Abstract
Vaccination against AIDS is hampered by great diversity between human immunodeficiency virus (HIV) strains. Heterologous B-subtype-based simian-human immunodeficiency virus (SHIV) DNA prime and poxvirus boost vaccine regimens can induce partial, T-cell-mediated, protective immunity in macaques. We analyzed a set of DNA, recombinant fowlpox viruses (FPV), and vaccinia viruses (VV) expressing subtype AE HIV type 1 (HIV-1) Tat, Rev, and Env proteins and SIV Gag/Pol in 30 pigtail macaques. SIV Gag-specific CD4 and CD8 T-cell responses were induced by sequential DNA/FPV vaccination, although lower FPV doses, VV/FPV vaccination, and DNA vaccines alone were not as consistently immunogenic. The SHIV AE DNA prime, FPV boost regimens were significantly less immunogenic than comparable B-subtype SHIV vaccination. Peak viral load was modestly (0.4 log10 copies/ml) lower among the AE subtype SHIV-immunized animals compared to controls following the virulent B subtype SHIV challenge. Protection from persistent high levels of viremia and CD4 T-cell depletion was less in AE subtype compared to B subtype SHIV-vaccinated macaques. Gag was highly immunodominant over the other AE subtype SHIV vaccine proteins after vaccination, and this immunodominance was exacerbated after challenge. Interestingly, the lower level of priming of immune responses did not blunt postchallenge Gag-specific recall responses, despite more modest protection. These studies suggest priming of T-cell immunity to prevent AIDS in humans is possible, but differences in the immunogenicity of various subtype vaccines and broad cross-subtype protection are substantial hurdles.
The efficiency of T-cell-mediated clearance of human immunodeficiency virus (HIV)-infected cells by candidate prophylactic vaccines is currently being assessed (22). Vaccine-induced T-cell responses correlate with partial protection from disease in macaques following various virulent primate lentivirus challenge experiments (5). Heterologous prime and boost HIV vaccine strategies involving priming by DNA vaccination and boosting with recombinant attenuated poxvirus vectors (such as fowlpox virus [FPV]) encoding common HIV or simian immunodeficiency virus (SIV) antigens reliably induce T-cell immune responses in outbred nonhuman primates. Reductions in viral levels and retention of CD4 T cells in macaques challenged with SHIV strains closely related to the vaccine antigens have been reported (3, 5, 10). Some but not all SIV vaccination studies using similar heterologous prime/boost vaccine modalities have also shown positive results using challenge strains closely matched to vaccine antigens (17, 27). Heterologous prime/boost regimens have to date had mixed success in human studies (3, 16, 17, 21). The ability of T-cell-based vaccine regimens to protect macaques or humans from more diverse primate lentivirus strains remains to be defined.
The global HIV type 1 (HIV-1) epidemic consists primarily of non-B-subtype group M HIV-1 strains. Despite this, the majority of vaccines evaluated in preclinical and clinical trials are based on subtype B forms present in developed countries within Europe, North America, and Australia. The circulating recombinant subtype AE is common in many Southeast Asian countries, including Thailand, although no preclinical efficacy studies or clinical trials with subtype AE vaccines have been reported. We previously demonstrated the safety and immunogenicity of subtype AE HIV-1 DNA and FPV vaccines in macaques (12), modeled on our previous macaque and human work with subtype B HIV-1 and SHIV vaccines (8, 9, 18-20). In this study we analyzed a set of subtype AE SHIV vaccines expressing SIV Gag and Pol and HIV-1 subtype AE Env, Tat, and Rev for immunogenicity and efficacy in pigtail macaques.
A disadvantage of prime/boost vaccination strategies in the field is the requirement for multiple vaccinations and the relatively poor priming of immune responses with DNA vaccination alone. Recent studies suggest that dual poxvirus regimens, such as vaccinia virus (VV) priming and FPV boosting, are highly immunogenic (4, 23). Since both are live vector vaccines, a single immunization of each component may suffice. In addition, it is possible, based on suboptimal results from recent human studies, that increased doses of DNA and/or FPV vaccines could improve immunogenicity and efficacy (13).
In efforts to improve and further understand the limitations of DNA/poxvirus prime/boost regimens, we studied multigenic subtype AE SHIV DNA, vaccinia virus, and fowlpox virus vaccines for immunogenicity and protective efficacy in 30 pigtail macaques. Comparator regimens included DNA vaccination alone, a higher dose of the FPV booster vaccines, and a VV/FPV prime/boost regimen. The macaques were subsequently mucosally challenged with a heterologous B subtype SHIV. The macaques were stratified for the presence of the Mane-A*10 major histocompatibility complex (MHC) allele (the first such study in pigtail macaques), and CD8 T-cell immunogenicity in the subset of animals with this allele was followed using an MHC tetramer.
MATERIALS AND METHODS
Monkeys.
Juvenile Macaca nemestrina monkeys were free from HIV-1/SIV/simian retrovirus infection and anesthetized with ketamine (10 mg/kg of body weight, intramuscular [i.m.]) prior to procedures. All experiments were performed according to National Institutes of Health guidelines on the care and use of laboratory animals and were approved by the University of Melbourne and CSIRO Livestock Industries Animal Experimentation and Ethics committees.
DNA vaccinations.
The DNA vaccine, pHIS-SHIV-AE, was based on the previously described pHIS-HIV-AE. pHIS-SHIV-AE encodes full-length unmutated SIVmac239 Gag and Pol, HIV-193TH253 Tat, Rev, and Vpu, and the 5′- and 3′-thirds of HIV-193TH253 Env. Genes were inserted into the vector pHIS-64 (Coley Pharmaceutical Group, Wellesley, MA) behind the human cytomegalovirus immediate-early promoter. Plasmid vector pHIS-64 has kanamycin resistance, a bovine growth hormone poly(A) termination signal, and 64 CpG motifs in addition to those naturally present that are primate optimized. Empty vector plasmid DNA, pHIS, served as a control vaccine. Plasmid pHIS-SHIV-AE for immunization was prepared by QIAGEN (Hilden, Germany), and control DNA vaccine pHIS was prepared using the EndoFree plasmid Giga kit (QIAGEN). Plasmid DNA in normal saline was injected i.m. at 1 mg/ml at weeks 0 and 4 (Table 1).
TABLE 1.
Vaccination and challenge regimens
| Vaccine regimen | n | Animal nos.a | Subtype AE SHIV immunization at:
|
|||
|---|---|---|---|---|---|---|
| Week 0 | Week 4 | Week 8 | Week 22 (all groups) | |||
| Control | 6 | 6167, 6352, 5350, 6264, 5620, 6376 | Control pHIS-empty (1 mg, i.m.) | Control pHIS-empty (1 mg, i.m.) | Control FPV-M3 (3 × 108 PFU, i.m.) | |
| Two-DNA/FPV-hi | 6 | 5396, 5616, 6279, 5085, 5912, 5396 | DNA pHIS-SHIV-AE (1 mg, i.m.) | DNA (1 mg) pHIS-SHIV-AE | FPV-SHIV-AE (3 × 108 PFU, i.m.) | |
| Two-DNA/FPV | 6 | 6276, 6370, 5023, 6284, 6263, 6353 | DNA pHIS-SHIV-AE (1 mg, i.m.) | DNA pHIS-SHIV-AE (1 mg, i.m.) | FPV-SHIV-AE (5 × 107 PFU, i.m.) | SHIVmn229 (subtype B) challenge (105 TCID50, intrarectal) |
| Three-DNA | 6 | 5614, 6351, 5618, 6364, 6173, 6371 | DNA pHIS-SHIV-AE (1 mg, i.m.) | DNA pHIS-SHIV-AE (1 mg, i.m.) | DNA pHIS-SHIV-AE (1 mg, i.m.) | |
| VV/FPV | 6 | 6349, 6377, 6259, 6363, 6262, 6388 | VV-SIV Gag/Pol, VV-HIV-1AE Env, VV-HIV-1AE Tat/Rev (each, 108 PFU, i.m.) | FPV-SHIV-AE (3 × 108 PFU, i.m.) | ||
Animal numbers in bold indicate those that expressed the Mane-A*10 allele and were studied with the Mane-A*10/KP9 tetramer.
Recombinant fowlpox virus and vaccinia virus vaccines.
Construction of the FPV and VV vaccines has been described previously (7). A single FPV-SHIV-AE was constructed, modeled on an FPV-HIV-1-AE vaccine, which expressed almost all the antigens expressed by the pHIS-SHIV-AE DNA vaccine: SIV Gag/Pol from the F6,7,9 site, an HIV-193TH253 Tat/Rev fusion product from the REV site, and HIV-193TH253 Env, mutated to remove the middle third of the gene. Three separate VV recombinants were constructed encoding either wild-type SIV gag/pol, an HIV-193TH253 Tat/Rev fusion product, or the mutated HIV-193TH253 Env (Table 1).
Intracellular IFN-γ staining (ICS).
Induction of antigen-specific intracellular gamma interferon (IFN-γ) expression in CD3+ CD8+ or CD3+ CD4+ T lymphocytes was assessed by flow cytometry as previously described (11, 21). Briefly, 200 μl whole blood was incubated with 1 μg/ml overlapping 15-mer peptide pools in dimethyl sulfoxide or dimethyl sulfoxide alone and the costimulatory antibodies anti-CD28 and anti-CD49d (BD Biosciences Pharmingen [BD], San Diego, CA) for 7 h. Brefeldin A (10 μg/ml; Sigma) was included during the last 5 h of the incubation. Anti-CD3-phycoerythin, anti-CD4-fluorescein isothiocyanate, and anti-CD8-peridinin chlorophyll a protein antibodies (clones SP34, M-T477, and SK1, respectively; BD) were added to each well and incubated for 30 min. Red blood cells were lysed (FACS lysing solution; BD) and washed with phosphate-buffered saline, and the remaining cells were permeabilized (FACS permeabilizing solution 2; BD). Permeabilized cells were then incubated with anti-human IFN-γ-allophycocyanin antibody (clone B27; BD) prior to fixing with formaldehyde and acquisition (LSRII; BD). Acquisition data were analyzed using Flowjo version 6.3.2 (Tree Star, Ashland, OR). The percentage of antigen-specific gated lymphocytes expressing IFN-γ was assessed in both CD3+ CD4+ and CD3+ CD8+ lymphocyte subsets. Peptides used were all 15-mers overlapping by 11 amino acids (aa) and were either obtained from the NIH AIDS Research and Reference Reagent Program (SIV Gag and Pol) or purchased from Auspep (HIV-193TH253 Tat, Rev, and Env).
MHC typing and Mane-A*10/KP9 tetramer analyses.
Both reference strand-mediated conformational analysis and sequence-specific primer-PCR were used to identify Mane-A*10+ animals capable of responding to the SIV Gag KP9 epitope as previously described (26). Briefly, the reference strand-mediated conformation analysis method used an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) to heteroduplex a 677-bp fragment of pigtail macaque MHC class I cDNA product with labeled rhesus macaque alleles Mamu-A*15, Mamu-A*20, and Mamu-B*60 reference strands. Mane-A*10 heteroduplexes were identified by running clones with unique mobility patterns. Data were analyzed using DAx data acquisition and analysis software (Van Mierlo Software, Eindhoven, The Netherlands) (26). Mane-A*10-specific primers were used to confirm the Mane-A*10 status of pigtail macaques by sequence-specific primer-PCR on peripheral blood mononuclear cell cDNA as previously described using three separate primer sets (26). KP9-specific T-cell responses were monitored by flow cytometry using a KP9/Mane-A*10 tetramer as previously described (1, 26). The Mane-A*10 polypeptide, human β2-microglobulin, and KP9 peptide (GL Biochem, Shanghai, China) were complexed to form the KP9/Mane-A*10 tetramer, which was subsequently conjugated with phycoerythrin. Fresh whole blood (200 μl) was stained with the KP9/Mane-A*10 tetramer (1:200 to 1:400 dilution) and then counterstained with anti-CD3-fluorescein isothiocyanate (clone SP34; Becton-Dickinson) and anti-CD8-allophycocyanin (clone SK1; BD). Acquisition was performed on an LSR II flow cytometer (BD) and analyzed using Flowjo version 6.3.2 (Tree Star).
SHIV challenge of macaques.
To assess vaccine efficacy, all macaques were atraumatically inoculated intrarectally with SHIVmn229 (5 × 104 50% tissue culture infective doses [TCID50]/ml) in 0.5-ml doses over 2 days (total, 105 TCID50/ml) as previously described (11). The administered dose of 105 TCID50 represents ≥500 monkey infectious doses (MID). SHIV viremia was quantified by reverse transcriptase real-time PCR on an ABI 7700 machine as described above except a TaqMan probe was used instead of a molecular beacon to detect fluorescence (11). Depletion of peripheral CD4 T cells was monitored by flow cytometry as described elsewhere (6).
Power and statistical considerations.
The primary end points, set prior to starting the study, were differences in outcome of SHIV challenge (peak and set point SHIV viremia and set point peripheral CD4 T-cell levels) between vaccinated and control groups. Based on previous studies with this challenge stock (11), six macaques/group powered the study (80%; α = 0.05) to detect 0.5 log10 differences in both peak (week 2) and set point (mean of weeks 4 to 11) plasma SHIV RNA and 5% set point CD4 T lymphocytes between groups. Week 11 was chosen as the end of the set point time period based on previous studies (11), as all macaques were likely to be alive and contributing data points up to that time but at least some control macaques were likely to be euthanized shortly thereafter. Statistical comparisons of immunogenicity and efficacy across vaccine groups utilized a Kruskal-Wallis analysis of variance (ANOVA) test for overall significance, with a Dunn's multiple comparisons test between individual groups. A nonparametric two-way ANOVA test (on ranks) was used for statistical comparisons across vaccine groups of both the T-cell responses to the various peptides and the effectiveness of the SHIV vaccination subtype (AE or B), and a Bonferroni posttest was used to compare between individual peptides or between subtypes within each vaccine group. For comparisons between two groups (e.g., all vaccinated animals versus controls), we used a Mann-Whitney test.
RESULTS
Vaccine groups.
Our previous work with subtype B SHIV-based DNA prime/FPV boost regimens demonstrated high levels of T-cell immunogenicity and partial protection from higher viral loads and CD4 T-cell depletion following a subtype B SHIV challenge (10). To both assist evaluating the efficacy of subtype AE regimens suitable for use in Southeast Asia and assess protection across diverse subtypes, we studied four separate subtype AE SHIV vaccine regimens in groups of six macaques (Table 1). We included arms with (i) higher doses on the FPV boost vaccination (based on earlier work with FPV doses in macaques (12), (ii) DNA vaccination only (based on surprisingly good efficacy despite limited immunogenicity with this arm in the previous subtype-B SHIV trial [9]), and (iii) VV prime/FPV boost (based on our own and previous reports demonstrating strong T-cell immunogenicity with this approach [4, 23]). The vaccine regimens are shown in Table 1.
T-cell responses to Gag postvaccination.
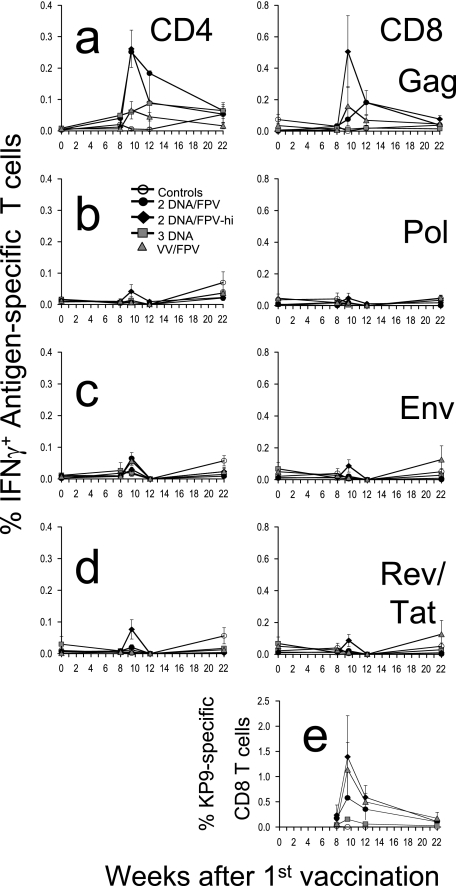
The kinetics, magnitude, and phenotype of the cellular response induced by vaccination were quantified by antigen-specific CD4 and CD8 T-cell expression of IFN-γ by ICS assays in all 30 macaques at multiple time points after vaccination (Fig. 1). The high-dose DNA/FPV vaccine regimen was overall the most immunogenic, inducing a mean response to a pool of SIV Gag peptides in CD8 T cells of 0.51% (range, 0.05 to 1.35%) and in CD4 T cells of 0.26% (range, 0.07 to 0.46) 1.5 weeks after the final vaccination (Fig. 1a). The mean SIV Gag-specific CD8 T-cell responses at weeks 1.5 to 4 postvaccination in the DNA/FPV high-dose FPV regimen were significantly greater than controls (by Kruskal-Wallis across all groups, ANOVA, P < 0.004; Dunn's multiple comparison test, P < 0.01). The mean SIV Gag-specific CD4 T-cell responses postvaccination in both the DNA/FPV high-dose and low-dose regimens were also significantly greater than controls (Kruskal-Wallis, P = 0.001; Dunn's multiple comparison test, P < 0.01).
FIG. 1.
Time course of mean cellular immune responses and standard error following vaccination. Groups of macaques were immunized with two-DNA/FPV-low-dose FPV (black circles), two-DNA/FPV-high-dose FPV (black triangles), three-DNA (gray squares), VV/FPV (gray triangles), or control vaccines (open circles) as shown in Table 1. (A to D) CD4 and CD8 T-cell responses are shown in the left and right panels, respectively, in response to stimulation with SIV Gag 15-mer peptide pool (A), SIV Pol 15-mer peptide pool (B), HIV-1 subtype AE Env 15-mer peptide pool (C), or a combined pool of HIV-1 subtype AE Tat and Rev 15-mer peptide pools (D). (E) CD8 T-cell responses to the SIV Gag epitope KP9 were measured in the subset of macaques that were Mane-A*10+ by MHC tetramer staining.
Compared to the DNA/FPV group with the high dose of FPV (DNA/FPV-hi), the other vaccination regimens induced lower mean levels of antigen-specific T cells, although there was significant variability between the outbred animals and there were no statistically significant differences between the other groups of six animals. CD8 T-cell responses 1.5 weeks after the final vaccination in the DNA/FPV low-dose regimen were 0.08% (±0.06% standard error [SE]; range, 0.02 to 0.15%), in the VV/FPV regimen they were 0.16% (±0.12% SE; range, 0.00 to 0.77%), in the DNA-only regimen they were 0.01% (±0.01% SE; range, 0.00 to 0.03%), and in the controls they were all 0.00%. Utilizing VV instead of DNA as the priming vaccination prior to the FPV booster did not elevate responses relative to the DNA/FPV-hi regimen. Three DNA vaccinations alone without FPV boosting induced much lower levels of SIV Gag-specific IFN-γ-expressing CD4 and CD8 T cells that were indistinguishable from controls.
T-cell responses to other antigens postvaccination.
All the DNA, VV, and FPV vaccines expressed HIV-1 subtype AE Env, Tat, and Rev antigens and SIVmac251 Pol. A set of 15-mer peptides overlapping by 11 amino acids to the homologous HIV-193TH253 Env, Tat, and Rev was constructed and used to assess T-cell immunogenicity to these antigens. Responses to the overlapping peptides spanning the HIV-1 Env, a combined pool of HIV-1 Tat/Rev, and polymerase of SIV, were minimal (Fig. 1a to d), with the CD8 T-cell responses being significantly lower than those observed for SIV Gag (P <0.008, nonparametric two-way ANOVA). Peak CD8 or CD4 T-cell responses after the last vaccination to Pol, Env, and Tat/Rev were only ≤0.20, ≤0.13, and ≤0.23%, respectively, and were not significantly different from controls.
Responses to immunodominant KP9 epitope postvaccination.
A subset of 11 of the 30 pigtail macaques expressed the Mane-A*10 MHC class I allele as identified by reference strand conformational analysis and sequence-specific PCR (data not shown). The Mane-A*10 molecule presents the dominant SIV Gag epitope KP9, and we recently developed and validated a Mane-A*10/KP9 tetramer for use in sensitively detecting KP9-specific T cells (25, 26). The Mane-A*10+ macaques were randomly distributed across the five groups of vaccinated and control macaques. The CD8 T-cell responses detected by Mane-A*10/KP9 tetramer staining were higher than those detected by the functional production of IFN-γ in response to Gag in the ICS assay, peaking at 2.2% in one animal in the DNA/FPV-hi regimen 1.5 weeks after the FPV boost. The mean response across all the prime/boost groups (DNA/FPV or VV/FPV) was 0.76% 1.5 to 4 weeks after the last vaccination, compared to a mean response in controls of 0.00% or of macaques vaccinated with DNA only of 0.11% (P = 0.025 by Kruskal-Wallis; P < 0.05 by Dunn's multiple comparison post test).
SHIV challenge.
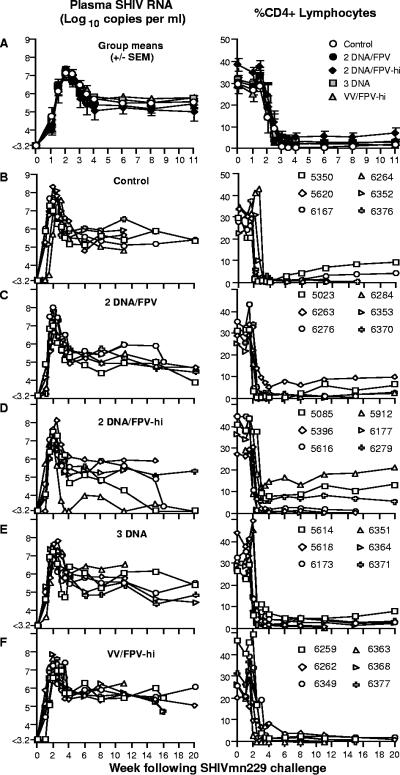
The HIV-1HXB2-derived SHIVmn229 challenge stock was inoculated intrarectally (105 TCID50 or ≥500 MID) into all 30 macaques 14 weeks after the last vaccination. CD4 T cells and plasma SHIV RNA were followed for 20 weeks, unless macaques were euthanized to avoid AIDS-related illnesses (Fig. 2 and 3). The six control macaques receiving DNA and FPV vaccines not expressing SHIV antigens had high peak levels of SHIV RNA (mean, 7.77 log10 copies/ml; range, 7.29 to 8.32) following inoculation and at set point (weeks 4 to 11; mean, 5.6 log10 copies/ml). All six control macaques lost peripheral CD4 T cells precipitously, declining to a mean of only 0.6% of total lymphocytes (range, 0.2 to 1.2%) 4 weeks after challenge. Four of the six macaques were euthanized within 15 weeks of inoculation with incipient AIDS.
FIG. 2.
Outcome of SHIVmn229 challenge. Each macaque was challenged intrarectally with a MID of ≥500 SHIVmn229 14 weeks post-final immunization. Blood samples were analyzed for plasma SHIV RNA by real-time PCR (left panel) and for peripheral CD4 T-cell loss (right panel). (A) Mean (± SE of the mean) plasma SHIV RNA and CD4 T cells for each vaccine group. (B to F) Plasma SHIV RNA and CD4 T-cell responses for individual macaques grouped for each vaccine regimen.
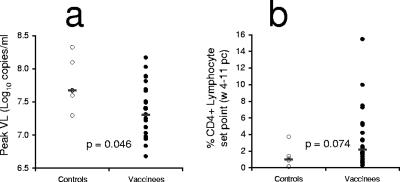
FIG. 3.
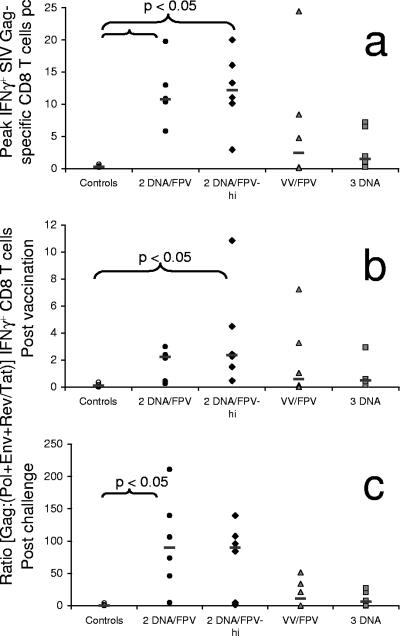
Outcome of challenge. All 24 vaccinated animals were compared to the six controls for peak viral load (VL) (A) and set point CD4 T-cell levels (B) (mean, weeks 4 to 11). P values are for Mann-Whitney tests. pc, postchallenge.
Effects of vaccination on viral load postchallenge.
All 24 vaccinated macaques became infected, and there was very limited protection afforded by the SHIV-AE vaccine regimens from high levels of the subtype B SHIV replication or CD4 T-cell depletion. The mean peak SHIV viral load across all 24 vaccinated macaques was significantly but modestly lower (0.41 log10 copies/ml) than in the control animals (Fig. 2 and 3) (P = 0.046, Mann-Whitney). Mean peak viral load in the control animals was 7.77 log10 copies/ml, compared to 7.39 log10 copies/ml in the DNA/FPV, 7.36 log10 copies/ml in the DNA/FPV-hi, 7.26 log10 copies/ml in the DNA only, and 7.43 log10 copies/ml in the VV/FPV group (overall range, 6.68 to 8.03). Set point viral load was defined prior to challenge as the mean viral load over weeks 4 to 11 following challenge, when viral load is relatively stable and all 30 animals were likely to be still alive. Set point SHIV viral load was only marginally and not statistically significantly lower than controls in the vaccine groups vaccinated with the DNA/FPV-hi and DNA/FPV arms (0.47 and 0.24 log10 copies/ml lower, respectively).
Effects of vaccination on CD4 T-cell depletion postchallenge.
There was a nonsignificant trend towards retention of total peripheral CD4 T cells with vaccines compared to controls (P = 0.074, Mann-Whitney). Although the group immunized with the two-DNA/FPV-hi regimen had higher set point CD4 T-cell levels between weeks 4 and 11 (mean, 6.05% CD4 T lymphocytes; range, 0.63 to 15.5%) compared to controls (1.23%; range, 0.15 to 3.71%), there were no significant differences between any of the vaccine groups (P = 0.3, Kruskal-Wallis ANOVA). The retention of CD4 T cells in the two-DNA/FPV-hi group was primarily driven by three animals (6279, 5085, 5912) (Fig. 2D) which had recovered total CD4 T cells of 8.3, 12.8, and 17.9%, respectively, by week 11. The mean set point levels of peripheral CD4 T cells in the two-DNA/FPV-vaccinated group (2.56%), the three-DNA group (3.07%), and the VV/FPV group (2.04%) were not significantly different from controls (1.23%).
The 30 animals were followed for up to 20 weeks following SHIV challenge (Fig. 2B to F). Overall, half (15 of 30) of the animals were euthanized with incipient AIDS between weeks 11 and 20. These included four animals in control group, two in the DNA/FPV group, three in the DNA/FPV-hi group, two in the three-DNA group, and four in the VV/FPV group.
Immune responses to Gag following challenge.
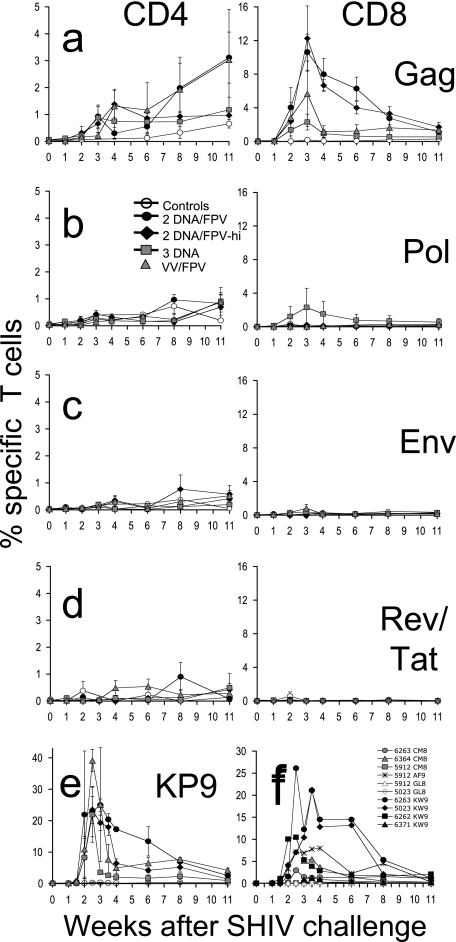
Gag-specific CD8 T cells expressing IFN-γ were dramatically boosted after challenge in vaccinated macaques (Fig. 4a, right panel). Control macaques did not mount large Gag-specific CD8 T-cell responses postchallenge (mean response 3 weeks after challenge was 0.2%; range, 0.0 to 0.4%). Significantly higher mean peak Gag-specific CD8 T-cell responses postchallenge were observed in DNA/FPV-hi- and DNA/FPV-vaccinated animals over controls (P = 0.003, Kruskal-Wallis ANOVA; P < 0.05, Dunn's multiple comparison test) (Fig. 5a). Mean Gag-specific CD8 T cells peaked 3 weeks following challenge at 12.2% (range, 2.4 to 20.0%) in the DNA/FPV-hi group. The DNA/FPV low-dose vaccine group had a similar mean postchallenge CD8 T-cell response (10.6%; range, 5.4% to 19.8%).
FIG. 4.
Immune responses detected post-SHIVmn229 challenge. (A to D) Time courses of the mean (± SE of the mean) CD4 T-cell (left panel) and CD8 T-cell (right panel) immune responses postchallenge are shown for each vaccine group by IFN-γ ICS to stimulation with SIV Gag 15-mer peptide pool (A), SIV Pol 15-mer peptides (B), HIV-1 subtype AE Env 15-mer peptide pool (C), or a combined pool of HIV-1 subtype AE Tat and Rev 15-mer peptide pools (D). (E) CD8 T-cell responses to the SIV Gag epitope KP9 were measured in the subset of macaques that were Mane-A*10+ by MHC tetramer. (F) Some monkeys mounted responses to other known pigtail macaque Gag epitopes (KW9, CM8, AF9, and GL8) prechallenge, and these were followed postchallenge by IFN-γ ICS.
FIG. 5.
Immunodominant Gag cytotoxic T-lymphocyte responses postchallenge (pc). (A) Peak Gag-specific CD8 T-cell responses postchallenge are shown for each animal in each vaccine group. (B and C) Gag immunodominance (measured as the ratio of Gag to [Env plus Pol plus Tat/Rev] specific CD8 T-cell responses) for each animal in each vaccine group after vaccination (mean, weeks 1.5 to 14 post-final vaccination) (B) and after challenge (mean, weeks 1 to 11) (C). The P values for differences between vaccine groups were determined using Dunn's multiple comparison posttest to a Kruskal-Wallis ANOVA.
A slow postchallenge increase in IFN-γ-expressing Gag-specific CD4 T cells was also observed in some vaccinated macaques after SHIV challenge despite depletion of total CD4 T cells in all animals (Fig. 4a, left panel). Data were not available from all macaques at time points because of the massive loss of CD4 T cells in many animals. Data were censored where there were too few macaques (<3 out of 6 within the group) with sufficient peripheral CD4 T cells (>1%) to estimate the mean number of SHIV-specific CD4 T cells. Despite these limitations, a proportion of CD4 T cells expressed IFN-γ in response to Gag stimulation in the vaccinated animals, reaching a mean of 2.1% by 11 weeks after SHIV challenge. Anamnestic CD4 and CD8 T-cell responses to Pol, Env, and a combined Tat/Rev peptide pool were low, sporadic among the vaccine groups, and not significantly greater than controls.
Responses to immunodominant Gag epitopes postchallenge.
In the subset of animals expressing the Mane-A*10 allele, KP9-specific CD8 T-cell responses were also analyzed following the SHIV challenge in Mane-A*10/KP9 tetramer studies (Fig. 4e). A rise in KP9-specific CD8 T cells from <0.3% on the day of challenge to up to 43% of all CD8 T cells within 3 weeks postchallenge was observed in the vaccinated animals, compared to <0.7% in controls. All of the vaccinated Mane-A*10 animals had a massive rise in KP9-specific responses, although peak responses were variable and not significantly different across the small numbers of Mane-A*10+ animals (two to three/group) within the four vaccine groups.
The KP9 epitope was presented by just over one-third of the animals, those that expressed Mane-A*10. We previously identified a number of other strong SIV Gag CD8 T-cell epitopes presented by pigtail macaques, including KW9 (aa 28 to 36 of SIVmac239 Gag), CM8 (Gag aa 417 to 424), AF9 (Gag aa 371 to 379), and GL8 (Gag aa 142 to 149) (15). Early after vaccination we screened all 24 vaccinated macaques for responses at these epitopes by IFN-γ ICS and identified 6 animals responding to one or more of these epitopes. We followed these macaques for epitope-specific responses by IFN-γ ICS after challenge (Fig. 4f). Overall, the pattern of anamnestic CD8 T-cell responses to these epitopes was similar to that observed for total Gag responses by ICS or KP9-specific responses by MHC tetramer, except that patterns of immunodominance could be discerned. KW9 was the strongest response studied, with three of the four responders reaching ICS responses of over 10%. In animal 6263, which had responses to both KW9 and CM8, the KW9 response was immunodominant. Similarly, animal 5023 had responses to both the KW9 and GL8 epitopes, and the KW9 epitope was also immunodominant. Animal 5912 had responses to three epitopes, AF9, CM8, and GL8, and the AF9 response was the immunodominant response. In a post hoc analysis, we also studied whether responding to one or more of these Gag epitopes improved outcome of infection. The six vaccinated animals that responded to at least one of these other defined Gag epitopes had significantly lower set point viral load (P = 0.001, nonparametric two-way ANOVA) and higher set point CD4 T-lymphocyte levels (P = 0.012) than the 18 vaccinated animals not responding to these epitopes (P = 0.001 and 0.012, respectively; nonparametric two-way ANOVA).
Vaccination enhances Gag immunodominance.
The vaccine regimens resulted in an increase in the mean postvaccination Gag-specific responses over responses to the other vaccine antigens (Fig. 5b). After challenge, however, a much greater mean increase in the Gag responses over the other antigens was observed in all vaccine groups (Fig. 5c) (P = 0.006, Kruskal-Wallis ANOVA), and there was nearly a 100-fold mean increase in Gag immunodominance in the DNA/FPV group that was significantly greater than in the controls (P < 0.05, Dunn's multiple comparison test). This suggests that the Gag-specific responses primed by vaccination were highly immunodominant on recall following challenge and that the vaccines did not efficiently prime broad responses to the other vaccine antigens.
Comparison of B subtype and AE subtype vaccination.
The genetic diversity of HIV-1 remains a major obstacle to HIV vaccination. However, the relative efficacy of SHIV vaccination with one subtype and protection against another subtype has not previously been studied. We recently performed and reported a separate B subtype SHIV DNA prime/fowlpox virus boost vaccine study using the same B subtype SHIVmn229 intrarectal challenge stock as used in this AE subtype SHIV study (9). Two of the vaccine groups in both studies received identical vaccine regimens (the DNA/FPV low-dose and three-DNA groups). We therefore compared the immunogenicity and protective efficacy of these regimens, where the difference was the subtype of SHIV vaccination (Fig. 6).
FIG. 6.
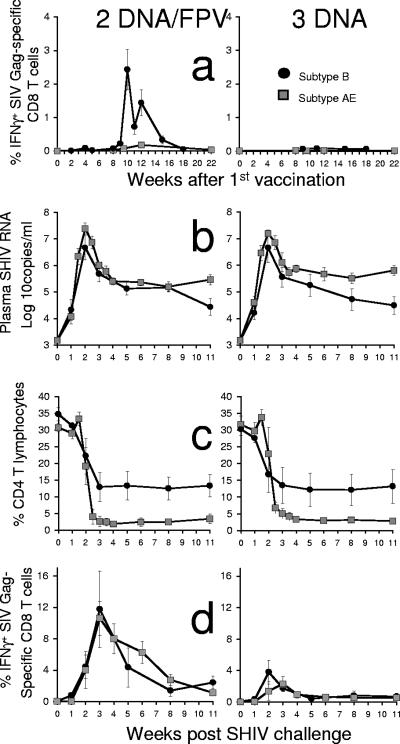
Comparison of the immunogenicity and efficacy of subtype B (black circles) and subtype AE (gray squares) SHIV vaccination of groups of six pigtail macaques. Two vaccine regimens, DNA/FPV-low-dose FPV (left panel) and DNA only (right panel), were identical between the B and AE subtype SHIV vaccinations, and both were challenged intrarectally with the same dose, route, and stock of SHIVmn229. (A) Vaccine-induced SIV Gag-specific CD8 T-cell responses by IFN-γ ICS. (B) Plasma SHIV RNA by real-time PCR after challenge. (C) Peripheral CD4 T cells after challenge. (D) Postchallenge SIV Gag-specific CD8 T-cell responses by IFN-γ ICS.
There was a lower effectiveness of the AE subtype SHIV vaccination strategies compared to the B subtype SHIV strategy. The AE subtype DNA/FPV regimen was considerably less immunogenic than the B subtype SHIV DNA/FPV regimen for Gag-specific T-cell responses. The mean proportion of CD8 T cells responding to Gag at 4 weeks after the last vaccination in the B subtype SHIV vaccination was 1.4% (range, 0.4 to 2.6%) compared to 0.2% in the AE SHIV vaccination (range, 0 to 0.6%) (P = 0.009, Mann-Whitney) (Fig. 6a). Both the AE and B subtype DNA vaccination-only regimens were equally poorly immunogenic (mean responses postvaccination of <0.1%) at inducing detectable responses by IFN-γ ICS.
The B subtype vaccination also produced a superior outcome following the B subtype SHIV challenge compared to the AE subtype SHIV vaccination (Fig. 6b and c). There were lower set point plasma SHIV RNA levels in both the subtype B DNA/FPV and DNA vaccination regimens (4.86 log10 copies/ml) compared to the corresponding AE subtype regimens (5.53 log10 copies/ml, respectively; P = 0.005, nonparametric two-way ANOVA). The control of viremia by the subtype B regimens resulted in a trend towards greater retention of set point CD4 T cells in the subtype B DNA/FPV and DNA vaccination regimens (12.8%) compared to the corresponding AE subtype regimens (2.8%; P = 0.064, nonparametric two-way ANOVA).
We were also interested in comparing the anamnestic CD8 T-cell response to Gag after challenge. Given the poorer immunogenicity after vaccinations and outcome of challenge, we assumed that the recall response to infection would be poorer. We were surprised to find that Gag-specific CD8 T-cell responses after challenge were nearly identical in the subtype B and AE regimens with DNA/FPV and DNA-only vaccine regimens (Fig. 6d).
DISCUSSION
Containment of HIV-1 replication by prophylactic vaccination is a major global scientific and public health challenge. The great diversity of HIV-1 strains and difficulty in inducing broad neutralizing antibodies makes this an even more formidable challenge. Subtype-specific HIV-1 vaccines expressing a broad array of HIV-1 antigens may be required for maximal effectiveness. Inducing protective immunity to various non-B subtypes of HIV-1 present in less developed countries is, however, poorly studied.
We previously performed a macaque study showing that subtype AE HIV-1 DNA and FPV vaccines expressing Gag, Pol, Env, Tat, and Rev were safe and induced significant CD4 and CD8 T-cell responses to subtype AE overlapping peptide sets spanning Gag, Pol, Env and, to a lesser extent, Tat and Rev (12). To further determine the preclinical efficacy of similar subtype AE vaccines in a heterologous challenge system, we constructed subtype AE SHIV DNA, FPV, and VV vaccines expressing SIV Gag and Pol and HIV-1 AE subtype Env, Tat, and Rev and evaluated their efficacy against a subtype B SHIV, heterologous for the HIV-1 genes. The DNA and FPV vaccines expressed all genes within a single construct. Gag-specific CD4 and CD8 T-cell responses were induced by vaccination, but minimal Pol, Env, Tat, or Rev responses were detected. Further, the levels of Gag-specific CD8 T-cell responses induced by the AE subtype SHIV were less than those observed in previous studies of subtype B SHIV vaccination.
The reasons for the poorer immunogenicity were not clear. Although the B and AE subtype DNA vaccines were very similarly constructed, the subtype B SHIV FPV vaccines expressed either SIV Gag or Pol or HIV-1 Env from separate FPV recombinants. There were modestly reduced levels of Gag expression from the AE subtype SHIV FPV compared to the B subtype SHIV B FPV in vitro (8). It is possible that the reduced expression of Gag or other genes in the AE subtype FPV SHIV recombinant expressing all five SHIV genes compromised immunogenicity, although the similar pure HIV-1 FPV recombinant constructed in an identical manner was broadly immunogenic in macaques (12). Further, the VV vaccines used in the VV/FPV group did express the SHIV genes from separate recombinants and were still only moderately immunogenic. This study illustrates the potential unforeseen difficulties in translating results from HIV-1 to SHIV multigenic vector vaccines and vice versa.
The poorer immunogenicity of the subtype AE vaccines was also followed by poorer efficacy after the subtype B SHIV challenge compared to a homologous B subtype challenge. There was a modest reduction in peak viral load but no significant differences in set point viral load or CD4 T-cell count. The group of six macaques with the best immunogenicity (DNA/FPV-hi) had two animals (5912 and 5085) controlling viral load to below the limits of quantitation by week 20, which resulted in preservation of total CD4 T cells of >10%; these were the only animals to do so (Fig. 2D). Interestingly, animal 5912 was also the only animal that recognized three previously defined strong Gag CD8 T-cell epitopes (AF9, GL8, and CM9), suggesting anecdotally that broad recognition of CD8 T-cell epitopes may be helpful in controlling viremia (Fig. 4f). Although we were not able to consistently induce broad responses to other SHIV proteins by these vaccines, our results lend support to the concept that this should be advantageous (2). It would ultimately also be insightful to immunize macaques against a B subtype SHIV and then challenge with a subtype AE SHIV; however, such virus constructs are not available at present.
Given the poor virologic outcome, there was a remarkably robust anamnestic Gag CD8 T-cell response postchallenge. For example, by KP9 MHC tetramer studies the vaccinated Mane-A*10+ animals had a mean KP9-specific CD8 T-cell response postchallenge of 26% of all CD8 T cells by 2.5 weeks after challenge, compared to <0.3% in the controls. This is similar to that observed in vaccinated Mamu-A*01+ rhesus macaques studied for CM9-specific responses by MHC tetramer after SHIV challenge (3, 5, 24). Indeed, the anamnestic Gag-specific CD8 T-cell response by IFN-γ ICS in the subtype AE SHIV DNA/FPV- or DNA-vaccinated animals was virtually identical to that in the subtype B SHIV DNA/FPV- or DNA-vaccinated animals (Fig. 6d), even though these animals had a much better virologic and immunologic outcome for challenge with the same SHIV virus.
There are several potential explanations for this intriguing observation of postchallenge immunogenicity. First, the quality of the CD8 or CD4 T-cell immunity induced by the subtype B and AE SHIV vaccinations may have been different even though the magnitude of the anamnestic response to the challenge virus was the same. For example, the greater levels of responses induced by vaccination may have primed T-cell responses with higher avidity, affinity, or the ability to secrete multiple cytokines/chemokines that were more effective at clearing virus-infected cells at the same total levels of responses (14). Second, the B subtype SHIV vaccination may have primed a broader response than the subtype AE SHIV vaccination, as discussed above. Third, partial earlier control of the B subtype SHIV challenge virus resulting in less-profound CD4 T-cell depletion may have facilitated a broader recognition of nonvaccine SHIV antigens, although we previously showed this was not related to early neutralizing antibody responses (9). The subtype AE SHIV-vaccinated animals were remarkable for their very poor recognition of non-Gag antigens (Fig. 3b to d and 5b and c), suggesting the strong dominance of the primed Gag-specific responses to the detriment of a broader array of responses. The breadth of the response is likely to be crucial when faced with a heterologous challenge.
In conclusion, this large macaque study of subtype AE SHIV vaccines found that although Gag-specific CD8 and CD4 T-cell responses were primed by vaccination, the responses induced were narrow and insufficient to result in consistent control of a heterologous B subtype SHIV challenge. Further refinements in vaccine design or delivery to induce broader cross-subtype T-cell responses to non-B subtype HIV antigens will likely be required to facilitate more effective control of HIV replication in less-developed countries.
Acknowledgments
This work was supported by the NIH HIV Vaccine Design and Development Team award N01-AI-05395, Australian National Health and Medical Research Council grants 299907, 251653, 350841, and 251654, a James S. McDonnell Foundation Award, a Sylvia and Charles Viertel fellowship, and the Australian Centre for HIV and Hepatitis Research.
We thank all members of the Australian Thai HIV Vaccine Consortium. We thank Sheilajen Alcantara, Jeanette Reece, Erin Taylor, Liyen Loh, Rachel Amos-Ritchie, Mary Ann Anderson, Rhonda Voysey, Jie Lin, and Andrew Brooks for expert technical assistance and Kim Szalnowski, Andrew Sydenham, and Leah Protyniak for expert animal care.
Footnotes
Published ahead of print on 18 October 2006.
REFERENCES
- 1.Altman, J. D., P. Moss, P. Goulder, D. H. Barouch, W. M. McHeyzer, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., J. M. Smith, S. I. Staprans, D. C. Montefiori, F. Villinger, J. D. Altman, S. P. O'Neil, N. L. Kozyr, Y. Xu, L. S. Wyatt, P. L. Earl, J. G. Herndon, J. M. McNicholl, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J. Virol. 76:6138-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. O'Neil, S. Staprans, D. Montefiori, Y. Xu, J. Herndon, L. Wyatt, M. Candido, N. Kozyr, P. Earl, J. Smith, H. Ma, B. Grimm, M. Hulsey, J. Miller, H. McClure, J. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, R. J., C. M. Hannan, S. C. Gilbert, S. M. Laidlaw, E. G. Sheu, S. Korten, R. Sinden, G. A. Butcher, M. A. Skinner, and A. V. Hill. 2004. Enhanced CD8+ T cell immune responses and protection elicited against Plasmodium berghei malaria by prime boost immunization regimens using a novel attenuated fowlpox virus. J. Immunol. 172:3094-3100. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 6.Batten, C. J., R. D. Rose, K. M. Wilson, M. B. Agy, S. Chea, I. Stratov, D. C. Montefiori, and S. J. Kent. 2006. Comparative evaluation of simian, simian-human, and human immunodeficiency virus infections in the pigtail macaque (Macaca nemestrina) model. AIDS Res. Hum. Retrovir. 22:580-588. [DOI] [PubMed] [Google Scholar]
- 7.Boyle, D. B., M. Anderson, R. Amos, R. Voysey, and B. E. H. Coupar. 2004. Construction of recombinant fowlpox viruses carrying multiple vaccine antigens and immunomodulatory molecules. BioTechniques 37:104-111. [DOI] [PubMed] [Google Scholar]
- 8.Coupar, B. E., D. F. Purcell, S. A. Thomson, I. A. Ramshaw, S. J. Kent, and D. B. Boyle. 2006. Fowlpox virus vaccines for HIV and SHIV clinical and pre-clinical trials. Vaccine 24:1378-1388. [DOI] [PubMed] [Google Scholar]
- 9.Dale, C. J., R. De Rose, I. Stratov, S. Chea, D. Montefiori, S. A. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, M. Law, and S. J. Kent. 2004. Efficacy of DNA and fowlpox virus prime/boost vaccines for simian/human immunodeficiency virus. J. Virol. 78:13819-13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dale, C. J., R. De Rose, K. Wilson, H. Croom, S. A. Thomson, B. E. Coupar, A. J. Ramsay, D. F. Purcell, R. Ffrench, M. Law, S. Emery, D. A. Cooper, I. A. Ramshaw, D. B. Boyle, and S. J. Kent. 2004. Evaluation in macaques of HIV-1 DNA vaccines containing primate CpG motifs and fowlpoxvirus vaccines co-expressing IFN-γ or IL-12. Vaccine 23:188-197. [DOI] [PubMed] [Google Scholar]
- 11.Dale, C. J., X. S. Liu, R. De Rose, D. F. Purcell, J. Anderson, Y. Xu, G. R. Leggatt, I. H. Frazer, and S. J. Kent. 2002. Chimeric human papilloma virus-simian/human immunodeficiency virus virus-like-particle vaccines: immunogenicity and protective efficacy in macaques. Virology 301:176-187. [DOI] [PubMed] [Google Scholar]
- 12.De Rose, R., S. Chea, C. J. Dale, J. Reece, C. S. Fernandez, K. M. Wilson, S. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, M. T. Sullivan, and S. J. Kent. 2005. Subtype AE HIV-1 DNA and recombinant fowlpox virus vaccines encoding five shared HIV-1 genes: safety and T cell immunogenicity in macaques. Vaccine 23:1949-1956. [DOI] [PubMed] [Google Scholar]
- 13.De Rose, R., M. T. Sullivan, C. J. Dale, A. Kelleher, S. Emery, C. D. A., I. A. Ramshaw, D. Boyle, and S. J. Kent. 2006. Dose-response relationship of DNA and recombinant fowlpox virus prime-boost vaccines: implications for future trials. Hum. Vaccines 2:134-136. [DOI] [PubMed] [Google Scholar]
- 14.Estcourt, M. J., A. J. Ramsay, A. Brooks, S. A. Thomson, C. J. Medveckzy, and I. A. Ramshaw. 2002. Prime-boost immunization generates a high frequency, high-avidity CD8+ cytotoxic T lymphocyte population. Int. Immunol. 14:31-37. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez, C. S., I. Stratov, R. De Rose, K. Walsh, C. J. Dale, M. Z. Smith, M. B. Agy, S. L. Hu, K. Krebs, D. I. Watkins, H. O'Connor, D., M. P. Davenport, and S. J. Kent. 2005. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J. Virol. 79:5721-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanke, T., T. J. Blanchard, J. Schneider, C. M. Hannan, M. Becker, S. C. Gilbert, A. V. Hill, G. L. Smith, and A. McMichael. 1998. Enhancement of MHC class I-restricted peptide-specific T cell induction by a DNA prime/MVA boost vaccination regime. Vaccine 16:439-445. [DOI] [PubMed] [Google Scholar]
- 17.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelleher, A. D., R. L. Puls, M. Bebbington, D. Boyle, R. Ffrench, S. J. Kent, S. Kippax, D. F. Purcell, S. Thomson, H. Wand, D. A. Cooper, and S. Emery. 2006. A randomized, placebo-controlled phase I trial of DNA prime, recombinant fowlpox virus boost prophylactic vaccine for HIV-1. AIDS 20:294-297. [DOI] [PubMed] [Google Scholar]
- 19.Kent, S. J., C. J. Dale, C. Ranasinghe, I. Stratov, R. De Rose, S. Chea, D. Montefiori, S. Thomson, I. A. Ramshaw, B. E. Coupar, D. B. Boyle, M. Law, K. M. Wilson, and A. J. Ramsay. 2005. Mucosally-administered human-simian immunodeficiency virus DNA and fowlpox virus-based recombinant vaccines reduce acute phase viral replication in macaques following vaginal challenge with CCR5-tropic SHIVSF162P3. Vaccine 23:5009-5021. [DOI] [PubMed] [Google Scholar]
- 20.Kent, S. J., A. Zhao, S. Best, J. D. Chandler, D. B. Boyle, and I. A. Ramshaw. 1998. Enhanced T cell immunogenicity and protective efficacy from a HIV-1 vaccine regimen consisting of consecutive priming with DNA and boosting with recombinant fowlpox virus. J. Virol. 72:10180-10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maecker, H. T., H. S. Dunn, M. A. Suni, E. Khatamzas, C. J. Pitcher, T. Bunde, N. Persaud, W. Trigona, T. M. Fu, E. Sinclair, B. M. Bredt, J. M. McCune, V. C. Maino, F. Kern, and L. J. Picker. 2001. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol. Methods 255:27-40. [DOI] [PubMed] [Google Scholar]
- 22.Nabel, G. J. 2001. Challenges and opportunities for development of an AIDS vaccine. Nature 410:1002-1007. [DOI] [PubMed] [Google Scholar]
- 23.Ranasinghe, C., J. C. Medveczky, D. Woltring, K. Gao, S. Thomson, B. E. Coupar, D. B. Boyle, A. J. Ramsay, and I. A. Ramshaw. 2006. Evaluation of fowlpox-vaccinia virus prime-boost vaccine strategies for high-level mucosal and systemic immunity against HIV-1. Vaccine 24:5881-5895. [DOI] [PubMed] [Google Scholar]
- 24.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 25.Smith, M. Z., C. J. Dale, R. De Rose, I. Stratov, C. S. Fernandez, A. G. Brooks, J. T. Weinfurter, K. Krebs, C. Riek, D. I. Watkins, D. H. O'Connor, and S. J. Kent. 2005. Analysis of pigtail macaque major histocompatibility complex class I molecules presenting immunodominant simian immunodeficiency virus epitopes. J. Virol. 79:684-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, M. Z., C. S. Fernandez, A. Chung, C. J. Dale, R. De Rose, J. Lin, A. G. Brooks, K. C. Krebs, D. I. Watkins, D. H. O'Connor, M. P. Davenport, and S. J. Kent. 2005. The pigtail macaque MHC class I allele Mane-A*10 presents an immunodominant SIV Gag epitope: identification, tetramer development and implications of immune escape and reversion. J. Med. Primatol. 34:282-293. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, N. A., J. Reed, G. S. Napoe, S. Piaskowski, A. Szymanski, J. Furlott, E. J. Gonzalez, L. J. Yant, N. J. Maness, G. E. May, T. Soma, M. R. Reynolds, E. Rakasz, R. Rudersdorf, A. B. McDermott, H. O'Connor, D., T. C. Friedrich, D. B. Allison, A. Patki, L. J. Picker, D. R. Burton, J. Lin, L. Huang, D. Patel, G. Heindecker, J. Fan, M. Citron, M. Horton, F. Wang, X. Liang, J. W. Shiver, D. R. Casimiro, and D. I. Watkins. 2006. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J. Virol. 80:5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]