catalytic triad that was observed in HAV 3Cpro, the first picornaviral 3C protease for which the structure was reported (1); in that case, only the Asp residue of the triad was directed away from the active site. At the time, this finding was unprecedented, and since the deformed triad was observed in subsequent independent structure determinations (3, 4), this led to the formulation of a dyad hypothesis for the catalytic mechanism for picornaviral 3C proteases which asserted that the Cys-His dyad is sufficient for activity (34). This hypothesis has only lately been challenged as a result of recent structural analyses of FMDV 3Cpro (6), HAV 3Cpro (37), and the 3C-like NIa protease from tobacco etch virus (TEV) (31), which observed Cys-His-Asp triads in these enzymes that closely mimicked the Ser-His-Asp triads found in serine proteases.
Collectively, the observations from crystallographic analyses of FMDV 3Cpro and HAV 3Cpro emphasize that protein flexibility can make it difficult to draw firm conclusions regarding enzyme function from structural data alone. Although crystallographic methods can yield very detailed information and are often viewed as a “gold standard,” interpretation of the structure should, if possible, be supported by comparative structural analysis or, better still, complementary functional investigations (e.g., mutagenesis). Our study shows that once an initial structure determination has been made, it can be relatively easy to engineer alternative crystal forms and that these may be valuable in helping to test structure-based hypotheses.
It is somewhat puzzling to observe that distorted catalytic triads have, to the best of our knowledge, not been observed for any bona fide chymotrypsin-like serine proteases. In part, this may reflect the fact that many of the structures of this class of enzyme were solved with bacterial or secreted eukaryotic proteins containing stabilizing internal disulfide bridges, which are not found in the picornaviral 3C proteases. Alternatively, the inherent flexibility may derive in part from the fact that 3Cpro may be cleaved in cis (16, 17, 29, 32) from the polyprotein precursor or from the fact that the protease has additional functions (e.g., as part of the 3CD precursor needed for initiation of RNA replication) (27). It is interesting that HRV 2Apro, another chymotrypsin-like Cys-protease, also exhibits flexibility in the active site (30).
Comparison of β-ribbon structures.
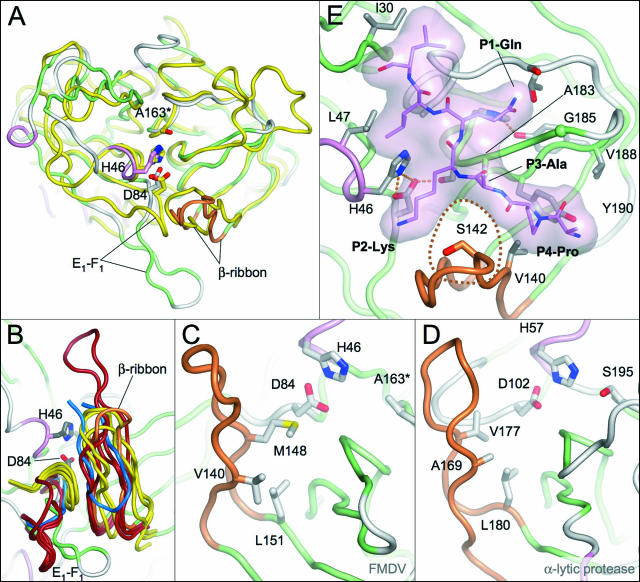
The 13-residue β-ribbon in FMDV 3Cpro is most similar in length and conformation to the corresponding features found in HRV 3Cpro and PV 3Cpro—both 12 residues long—but is also analogous to the equivalent 14-residue β-ribbons observed in several bacterial serine proteases such as the α-lytic protease (13), Streptomyces griseus proteases A (18) and B (14), and a Glu-specific protease from the same organism (28) (Fig. 4A and B). A significantly longer version is found in HAV 3Cpro (21 residues) (1, 3, 4, 37). Intriguingly, there is a β-ribbon in a similar position in the 3C-like NIa TEV protease (31), although in this case the residues forming the loop come from a C-terminal extension and the orientation of the β-ribbon is inverted (Fig. 4B).
FIG. 4.
Comparison of β-ribbon structures from FMDV 3Cpro and related proteases. (A) Superposition of FMDV 3Cpro (β-strands, green; α-helices, pink; β-ribbon, orange) and α-lytic protease (yellow). The side chains of active-site residues are shown as sticks. Only residues from FMDV 3Cpro are labeled). (B) Superposition of the β-ribbon and the adjacent E1-F1 loop from FMDV 3Cpro (colored as in panel A) and other picornaviral 3C proteases. PV (Protein Database [PDB] identification [ID], 1l1n), HRV (PDB ID, 1cqq), and HAV (PDB ID, 1hav) are dark red; the related serine proteases α-lytic protease (PDB ID, 2alp), S. griseus protease A (PDB ID, 5sga), and S. griseus Glu-specific protease (PDB ID, 1hpg) are yellow; and the TEV NIa protease (PDB ID, 1lvb) is blue. Note that in the TEV NIa protease the β-ribbon is inverted and derives from a C-terminal extension of the protein. Close-up views of the β-ribbon in FMDV 3Cpro (C) and the α-lytic protease (D) show residues that contribute to stabilization of this structural feature. (E) Model of the complex of FMDV 3Cpro with a PAKQ|LL peptide based on homology to the TEVpro-peptide complex (31). The peptide is depicted with lilac carbon atoms and a semitransparent surface. Key residues discussed in the text are labeled. The position of residue 142 (Ser in the structure), which is inserted between the P2 and P4 positions of the peptide substrate, is highlighted by a dotted line.
Given that our structural studies of FMDV 3Cpro seem to indicate that the loop is highly flexible—since it has been trapped in two very dissimilar conformations in two crystal forms—it is perhaps surprising that that the β-ribbon structures superpose so well. The structural similarity of the β-ribbons from this diverse set of enzymes is also all the more remarkable since they are stabilized by hydrophobic contacts with the adjacent E1-F1 loops from the N-terminal β-barrel and these are quite dissimilar in structure in different proteases (Fig. 4B). One factor apparently contributing to the structural homology of the β-ribbons is the conservation of a hydrophobic residue at the position equivalent to L151 in FMDV 3Cpro at the base of the ribbon which packs underneath a pair of hydrophobic residues on adjacent strands (V140 and M148 in FMDV 3Cpro; Fig. 4C and D); these three hydrophobic residues are generally conserved in the other proteases as hydrophobic residues (Table 3). Differences between the β-ribbons are more apparent at the apical tip, which is sometimes associated with relatively high B factors (as in FMDV 3Cpro). However, some of this variation may well be due to different packing environments and it will be of interest to directly probe the dynamic nature of this feature in solution (e.g., by nuclear magnetic resonance methods) in the presence and absence of substrates. In any case, even at the tip of the β-ribbon there are important conserved features (see below).
TABLE 3.
Structure-based sequence alignment of residues involved in β-ribbon stabilization and formation of the P4 pocket
| Enzyme | Residue involved in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| β-Ribbon stabilization | P4 pocket formation | |||||||
| FMDV 3Cpro | V140 | M148 | L151a | V140 | S142c | A183 | G185 | Y190 |
| HRV2 3Cpro | I125 | T132b | M135 | I125 | L127 | G163 | N165 | F170 |
| HRV14 3Cpro | I124 | T131b | M134 | I124 | L126 | G162 | N164 | F169 |
| PV 3Cpro | I125 | T132b | T135b | I125 | L127 | G163 | N165 | F170 |
| HAV 3Cpro | I141 | V157 | A160 | I141 | V143 | A163 | G195 | V200 |
| TEV 3C-like protease | H214 | W211 | F139 | H214 | V216 | A169 | N171 | Y178 |
| Bacterial serine proteases | ||||||||
| α-Lytic protease | A169 | V177 | L180 | A169 | Y171 | G215 | N217 | L227 |
| S. griseus protease A | V169 | V177 | M180 | V169 | Y171 | G215 | S217 | F227 |
| S. griseus Glu-specific protease | V169 | V177 | M180 | V169 | Y171 | G215 | S217 | I227 |
This residue, which sits at the base of the loop, is conserved as hydrophobic even in serine proteases with a β-ribbon feature. In FMDV 3Cpro, V140 and M148, which are in adjacent positions on separate strands of the β-ribbon, both contact L151 (Fig. 4C) and appear to be particularly important for stabilization.
In each case, the methyl group of the side chain provides the required hydrophobic pocket function (equivalent to L151 in FMDV 3Cpro).
Ser in crystal structure but Cys in wild-type 3Cpro (and Leu in most active mutant enzymes; see Table 2).
Functional role of the β-ribbon.
Whatever the precise factors involved in stabilization of this β-ribbon feature, it clearly has a role in substrate recognition. The ribbon is positioned close to the side of the peptide binding cleft that accommodates the N-terminal portion of the peptide substrate; in particular, it makes contacts with the residues in the P2-to-P4 positions of the substrate (labeled using the scheme of Schechter and Berger [33]). Perhaps surprisingly, in those cases where comparisons have been made between the enzyme structures in the absence and presence of a peptide substrate or peptidomimetic inhibitor (e.g., HRV 3Cpro [23], α-lytic protease [7], and S. griseus protease A [18]), only negligible changes in the conformation of the β-ribbon were found to occur upon ligand binding. Most tellingly for our present purposes, the structural details revealed by these cocrystallization studies reveal a common mode of peptide recognition among these different proteases. In particular, it is clear that residue L127 at the tip of the β-ribbon HRV2 3Cpro has a central role in substrate binding. The cocrystal structure of HRV2 3Cpro complexed with the peptidomimetic inhibitor AG7088 (23) shows that L127 is positioned directly underneath the main chain atoms of the P3 residue and can make hydrophobic contacts on either side with the P2 and P4 side chains, both of which point down into the peptide binding cleft and are therefore important for substrate recognition. Superposition of FMDV 3Cpro onto the HRV2 3Cpro:inhibitor complex reveals that residue 142 from the FMDV protein (Ser in our present structure but Cys in wild-type 3Cpro) maps closely to the position occupied by L127 in HRV2 3Cpro. This suggests that relatively apolar C142 in FMDV 3Cpro plays a role similar to that of L127 in HRV2 3Cpro. Remarkably, a hydrophobic functional group is conserved at this position in all of the homologous structures listed in Table 3, suggesting that the interaction of the side chain at this position with the P2 and P4 side chains of the peptide substrate is important for substrate binding.
This hypothesis is strongly supported by our comparative analysis of FMDV mutant 3Cpro enzymes containing substitutions at position 142 (Fig. 3). Wild-type FMDV 3Cpro (C142) exhibited good activity in our assays (in spite of instability problems attributed to the relatively low solubility of the protein) (5, 6), but the C142S substitution, which introduces a polar side chain, drastically reduced enzyme activity. In stark contrast, the C142L substitution, which inserts the same residue as found at this position in HRV 3Cpro and PV 3Cpro, yielded essentially wild-type levels of activity. Substitution of a less apolar residue at position 142 (Ala, Val, or Thr) gave an intermediate level of activity. These data, which provide the first functional test of the importance of residues in the β-ribbon for protease activity, are consistent with the idea that C142 and its equivalents in homologous proteases make key interactions with the peptide substrate. They contribute not only to substrate specificity but also to catalytic efficiency, presumably by helping to position the substrate correctly in relation to the active site.
Consistent with the proposal that C142 in FMDV 3Cpro makes important contacts with the P2 and P4 residues of the substrate, we previously showed that Ala substitutions at these positions abrogated peptide cleavage by using an HPLC-based assay (6). Recent preliminary investigations using the more sensitive fluorometric assay to examine the effect of mutations in the VP1-2A peptide used in the present study (see Materials and Methods) found that replacement of P2-Lys with Thr prevented cleavage, while P2-Arg was cleaved but at a much reduced rate; replacement of P4-Pro with Val, Leu, or Ile blocked cleavage completely (unpublished data). Together, these results confirm the importance of interactions with the P2 and P4 side chains for effective proteolysis. Intriguingly, however, the substitutions chosen correspond to residues that occur in these positions within other FMDV polypeptide cleavage junctions that are obviously cleaved in vivo, suggesting that the effects of particular substitutions may depend on the overall sequence context.
A more direct test of the hypothetical role of the β-ribbon in substrate recognition requires structural analysis of FMDV 3Cpro with bound peptides, and work toward this goal is under way in our laboratory. In the meantime, the revised structure of the enzyme reported here allows us to reassess our model of the FMDV 3Cpro-peptide complex, which incorporates the VP1-2A peptide sequence IAPAKQ|LLNFD (vertical line indicates cleavage junction) (6). The revised model was constructed with the homologous structures from HRV 3Cpro and the TEV NIa protease, which have been cocrystallized with peptides (31) or peptidomimetic inhibitors (23), and provides fresh insights into substrate specificity that will be important for the design of inhibitory antiviral compounds. There are no evident changes in the mode of recognition of the P1 side chain, but the presence of the β-ribbon clearly affects the mode of interaction of the protease with its substrate in the P2-to-P4 region (Fig. 4E). We previously surmised that the P2 residue (Lys in the model) might be recognized by interaction with the side chain of Y190, but in the new structure this side chain is located about 12 Å from P2-Lys. Instead, the end of the P2 side chain may be exposed to solvent, which may account for the relative lack of conservation at this position in FMDV polyprotein junctions cleaved by 3Cpro, although for most of the residues that do occur at P2 (Lys, Thr, Gln, Ala, His, Arg, and Pro) (8) there is conservation of the ability to make apolar contacts with C142 at the tip of the β-ribbon. The P3 side chain is generally oriented toward solvent and makes little contribution to substrate specificity. The P4 side chain is accommodated in a pocket (S4) that is formed in part from residues of the β-ribbon (in addition to C142). In fact, the new structure of FMDV 3Cpro suggests that the S4 pocket is largely conserved in apolar character among all of the proteases shown to possess a β-ribbon feature. In FMDV 3Cpro, the key contact residues in the pocket are V140, C142, A183, G185, and Y190 (Fig. 4E); as shown in Table 3, the hydrophobic functionality of these residues is largely conserved (with the possible exception of the substitution of Asn at the position occupied by G185 in FMDV 3Cpro). This readily accounts for the general preference for small apolar residues at the P4 position of substrates for all of these enzymes.
Of all of the picornaviral 3C protease structures that have been analyzed to date, only FMDV 3Cpro exhibits disordering of the β-ribbon, suggesting that this loop may have greater flexibility in FMDV than in other picornaviral 3C proteases. As suggested previously, this enhanced flexibility may help to account for the more relaxed peptide specificity observed for FMDV 3Cpro (6). Nevertheless, it is also clear that contacts between the β-ribbon and the peptide substrate form important specificity determinants. Full resolution of the conundrum that the β-ribbon of FMDV 3Cpro confers specificity via direct contact and yet retains sufficient flexibility to allow the cleavage of a more diverse set of peptide sequences than other picornavirus 3C proteases requires more detailed investigation.
Acknowledgments
This work was funded by BBSRC grant support awarded to S.C. and R.J.L. N.R.-R. was funded by a Marie Curie Host Fellowship for Early Stage Research Training. We are grateful for access to station 14.1 at the Synchrotron Radiation Source, Daresbury, United Kingdom.
We thank Patricia Zunszain for careful reading of the manuscript.
Footnotes
Published ahead of print on 25 October 2006.
REFERENCES
- 1.Allaire, M., M. M. Chernaia, B. A. Malcolm, and M. N. James. 1994. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature 369:72-76. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, E., and R. C. Sheppard (ed.). 1989. Solid phase peptide synthesis. Oxford University Press, Oxford, United Kingdom.
- 3.Bergmann, E. M., M. M. Cherney, J. McKendrick, S. Frormann, C. Luo, B. A. Malcolm, J. C. Vederas, and M. N. James. 1999. Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV. Virology 265:153-163. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, E. M., S. C. Mosimann, M. M. Chernaia, B. A. Malcolm, and M. N. James. 1997. The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition. J. Virol. 71:2436-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birtley, J. R., and S. Curry. 2005. Crystallization of foot-and-mouth disease virus 3C protease: surface mutagenesis and a novel crystal-optimization strategy. Acta Crystallogr. D. 61:646-650. [DOI] [PubMed] [Google Scholar]
- 6.Birtley, J. R., S. R. Knox, A. M. Jaulent, P. Brick, R. J. Leatherbarrow, and S. Curry. 2005. Crystal structure of foot-and-mouth disease virus 3C protease: new insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 280:11520-11527. [DOI] [PubMed] [Google Scholar]
- 7.Bone, R., D. Frank, C. A. Kettner, and D. A. Agard. 1989. Structural analysis of specificity: alpha-lytic protease complexes with analogues of reaction intermediates. Biochemistry 28:7600-7609. [DOI] [PubMed] [Google Scholar]
- 7a.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Carrillo, C., E. R. Tulman, G. Delhon, Z. Lu, A. Carreno, A. Vagnozzi, G. F. Kutish, and D. L. Rock. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 79:6487-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collaborative Computer Project No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Delano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 11.Dragovich, P. S., S. E. Webber, R. E. Babine, S. A. Fuhrman, A. K. Patick, D. A. Matthews, S. H. Reich, J. T. Marakovits, T. J. Prins, R. Zhou, J. Tikhe, E. S. Littlefield, T. M. Bleckman, M. B. Wallace, T. L. Little, C. E. Ford, J. W. Meador III, R. A. Ferre, E. L. Brown, S. L. Binford, D. M. DeLisle, and S. T. Worland. 1998. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J. Med. Chem. 41:2819-2834. [DOI] [PubMed] [Google Scholar]
- 12.Fersht, A. 1984. Enzyme structure and mechanism, 2nd ed. W. H. Freeman & Co. Ltd., New York, N.Y.
- 13.Fujinaga, M., L. T. Delbaere, G. D. Brayer, and M. N. James. 1985. Refined structure of alpha-lytic protease at 1.7 Å resolution. Analysis of hydrogen bonding and solvent structure. J. Mol. Biol. 184:479-502. [DOI] [PubMed] [Google Scholar]
- 14.Fujinaga, M., R. J. Read, A. Sielecki, W. Ardelt, M. Laskowski, Jr., and M. N. James. 1982. Refined crystal structure of the molecular complex of Streptomyces griseus protease B, a serine protease, with the third domain of the ovomucoid inhibitor from turkey. Proc. Natl. Acad. Sci. USA 79:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubman, M. J., and B. Baxt. 2004. Foot-and-mouth disease. Clin. Microbiol. Rev. 17:465-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanecak, R., B. L. Semler, H. Ariga, C. W. Anderson, and E. Wimmer. 1984. Expression of a cloned gene segment of poliovirus in E. coli: evidence for autocatalytic production of the viral proteinase. Cell 37:1063-1073. [DOI] [PubMed] [Google Scholar]
- 17.Harmon, S. A., W. Updike, X. Y. Jia, D. F. Summers, and E. Ehrenfeld. 1992. Polyprotein processing in cis and in trans by hepatitis A virus 3C protease cloned and expressed in Escherichia coli. J. Virol. 66:5242-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, M. N., A. R. Sielecki, G. D. Brayer, L. T. Delbaere, and C. A. Bauer. 1980. Structures of product and inhibitor complexes of Streptomyces griseus protease A at 1.8 Å resolution. A model for serine protease catalysis. J. Mol. Biol. 144:43-88. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these maps. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Knowles, N. J., and A. R. Samuel. 2003. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 91:65-80. [DOI] [PubMed] [Google Scholar]
- 21.Leatherbarrow, R. J. 2003. GraFit, 5.0 ed. Erithacus Software Limited, London, United Kingdom.
- 22.Li, W., N. Ross-Smith, C. G. Proud, and G. J. Belsham. 2001. Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: identification of the eIF4AI cleavage site. FEBS Lett. 507:1-5. [DOI] [PubMed] [Google Scholar]
- 23.Matthews, D. A., P. S. Dragovich, S. E. Webber, S. A. Fuhrman, A. K. Patick, L. S. Zalman, T. F. Hendrickson, R. A. Love, T. J. Prins, J. T. Marakovits, R. Zhou, J. Tikhe, C. E. Ford, J. W. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, M. A. Brothers, D. M. DeLisle, and S. T. Worland. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 96:11000-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews, D. A., W. W. Smith, R. A. Ferre, B. Condon, G. Budahazi, W. Sisson, J. E. Villafranca, C. A. Janson, H. E. McElroy, C. L. Gribskov, and S. Worland. 1994. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77:761-771. [DOI] [PubMed] [Google Scholar]
- 25.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61:458-464. [DOI] [PubMed] [Google Scholar]
- 26.Mosimann, S. C., M. M. Cherney, S. Sia, S. Plotch, and M. N. James. 1997. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 273:1032-1047. [DOI] [PubMed] [Google Scholar]
- 27.Nayak, A., I. G. Goodfellow, and G. J. Belsham. 2005. Factors required for the uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J. Virol. 79:7698-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nienaber, V. L., K. Breddam, and J. J. Birktoft. 1993. A glutamic acid specific serine protease utilizes a novel histidine triad in substrate binding. Biochemistry 32:11469-11475. [DOI] [PubMed] [Google Scholar]
- 29.Palmenberg, A. C., and R. R. Rueckert. 1982. Evidence for intramolecular self-cleavage of picornaviral replicase precursors. J. Virol. 41:244-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen, J. F., M. M. Cherney, H. D. Liebig, T. Skern, E. Kuechler, and M. N. James. 1999. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 18:5463-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan, J., A. Zdanov, A. G. Evdokimov, J. E. Tropea, H. K. Peters III, R. B. Kapust, M. Li, A. Wlodawer, and D. S. Waugh. 2002. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 277:50564-50572. [DOI] [PubMed] [Google Scholar]
- 32.Richards, O. C., L. A. Ivanoff, K. Bienkowska-Szewczyk, B. Butt, S. R. Petteway, Jr., M. A. Rothstein, and E. Ehrenfeld. 1987. Formation of poliovirus RNA polymerase 3D in Escherichia coli by cleavage of fusion proteins expressed from cloned viral cDNA. Virology 161:348-356. [DOI] [PubMed] [Google Scholar]
- 33.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 34.Seipelt, J., A. Guarne, E. Bergmann, M. James, W. Sommergruber, I. Fita, and T. Skern. 1999. The structures of picornaviral proteinases. Virus Res. 62:159-168. [DOI] [PubMed] [Google Scholar]
- 35.Sutmoller, P., S. S. Barteling, R. C. Olascoaga, and K. J. Sumption. 2003. Control and eradication of foot-and-mouth disease. Virus Res. 91:101-144. [DOI] [PubMed] [Google Scholar]
- 36.Taliani, M., E. Bianchi, F. Narjes, M. Fossatelli, A. Urbani, C. Steinkuhler, R. De Francesco, and A. Pessi. 1996. A continuous assay of hepatitis C virus protease based on resonance energy transfer depsipeptide substrates. Anal. Biochem. 240:60-67. [DOI] [PubMed] [Google Scholar]
- 37.Yin, J., E. M. Bergmann, M. M. Cherney, M. S. Lall, R. P. Jain, J. C. Vederas, and M. N. James. 2005. Dual modes of modification of hepatitis A virus 3C protease by a serine-derived beta-lactone: selective crystallization and formation of a functional catalytic triad in the active site. J. Mol. Biol. 354:854-871. [DOI] [PMC free article] [PubMed] [Google Scholar]