Abstract
In the past several years, a number of cellular proteins have been identified as candidate entry receptors for hepatitis C virus (HCV) by using surrogate models of HCV infection. Among these, the tetraspanin CD81 and scavenger receptor B type I (SR-BI), both of which localize to specialized plasma membrane domains enriched in cholesterol, have been suggested to be key players in HCV entry. In the current study, we used a recently developed in vitro HCV infection system to demonstrate that both CD81 and SR-BI are required for authentic HCV infection in vitro, that they function cooperatively to initiate HCV infection, and that CD81-mediated HCV entry is, in part, dependent on membrane cholesterol.
Hepatitis C virus (HCV) is a member of the Flaviviridae family of viruses and is a major cause of chronic hepatitis and hepatocellular carcinoma (3, 15). The HCV genome is a positive-strand ∼9.6-kb RNA molecule consisting of a single open reading frame which is flanked by 5′ and 3′ untranslated regions (UTR). The HCV 5′ UTR contains a highly structured internal ribosome entry site (14, 50, 64, 78, 79, 86), while the 3′ UTR is essential for replication (32, 88). The HCV open reading frame encodes a single polyprotein of 3,008 to 3,037 amino acids in length that is posttranslationally processed by host and viral proteases to produce at least 10 different proteins: core protein, envelope proteins E1 and E2, p7, and nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (6, 15, 64).
The host-virus interactions required during the initial steps of HCV infection are not completely understood. Prior to the development of an in vitro infectious HCV system, HCV pseudotyped retroviral particles (HCVpp), which comprise a reporter retrovirus enclosed in an envelope containing HCV glycoproteins E1 and E2, have allowed detailed analyses of the host-virus interactions required for HCVpp entry (9, 10, 16, 30, 31, 34, 39, 46, 81, 83). Using a combination of HCVpp and recombinant soluble E2 glycoprotein (sE2) binding assays, a number of cellular proteins have been identified as potential candidates for HCV entry receptors, including the tetraspanin protein CD81 (61), scavenger receptor B type I (SR-BI) (68), which is a cellular protein that binds high-density lipoprotein (HDL), the low-density lipoprotein receptor (LDL-R) (1, 52), the C-type lectins L-SIGN and DC-SIGN (25, 33, 48, 62), heparin sulfate (8), and the asialoglycoprotein receptor (67). The roles of CD81 and SR-BI as HCVpp receptors are well documented (2, 7, 9, 11, 39, 68, 91), and CD81 was recently shown to be required for cell culture-derived HCV infection (47, 85, 89, 92). However, the extent to which SR-BI is required for HCV infection and whether it functions cooperatively with CD81 are poorly understood.
Cellular cholesterol is critical for infection by many viruses (23, 26, 40, 53, 73, 87). Indeed, components of the receptor complex for dengue virus, another flavivirus, are reported to be localized to cholesterol-enriched microdomains called lipid rafts, such that depletion of cholesterol by methyl-β-cyclodextrin (MβCD) inhibits dengue virus entry (65). These cholesterol-enriched microdomains serve as distinct domains on the plasma membrane, where many cellular proteins, including viral receptors, are preferentially localized. Examples of such proteins include caveolins, flotillins, Shc, mitogen-activated protein kinase, and protein kinase C (60). Localization to cholesterol-enriched plasma membrane microdomains has been demonstrated for both CD81 (18, 71) and SR-BI (66). SR-BI has been reported to be localized in cholesterol-enriched plasma membrane microdomains called caveolae (5, 35). Cross-linking of CD81 and downstream Lck activation by HCV sE2 are disrupted by MβCD in T cells (71), demonstrating a functional role for localization of CD81 to cholesterol-enriched domains. Furthermore, CD81 has been demonstrated to physically interact with cholesterol (17). While the dependence of HCV infection on cellular cholesterol is currently unknown, HCVpp fusion with liposomes is enhanced by the presence of cholesterol in the target membrane (46). These data strongly suggest that plasma membrane cholesterol may be required for HCV entry.
In the current study, we used the recently reported in vitro JFH-1 HCV infection and HCVpp viral systems and antibodies specific for CD81 and SR-BI to demonstrate that CD81 and SR-BI function cooperatively to mediate HCV infection in Huh-7 cells. The depletion of membrane cholesterol using the cholesterol-depleting drug MβCD decreases HCV infection, in part by causing the mislocalization and decreased surface expression of CD81.
MATERIALS AND METHODS
Tissue culture and generation of stable HCV subgenomic replicon cell lines.
The JFH-1 (genotype 2a) subgenomic HCV replicon was obtained from Takaji Wakita (Tokyo Metropolitan Institute of Neuroscience, Tokyo, Japan) and passaged as previously described (27, 43, 44). HCV replicon-containing and parental Huh-7 cells were cultured in complete Dulbecco's modified Eagle's medium (DMEM) (supplemented with 10% fetal calf serum [FCS], 100 U/ml penicillin, 100 mg/ml streptomycin, 2 mM l-glutamine, and 0.1 mM nonessential amino acids). The HCV replicon cell lines were cultured in the presence of 500 μg/ml Geneticin (Invitrogen, San Diego, CA). Huh-7 cells were cultured as described previously (41, 42, 92).
Generation of JFH-1 virus stocks and infections.
JFH-1 virus stocks were generated by infecting Huh-7.5.1 cells as described previously (92). The multiplicity of infection (MOI) was calculated based on JFH-1 titer as measured by focus forming units/ml. For SR-BI and CD81 neutralization experiments, Huh-7 cells were pretreated with the respective antibodies as discussed below and infected with JFH-1 at an MOI of 0.3 in a volume of 50 μl complete DMEM. The virus inoculum was incubated with cells for 5 h, after which cells were washed twice with phosphate-buffered saline (PBS) and incubated with complete DMEM for the duration of the experiment. For all experiments involving depletion of cellular cholesterol by MβCD, infections were performed using an MOI of 0.02.
RNA extractions, reverse transcription, and real-time PCR analysis.
Total RNA was isolated using guanidine thiocyanate extraction as described previously (20). Reverse transcription (RT) reactions and real-time PCR analyses were performed as previously described (42, 92).
HCV neutralization assays.
Virus neutralization assays were performed as previously described (92). Antibodies directed against the extracellular loop of SR-BI were raised by genetic immunization of Wistar rats with a pcDNA expression vector containing the full-length human SR-BI cDNA (pcDNA SR-BI/CLA-1; a gift from T. Huby, INSERM, Dyslipoproteinemia and Atherosclerosis Research Unit, Hopital de la Pitié, Paris, France). Briefly, rats received four injections of 20 μg pcDNA SR-BI intraperitoneally at 2-week intervals. Preimmune control serum was collected from the same rat bled before immunization. To analyze the specificity of the produced anti-SR-BI polyclonal serum, CHO cells were transfected with pcDNA (control vector) or pcDNA SR-BI using liposome-mediated gene transfer (Lipofectamine; Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol. CHO cells were then incubated with anti-SR-BI polyclonal serum or preimmune control serum and analyzed for cell surface SR-BI expression by flow cytometry as described previously (7). Huh-7 cells (5 × 103/well) were seeded into 96-well plates (Corning Inc., Corning, NY), and the following day, cells were incubated with dilutions of antibodies specific for SR-BI or CD81 (5A6 clone; a gift from Shoshana Levy, Stanford University, Stanford, CA) for 1 h at 37°C. Cells were then infected with JFH-1 at an MOI of 0.3, and intracellular JFH-1 RNA and NS5A protein expression levels were monitored by RT-PCR and indirect immunofluorescence, respectively, 3 days later, as previously described (92). We define an effect of both antibodies as synergistic or cooperative if the change (n-fold) when using the two antibodies is equal to or greater than that of the product rather than the sum of the changes (n-fold) of HCV RNA replication detected when using the antibodies individually.
Cholesterol depletion experiments.
MβCD and cholesterol were obtained from Sigma (St. Louis, MO). Huh-7 cells (105/well) were seeded into 12-well BioCoat plates (BioCoat, Horsham, PA). The next day, cells were washed with PBS and incubated with 7.5 mM MβCD in incomplete DMEM (without FCS) for 30 min at 37°C. Cells were washed with PBS and then incubated in incomplete DMEM alone or containing 150 μg/ml cholesterol (to replenish cholesterol levels) for 1 hour at 37°C. Huh-7 cells were washed twice with PBS and then infected with JFH-1 virus at an MOI of 0.02 in DMEM containing 5% FCS. At various times postinfection, total RNA was isolated for RT-PCR analysis. MβCD treatment of preinfected Huh-7 cells or JFH-1 subgenomic (JFH-1 SG) replicon-containing cell lines was performed in a similar manner.
HCVpp generation and infection.
All pseudoviruses were generated by cotransfection of plasmids encoding (i) a provirus containing the desired reporter gene or transgene, (ii) human immunodeficiency virus (HIV) Gag-Pol, and (iii) an appropriate envelope glycoprotein. All HCVpp used in this study were generated using the H77 E1E2 sequence (residues 170 to 746). On the day prior to transfection, 8 × 105 293T cells were seeded in a 35-mm well. The following day, a total of 1.5 μg DNA was transfected using Lipofectamine (Invitrogen, Carlsbad, CA). The media were replaced after 6 h. Supernatants were harvested at 48 and 72 h posttransfection, pooled, and filtered using a 0.45-μm-pore-size filter. The plasmid combinations and ratios (by weight) used to generate luciferase reporter HCVpp are listed below. Equal amounts of pNL43.luc.R-.E- (encoding a provirus containing luciferase and HIV Gag-Pol) and plasmids expressing HCV E1E2, murine leukemia virus (MLV) Env, and empty vector were cotransfected, giving rise to HCVpp, MLVpp, and “no envelope” pseudoparticles, respectively, as previously reported (39). To generate green fluorescent protein (GFP) reporter HCVpp, plasmids encoding (i) an HIV provirus encoding GFP (12), (ii) HIV Gag-Pol, and (iii) H77 E1E2 or empty vector were transfected at a 1:1:4 ratio. Infection assays with luciferase reporter HCVpp were performed in a 96-well format using 104 target cells per well. Cells were infected with pseudovirus supernatants diluted in fresh media (1:5 for HCVpp and “no Env” pp; 1:500 for MLVpp). After 8 h, the media were changed and luciferase assays were performed at 72 h postinfection as previously described (7). Briefly, cells were lysed with 40 μl cell culture lysis (Promega, Madison, WI) and the expression of the luciferase reporter was measured after the addition of 50 μl luciferase substrate (Promega, Madison, WI) on a Centro LB960 luminometer (Berthold Technologies, Bad Wildbad, Germany). For infection assays with the GFP reporter, 3 × 104 HCVpp were plated in 48-well plates. The next day, the cells were infected with pseudovirus for 8 h, the media were changed, and cells were further cultured for an additional 48 h prior to harvesting and fixation with 1% (wt/vol) paraformaldehyde. GFP expression was quantified using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Indirect immunofluorescence staining of CD81.
The protocol for indirect immunofluorescence has been described previously (41, 92). Briefly, 3 × 104 Huh-7 cells were seeded into Lab-Tek eight-chamber coverglasses (Nalge Nunc International, Naperville, IL). The next day, the cells were treated with MβCD as described above. After treatment, Huh-7 cells were fixed with 4% paraformaldehyde for 20 min at room temperature and stained for CD81 expression by indirect immunofluorescence analysis. Cells were incubated with antibodies specific to CD81 (5A6 clone; a gift from Shoshana Levy, Stanford University, Stanford, CA) at a dilution of 1:100, followed by incubation with a secondary antibody conjugated to Alexa 555 fluorochrome (Invitrogen Corporation, Carlsbad, CA). Serial Z sections were analyzed using a deconvolution microscope as described previously (41).
Flow cytometric analysis of cell surface CD81 and SR-BI.
Huh-7 cells (8 × 105) were seeded into 60-mm dishes (Corning Inc., Corning, NY). The next day, cells were treated with MβCD as described above, trypsinized, and resuspended in staining buffer (1% bovine serum albumin in PBS). Cells were incubated with antibodies specific for CD81 (Serotec, Raleigh, NC) or SR-BI (Novus Biologicals, Littleton, CO) at dilutions of 1:100 for 30 min at 4°C. Incubation without primary antibodies gave no specific staining. Cells were washed three times with staining buffer and incubated with donkey anti-mouse (CD81) or anti-rabbit (SR-BI) antibody conjugated to phycoerythrin (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:100 dilution for 30 min at 4°C. Cells were washed three times, fixed in 2% paraformaldehyde, and analyzed by flow cytometry using a Becton Dickinson FACSArray. A total of 50,000 events were collected per sample. Mean fluorescence intensities (MFI) were measured by calculating the geometric mean for each histogram peak.
Quantitation of intracellular cholesterol levels.
Huh-7 cells (8 × 105/well) were seeded in 12-well plates (Corning Inc., Corning, NY). The next day, cells were treated with 7.5 mM MβCD as described above. The cells were washed twice with PBS, counted, and lysed in PBS containing 1% Triton X-100. Lysates generated from 4 × 105 Huh-7 cells were spun at 20,000 × g for 5 min to remove debris, and cholesterol levels were quantitated using an Amplex Red cholesterol assay kit (Molecular Probes, Eugene, OR) as per the manufacturer's recommendations. Fluorescence was measured using TECAN Safire II (Tecan Systems Inc., San Jose, CA). A standard curve using purified cholesterol was generated for each experiment.
Statistical analyses.
All statistical analyses were performed using Microsoft Excel software. All graphs represent means ± standard deviations (SDs). P values for all data were determined using the paired t test.
RESULTS
CD81 and SR-BI are required for HCV infection.
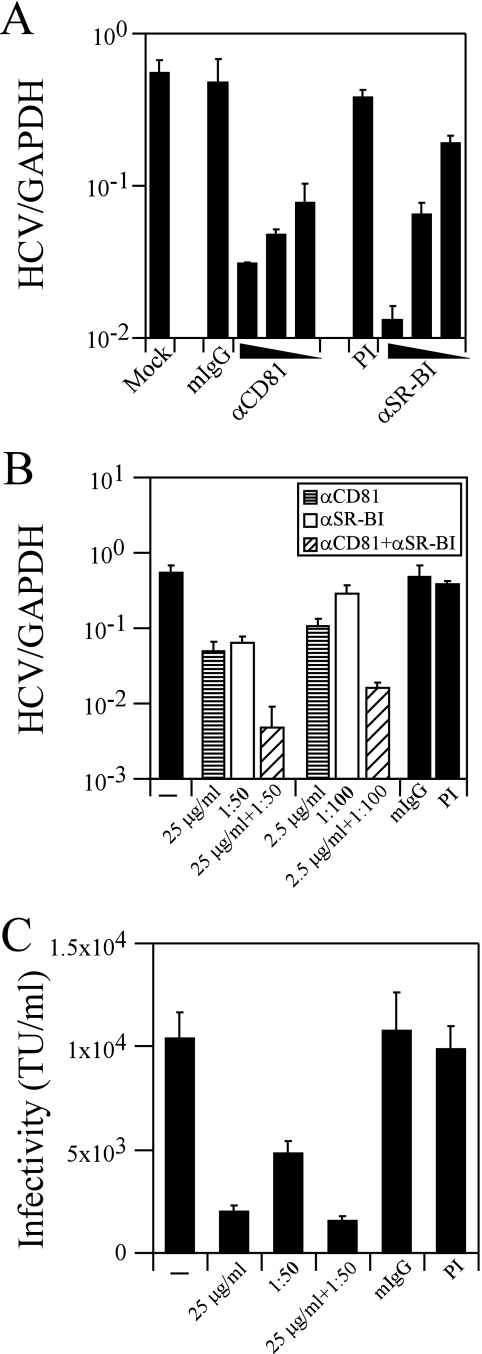
To determine whether CD81 and SR-BI are required for HCV infection, we infected Huh-7 human hepatoma cells with JFH-1 virus in the presence of antibodies specific for CD81 and SR-BI, individually and together (92). As shown in Fig. 1A, incubation of Huh-7 cells with various concentrations of a monoclonal mouse anti-CD81 antibody resulted in a dose-dependent inhibition of intracellular JFH-1 RNA at 3 days postinfection (∼17.8-fold [P = 0.014] and ∼11.6-fold [P = 0.016] at 25- and 10-μg/ml antibody concentrations, respectively). Similarly, the rat anti-SR-BI polyclonal antiserum resulted in ∼8.6-fold (P = 0.014) and ∼2.9-fold decreases in intracellular JFH-1 RNA levels at 3 days postinfection at dilutions of 1:50 and 1:100, respectively (Fig. 1A). No significant decrease in intracellular JFH-1 RNA was detected using either isotype-matched mouse immunoglobulin G (IgG) or preimmune (PI) rat serum (Fig. 1A). The number of NS5A-positive Huh-7 cells was also markedly decreased after incubation with anti-CD81 (compare Fig. 2A and D) or anti-SR-BI (compare Fig. 2A and E) antibody, consistent with the decrease in intracellular JFH-1 RNA. No significant change in cell proliferation was observed when the cells were incubated with either anti-CD81 or anti-SR-BI antibody (data not shown).
FIG. 1.
SR-BI is required for JFH-1 infection. Huh-7 cells were incubated with anti-CD81 or anti-SR-BI antibody and inoculated with JFH-1, and infectivity was analyzed by measuring intracellular JFH-1 RNA at 3 days postinfection. (A) Huh-7 cells were infected with JFH-1 in the presence of three dilutions each of anti-CD81 (250, 25, and 10 μg/ml) and anti-SR-BI (1:20, 1:50, and 1:100) antibodies. As controls, Huh-7 cells were incubated with isotype-matched mouse IgG (mIgG) and PI rat serum at 250-μg/ml and 1:50 dilutions, respectively. (B) Huh-7 cells were incubated with concentrations of anti-CD81 antibodies alone (bars with horizontal lines), anti-SR-BI antibodies alone (unfilled bars), or both antibodies (bars with diagonal lines), and intracellular JFH-1 RNA levels were measured 3 days later. Two dilutions each of anti-CD81 (25 and 2.5 μg/ml) and anti-SR-BI (1:50 and 1:100) antibodies were used. To demonstrate cooperativity, Huh-7 cells were incubated with combinations of anti-CD81 and anti-SR-BI antibodies. Cells were incubated with anti-CD81 and anti-SR-BI antibodies (25 μg/ml anti-CD81 plus 1:50 anti-SR-BI and 2.5 μg/ml anti-CD81 plus 1:100 anti-SR-BI). As controls for anti-CD81 and anti-SR-BI antibodies, Huh-7 cells were incubated with equivalent concentrations of isotype-matched mouse IgG and PI rat serum, respectively. HCV RNA copy numbers were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) copy numbers for all samples. These are representative data (means ± SDs) for three independent experiments. (C) Inhibition of HCVpp entry using antibodies specific to CD81 (25 mg/ml) and SR-BI (1:50) individually or in combination. TU, transducing units. These are representative data (means ± SDs) for three replicates.
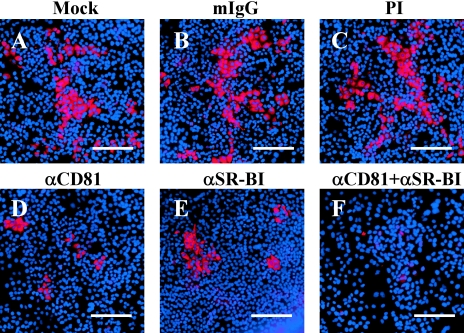
FIG. 2.
Anti-SR-BI antibodies inhibit JFH-1 NS5A protein expression. Indirect immunofluorescence analysis of NS5A protein expressed in JFH-1-infected Huh-7 cells alone (A) or in the presence of anti-CD81 (25 μg/ml) (D), anti-SR-BI (1:50) (E), or a combination of both anti-CD81 and anti-SR-BI antibodies (F). Huh-7 cells were incubated with equivalent concentrations of isotype-matched mouse IgG (mIgG) (B) and PI rat serum (C) as controls for the anti-CD81 and anti-SR-BI antibodies, respectively. The bars represent 400-μm distances. These are representative data for three independent experiments.
Cooperation between SR-BI and CD81 is required for HCV infection.
To determine whether CD81 and SR-BI function cooperatively to mediate HCV infection, Huh-7 cells were incubated with concentrations of anti-CD81 (25 and 2.5 μg/ml) and anti-SR-BI (1:50 and 1:100) antibodies and compared to cells incubated with anti-CD81 or anti-SR-BI antibody alone. While intracellular JFH-1 RNA levels were decreased ∼11.6-fold or ∼8.6-fold when Huh-7 cells were incubated with 25 μg/ml anti-CD81 or a 1:50 dilution of anti-SR-BI, respectively, coincubation of the Huh-7 cells with both anti-CD81 and anti-SR-BI antibodies at the same concentrations resulted in a significantly greater (∼116.6-fold; P = 0.012) decrease in intracellular JFH-1 RNA (Fig. 1B). This cooperativity between CD81 and SR-BI was also detected when Huh-7 cells were incubated with a lower dilution (1:100) of the anti-SR-BI antibody, which alone did not result in a statistically significant decrease (∼2-fold decrease; P = 0.19) in intracellular JFH-1 RNA (Fig. 1B). Using 10-fold less anti-CD81 antibody (2.5 μg/ml), intracellular JFH-1 RNA was decreased ∼5.1-fold (P = 0.013). However, in combination with a 1:100 dilution of anti-SR-BI antibody, intracellular JFH-1 RNA was decreased ∼34-fold (P = 0.014) (Fig. 1B). Similarly, fewer NS5A-positive Huh-7 cells were detected after coincubation with both anti-CD81 (25 μg/ml) and anti-SR-BI (1:50) antibodies (Fig. 2F) than when the cells were incubated with the antibodies individually (Fig. 2D and E). No significant change in cell proliferation was observed when the cells were incubated with a combination of anti-CD81 and anti-SR-BI antibodies. This synergistic effect suggests that SR-BI and CD81 function cooperatively during HCV infection, presumably at the level of entry.
To study this further, we evaluated the ability of the anti-CD81 and anti-SRBI antibodies, alone and in combination, to neutralize HCVpp entry into Huh-7 cells. Antibodies to CD81 (25 μg/ml) and SR-BI (1:50) neutralized HCVpp infectivity ∼5- and ∼2.1-fold, respectively, and ∼6.6-fold in combination (Fig. 1C). Similar results were observed at lower antibody concentrations (data not shown). These data suggest that while both CD81 and SR-BI are required for HCVpp entry, they do not function cooperatively in the HCVpp system.
MβCD decreases CD81 but increases SR-BI cell surface expression.
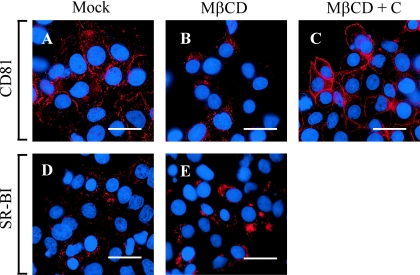
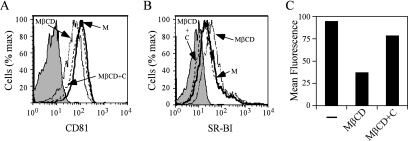
Since CD81 and SR-BI localize to specialized plasma membrane microdomains that are enriched in cholesterol (18, 66), we assessed whether depletion of cholesterol by MβCD alters the membrane expression of CD81 and SR-BI. Huh-7 cells were treated with MβCD, after which indirect immunofluorescence staining was performed using antibodies specific to CD81 and SR-BI. As shown in Fig. 3, M βCD treatment decreased plasma membrane localization of CD81 (compare Fig. 3A and B) and this could be reversed by the replenishment of cholesterol (Fig. 3C). In contrast, MβCD treatment did not appear to decrease SR-BI localization at the plasma membrane (data not shown). To confirm these observations, we used flow cytometric analysis to determine whether surface expression of CD81 and SR-BI was altered by MβCD treatment. Based on the MFI, MβCD treatment of Huh-7 cells resulted in an ∼2.5-fold decrease in cell surface expression of CD81 (Fig. 4A and C). Interestingly, cell surface expression of SR-BI was increased ∼2.4-fold (Fig. 4B). Furthermore, the changes in cell surface expression of CD81 (Fig. 4A) and SR-BI (Fig. 4B) were reversed when MβCD-treated cells were incubated with exogenous cholesterol. These results demonstrate that while depletion of cholesterol by MβCD results in a decrease of cell surface CD81 expression, SR-BI expression levels are induced after cholesterol depletion.
FIG. 3.
MβCD treatment of Huh-7 cells decreases CD81 plasma membrane expression. CD81 expression was analyzed in mock-treated Huh-7 cells (A) or in Huh-7 cells after treatment with 10 mM MβCD (B). CD81 expression was also determined in MβCD-treated cells to which exogenous cholesterol was added (C). SR-BI expression was also analyzed in mock-treated (D) and MβCD-treated (E) Huh-7 cells. The bars represent 60-μm distances. These are representative data for at least two independent experiments.
FIG. 4.
CD81 cell surface expression decreases after MβCD treatment. Cell surface expression of CD81 (A) and SR-BI (B) was analyzed in mock-treated Huh-7 cells (M), Huh-7 cells that were treated with 10 mM MβCD (MβCD), or MβCD-treated cells which were further incubated with cholesterol (MβCD+C). (C) MFI of CD81 surface expression after mock treatment or treatment with 10 mM MβCD in the presence (MβCD+C) or absence (MβCD) of exogenous cholesterol are plotted. These are representative data for at least two independent experiments.
MβCD depletion of cholesterol from Huh-7 cells inhibits HCV infection.
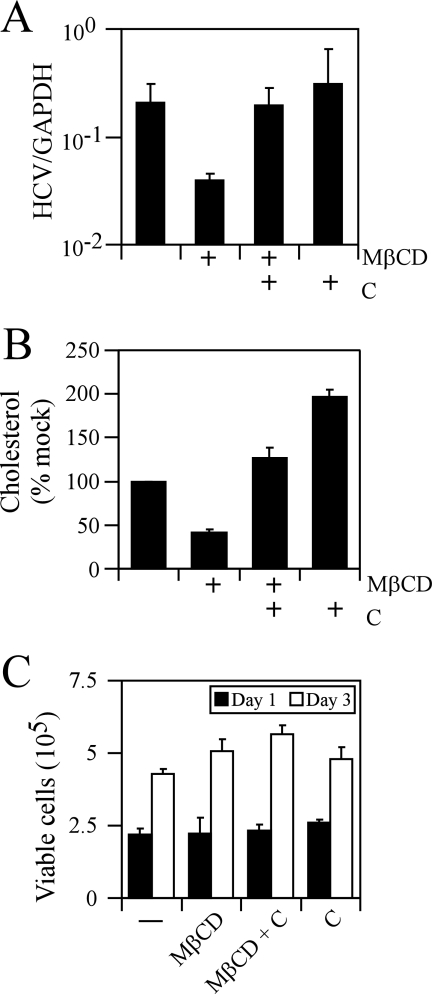
Since cholesterol depletion by MβCD led to a decrease in cell surface expression levels of CD81, we determined whether cellular cholesterol depletion inhibits JFH-1 infection in vitro. Accordingly, Huh-7 cells were pretreated with 7.5 mM MβCD and cholesterol was either added or not added back to the culture medium prior to infection. Cells were washed with PBS to remove MβCD and/or cholesterol before the cells were infected with JFH-1 at an MOI of 0.02. Infection of Huh-7 cells pretreated with MβCD resulted in an ∼6.2-fold decrease in intracellular JFH-1 RNA at 3 days postinfection (P = 0.03) (Fig. 5A). This decrease in JFH-1 RNA was reversed when MβCD-treated Huh-7 cells were incubated with exogenous cholesterol prior to infection (Fig. 5A), suggesting that the reduction in JFH-1 RNA is due to cellular cholesterol depletion. In order to determine whether MβCD treatment depletes cellular cholesterol levels, we quantitated the amount of cellular cholesterol before and after MβCD treatment. MβCD-treated Huh-7 cells reduced the total intracellular pools of cholesterol by ∼60% (P = 0.001) compared to mock-treated cells (Fig. 5B). Huh-7 cells treated with an inactive stereoisomer of MβCD, α-cyclodextrin (80), had no effect on intracellular JFH-1 RNA levels (data not shown). To determine whether MβCD treatment affected Huh-7 growth, we quantitated the number of viable Huh-7 cells at various times posttreatment by trypan blue exclusion. As can be seen in Fig. 5C, there was no significant change in the total number of viable Huh-7 cells during the course of the infection. Altogether, these data demonstrate that HCV infection in vitro is dependent on the cholesterol content of the cell.
FIG. 5.
Depletion of cholesterol by MβCD decreases JFH-1 infection in Huh-7 cells. Huh-7 cells were untreated or pretreated with 7.5 mM MβCD alone (MβCD) or MβCD plus cholesterol (MβCD + C) prior to infection with JFH-1. (A) Total RNA was harvested 3 days later, and levels of intracellular JFH-1 RNA were quantitated by RT-PCR. Relative HCV levels were measured by normalization to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) copy numbers for each sample. (B) Intracellular cholesterol levels were quantitated in mock-treated Huh-7 cells or cells that were treated with MβCD alone (MβCD) or MβCD plus cholesterol (MβCD + C). (C) Viable cells were quantitated by trypan blue exclusion after MβCD treatment (in the absence or presence of cholesterol) and compared to mock-treated Huh-7 cells.
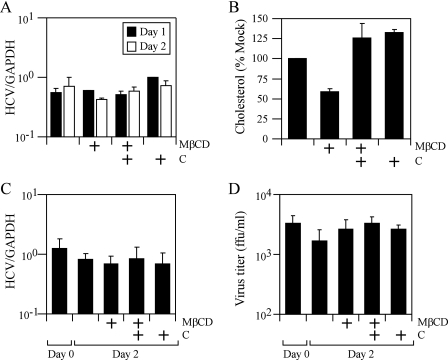
Cellular cholesterol depletion by MβCD does not affect HCV replicon RNA replication.
The decrease in intracellular JFH-1 RNA content after MβCD pretreatment could be due to an effect of MβCD either on JFH-1 RNA replication or on JFH-1 viral entry. To determine whether MβCD treatment affects HCV RNA replication, Huh-7 cells supporting replication of the JFH-1 SG replicon were treated with 7.5 mM MβCD and intracellular levels of JFH-1 RNA were measured at various times posttreatment. While treatment of JFH-1 SG replicon-containing cells with MβCD decreased levels of intracellular cholesterol ∼2-fold (Fig. 6B), it failed to significantly affect JFH-1 intracellular RNA levels at 2 days posttreatment (Fig. 6A). Similar results were obtained using the genotype 1b S1179I-adapted subgenomic replicon (data not shown).
FIG. 6.
Depletion of cholesterol by MβCD does not affect HCV RNA replication. (A) Huh-7 cells stably replicating the JFH-1 SG replicon were treated with 7.5 mM MβCD, and intracellular levels of JFH-1 RNA were measured at 1 and 2 days posttreatment. (B) Intracellular cholesterol levels in Huh-7 cells were quantitated after treatment with MβCD (MβCD) or MβCD plus cholesterol (MβCD+C). (C and D) Huh-7 cells were infected with JFH-1 for 9 days, after which cells were untreated or treated with 7.5 mM MβCD (MβCD) or MβCD plus cholesterol (MβCD+C). Intracellular JFH-1 RNA levels (C) and JFH-1 virus secretion (D) were quantitated before the start of treatment (day 0) and at 2 days posttreatment. These are representative data (means ± SDs) for three independent experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ffu, focus-forming units.
Depletion of cholesterol by MβCD does not affect the release of infectious HCV from infected cultures.
Since some viruses require cholesterol for virion assembly and/or egress from the cell (53-55, 57), we tested whether MβCD treatment affects HCV RNA replication and virus production during an ongoing JFH-1 infection. Huh-7 cells were infected with JFH-1 at an MOI of 0.02 for 9 days, by which time the majority of cells are infected with JFH-1 (determined by NS5A immunofluorescence staining) (data not shown) and secrete ∼103 to 104 focus-forming units/ml of infectious JFH-1 virus (day 0) (Fig. 6D). As shown in Fig. 6C, treatment of infected Huh-7 cells with MβCD did not significantly suppress JFH-1 RNA replication, consistent with our results for the JFH-1 SG replicon-containing cells (Fig. 6A). However, while MβCD treatment reduced intracellular cholesterol levels by 68% (data not shown), it failed to affect the secretion or infectivity of JFH-1 particles (Fig. 6D). These results suggest that HCV secretion is not dependent on cellular levels of cholesterol.
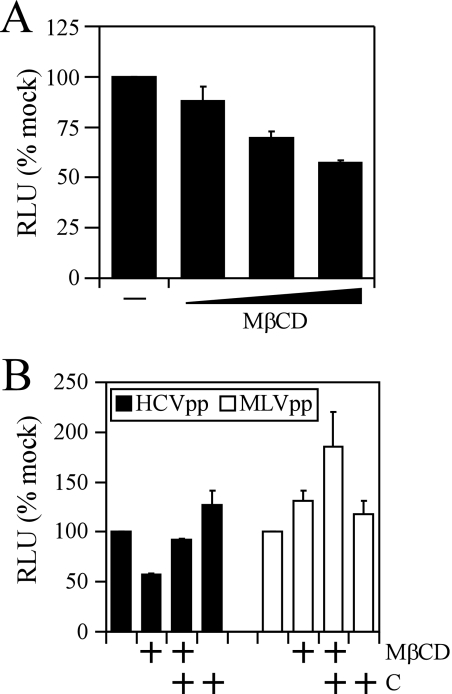
HCVpp entry is dependent on cellular cholesterol.
To specifically determine whether cholesterol depletion by MβCD inhibits HCV entry, we utilized the HCVpp system, which has been used extensively to address the molecular mechanisms of HCV entry into cells (9, 10, 16, 34, 46, 81, 83). Huh-7 cells were pretreated with MβCD in the presence or absence of supplementary cholesterol in the culture medium. The cells were infected with pseudotype particles expressing either HCV glycoproteins (HCVpp) or murine leukemia virus glycoproteins (MLVpp), and luciferase was quantitated at 72 h postinfection. As shown in Fig. 7A, there was a dose-dependent inhibition of HCVpp entry. Pretreatment of Huh-7 cells with 10 mM MβCD resulted in an ∼50% reduction (P = 0.007) of HCVpp entry (Fig. 7B). This inhibition could be reversed by the replenishment of cholesterol in the MβCD-treated cells prior to infection with HCVpp (Fig. 7). In comparison, entry of MLVpp was not significantly affected (P = 0.1) by the depletion of cholesterol (Fig. 7). Collectively, these data demonstrate that the cholesterol content of the cell influences the efficiency of viral entry during HCV infection.
FIG. 7.
Depletion of cholesterol by MβCD decreases HCVpp entry in Huh-7 cells. (A) Dose-dependent inhibition of HCVpp infection of Huh-7 cells after treatment with 2.5, 5, and 10 mM MβCD. (B) Huh-7 cells were untreated or treated with 10 mM MβCD (MβCD) or 10 mM MβCD plus cholesterol (MβCD+C) and infected with HCVpp or MLVpp. Relative light units (RLU) for each sample were calculated by normalization to mock-treated Huh-7 cells. These are representative data (means ± standard errors of the means) for two independent experiments.
DISCUSSION
The results reported herein demonstrate that SR-BI and CD81 function cooperatively to mediate HCV infection in vitro and indicate that cellular cholesterol is required for efficient HCV infection in vitro, most likely due to the effects of cholesterol on the cell surface expression level of CD81. The differences in the neutralizing activity of the anti-CD81 and anti-SRBI antibodies for HCVpp and infectious JFH-1 virus suggest that there are important differences in the mechanisms of action of the antibodies in the two viral systems. Anti-CD81 and anti-SRBI antibodies neutralize HCVpp entry but fail to show any synergistic effects in combination, suggesting that the cooperativity observed for inhibiting the initiation of JFH-1 infection may occur at a postentry level. Several reports have demonstrated an SR-BI-dependent activation of phosphatidylinositol 3-kinase (51, 69), where antibody engagement of SR-BI may activate signaling pathways that JFH-1 virus may utilize postentry. Furthermore, the mechanism by which SR-BI mediates HCV entry is not completely understood. While sE2 has been shown to bind SR-BI (7, 68), it has not been technically feasible to determine whether E1E2 glycoprotein heterodimers bind SR-BI. However, von Hahn et al. recently demonstrated that oxidized LDL, which is a natural ligand for SR-BI, can inhibit HCVpp and HCV infection, supporting a critical role for SR-BI in HCV entry (84). Furthermore, Heo et al. recently demonstrated that HCV E2 glycoprotein links soluble CD81 and SR-BI protein together (38). HCV in plasma from infected patients has been reported to associate with very-low-density lipoprotein (VLDL) and LDL (1, 4, 56, 75, 76), and a recent report suggests that virus-lipoprotein complexes interact with cells via SR-BI (49). Another candidate receptor for HCV, the LDL-R, has been reported to internalize plasma-derived virus by interacting with LDL and VLDL complexed with HCV particles (1, 4, 52). The LDL-R is responsible for binding and uptake of apolipoproteins B (ApoB0) and E (ApoE) (28), and since ApoB and ApoE have been demonstrated to be associated with HCV in the sera of infected patients (56), this has led to the suggestion that the LDL-R may internalize virus through binding of virion-associated lipoproteins. However, the experimental evidence to support a role of the LDL-R in HCV entry is lacking. Interestingly, oxidized low-density lipoprotein, a ligand for SR-BI, was shown to inhibit HCVpp and HCV infection (84). Since SR-BI can interact with HDL, VLDL, and native and chemically modified LDL (77), these results suggest that SR-BI may bind HCV particles through interactions with apolipoproteins. Interestingly, HDL was recently demonstrated to facilitate SR-BI-mediated HCVpp entry (81) and also decrease antibody-mediated neutralization of HCV (29, 82).
The current results demonstrate that HCV infection is dependent on the cholesterol content of the cell. Depletion of cholesterol from cellular membranes by MβCD has been demonstrated to inhibit infection by many viruses (19, 26, 63, 70). However, the extent to which flavivirus infections are sensitive to cellular cholesterol levels is still unclear. While infection by dengue virus is markedly inhibited by cholesterol depletion (65), infection by another flavivirus, tick-borne encephalitis virus, is only modestly affected (72). Our results demonstrate that depletion of cellular cholesterol by MβCD suppresses JFH-1 infection (Fig. 5A) and HCVpp entry (Fig. 7). However, the inhibition is incomplete and may reflect the partial depletion of cholesterol from plasma membranes. While many flaviviruses, including HCV, utilize clathrin-dependent receptor-mediated endocytosis to enter cells (13, 21, 22, 24, 36), other viruses enter cells through caveolae (58). Both caveolae and clathrin structures are specialized plasma membrane domains that are enriched in cholesterol, and both are disrupted by the depletion of cholesterol (45, 59). While membrane cholesterol may be directly required for receptor expression and/or localization to initiate HCV infection, it is possible that cholesterol is required for additional plasma membrane-associated protein-protein interactions that regulate the initiation of HCV infection. Alternatively, it is possible that HCV may utilize mechanisms for entry that do not depend solely on cellular membrane cholesterol levels, since cholesterol depletion does not completely inhibit HCV infection (Fig. 5A) or HCVpp entry (Fig. 7).
In contrast to its effect on CD81, MβCD depletion of cholesterol increases cell surface expression of SR-BI (Fig. 4B). These results are consistent with published data demonstrating that SR-BI mRNA and protein levels are increased after MβCD-mediated depletion of cholesterol (37, 74, 90). While our results suggest that the initial steps of HCV infection are dependent partially on membrane cholesterol, this could reflect a compensatory increase in SR-BI membrane expression in the context of reduced CD81 expression induced by cholesterol depletion. Since JFH-1 infection is inhibited under these circumstances, this may suggest that the SR-BI dependence of HCV infection may be secondary to CD81. This is supported by the observations that CD81-blocking antibodies strongly inhibit HCVpp entry (11) and that HepG2 cells, which express SR-BI but not CD81, are not permissive for HCVpp infection (11). On the other hand, these results could also be explained by the different levels of cell surface SR-BI and CD81 required for HCV infection.
In conclusion, we have demonstrated that HCV infection is dependent on a cooperative interaction between CD81 and SR-BI and that cellular cholesterol content has a significant impact on HCV entry, possibly by regulating cell surface expression and localization of CD81. Recent data suggest that HCV infectivity is dependent upon the density of SR-BI and CD81 in the target cell and that threshold amounts are required for efficient infection (A. E. Jennings, Z. Stamataki, J. Grove, P. Balfe, and J. A. McKeating, unpublished data). These results favor the existence of a multicomponent receptor-mediated mechanism of HCV entry in which CD81 and SR-BI play essential but different roles in the internalization of HCV. The results add to the growing knowledge of the importance of cholesterol and its synthesis in HCV infection and pathogenesis.
Acknowledgments
We thank Takaji Wakita for providing the JFH-1 subgenomic replicon and infectious clone, Michael Houghton and Dennis Burton for supplying us with the antibodies to NS5A and E2 proteins, respectively, and Natalia Reixach for help with the fluorescence microplate reader. We thank K. E. Hu for preparation of HCV pseudotype stocks, Adrian Thrasher (Institute of Child Health) for CSGW, and Paul Bienias (Rockefeller University, NY) for HIV Gag-Pol plasmids. We also thank Holly Maier for her helpful critique of the manuscript.
This study was supported by NIH grants CA108304 to F.V.C. and AI50798 to J.A.M. In addition, S.B.K. was supported by the American Cancer Society-Gloria Rosen Postdoctoral Research Fellowship.
Footnotes
Published ahead of print on 18 October 2006.
This is manuscript number 18188-MEM from The Scripps Research Institute.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, et al. 1992. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N. Engl. J. Med. 327:1899-1905. [DOI] [PubMed] [Google Scholar]
- 4.André, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitt, J., B. Trigatti, A. Rigotti, E. J. Smart, R. G. Anderson, S. Xu, and M. Krieger. 1997. Murine SR-BI, a high density lipoprotein receptor that mediates selective lipid uptake, is N-glycosylated and fatty acylated and colocalizes with plasma membrane caveolae. J. Biol. Chem. 272:13242-13249. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 7.Barth, H., R. Cerino, M. Arcuri, M. Hoffmann, P. Schürmann, M. I. Adah, B. Gissler, X. Zhao, V. Ghisetti, B. Lavezzo, H. E. Blum, F. von Weizsäcker, A. Vitelli, E. Scarselli, and T. F. Baumert. 2005. Scavenger receptor class B type I and hepatitis C virus infection of primary Tupaia hepatocytes. J. Virol. 79:5774-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 9.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 12.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley, D. W. 2000. Studies of non-A, non-B hepatitis and characterization of the hepatitis C virus in chimpanzees. Curr. Top. Microbiol. Immunol. 242:1-23. [DOI] [PubMed] [Google Scholar]
- 16.Callens, N., Y. Ciczora, B. Bartosch, N. Vu-Dac, F.-L. Cosset, J.-M. Pawlotsky, F. Penin, and J. Dubuisson. 2005. Basic residues in hypervariable region 1 of hepatitis C virus envelope glycoprotein e2 contribute to virus entry. J. Virol. 79:15331-15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charrin, S., S. Manie, C. Thiele, M. Billard, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33:2479-2489. [DOI] [PubMed] [Google Scholar]
- 18.Cherukuri, A., T. Shoham, H. W. Sohn, S. Levy, S. Brooks, R. Carter, and S. K. Pierce. 2004. The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts. J. Immunol. 172:370-380. [DOI] [PubMed] [Google Scholar]
- 19.Choi, K. S., H. Aizaki, and M. M. Lai. 2005. Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release. J. Virol. 79:9862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 21.Chu, J. J., P. W. Leong, and M. L. Ng. 2006. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology 349:463-475. [DOI] [PubMed] [Google Scholar]
- 22.Chu, J. J., and M. L. Ng. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78:10543-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung, C. S., C. Y. Huang, and W. Chang. 2005. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J. Virol. 79:1623-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codran, A., C. Royer, D. Jaeck, M. Bastien-Valle, T. F. Baumert, M. P. Kieny, C. A. Pereira, and J. P. Martin. 2006. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J. Gen. Virol. 87:2583-2593. [DOI] [PubMed] [Google Scholar]
- 25.Cormier, E. G., R. J. Durso, F. Tsamis, L. Boussemart, C. Manix, W. C. Olson, J. P. Gardner, and T. Dragic. 2004. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:14067-14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danthi, P., and M. Chow. 2004. Cholesterol removal by methyl-β-cyclodextrin inhibits poliovirus entry. J. Virol. 78:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Date, T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 28.Defesche, J. C. 2004. Low-density lipoprotein receptor—its structure, function, and mutations. Semin. Vasc. Med. 4:5-11. [DOI] [PubMed] [Google Scholar]
- 29.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P. E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F. L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285-18295. [DOI] [PubMed] [Google Scholar]
- 30.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 31.Drummer, H. E., and P. Poumbourios. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066-30072. [DOI] [PubMed] [Google Scholar]
- 32.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goffard, A., N. Callens, B. Bartosch, C. Wychowski, F.-L. Cosset, C. Montpellier, and J. Dubuisson. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400-8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graf, G. A., P. M. Connell, D. R. van der Westhuyzen, and E. J. Smart. 1999. The class B, type I scavenger receptor promotes the selective uptake of high density lipoprotein cholesterol ethers into caveolae. J. Biol. Chem. 274:12043-12048. [DOI] [PubMed] [Google Scholar]
- 36.Grummer, B., S. Grotha, and I. Greiser-Wilke. 2004. Bovine viral diarrhoea virus is internalized by clathrin-dependent receptor-mediated endocytosis. J. Vet. Med. B 51:427-432. [DOI] [PubMed] [Google Scholar]
- 37.Han, J., D. P. Hajjar, J. M. Tauras, and A. C. Nicholson. 1999. Cellular cholesterol regulates expression of the macrophage type B scavenger receptor, CD36. J. Lipid Res. 40:830-838. [PubMed] [Google Scholar]
- 38.Heo, T. H., S. M. Lee, B. Bartosch, F. L. Cosset, and C. Y. Kang. 2006. Hepatitis C virus E2 links soluble human CD81 and SR-B1 protein. Virus Res. 121:58-64. [DOI] [PubMed] [Google Scholar]
- 39.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang, H., Y. Li, T. Sadaoka, H. Tang, T. Yamamoto, K. Yamanishi, and Y. Mori. 2006. Human herpesvirus 6 envelope cholesterol is required for virus entry. J. Gen. Virol. 87:277-285. [DOI] [PubMed] [Google Scholar]
- 41.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 102:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 44.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laude, A. J., and I. A. Prior. 2004. Plasma membrane microdomains: organization, function and trafficking. Mol. Membr. Biol. 21:193-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavillette, D., B. Bartosch, D. Nourrisson, G. Verney, F. L. Cosset, F. Penin, and E. I. Pecheur. 2006. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J. Biol. Chem. 281:3909-3917. [DOI] [PubMed] [Google Scholar]
- 47.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 48.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 49.Maillard, P., T. Huby, U. Andreo, M. Moreau, J. Chapman, and A. Budkowska. 2006. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 20:735-737. [DOI] [PubMed] [Google Scholar]
- 50.McCaffrey, A. P., K. Ohashi, L. Meuse, S. Shen, A. M. Lancaster, P. J. Lukavsky, P. Sarnow, and M. A. Kay. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5:676-684. [DOI] [PubMed] [Google Scholar]
- 51.Mineo, C., I. S. Yuhanna, M. J. Quon, and P. W. Shaul. 2003. High density lipoprotein-induced endothelial nitric-oxide synthase activation is mediated by Akt and MAP kinases. J. Biol. Chem. 278:9142-9149. [DOI] [PubMed] [Google Scholar]
- 52.Monazahian, M., I. Bohme, S. Bonk, A. Koch, C. Scholz, S. Grethe, and R. Thomssen. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57:223-229. [DOI] [PubMed] [Google Scholar]
- 53.Nayak, D. P., and S. Barman. 2002. Role of lipid rafts in virus assembly and budding. Adv. Virus Res. 58:1-28. [DOI] [PubMed] [Google Scholar]
- 54.Nayak, D. P., and E. K. Hui. 2004. The role of lipid microdomains in virus biology. Subcell. Biochem. 37:443-491. [DOI] [PubMed] [Google Scholar]
- 55.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortegren, U., M. Karlsson, N. Blazic, M. Blomqvist, F. H. Nystrom, J. Gustavsson, P. Fredman, and P. Stralfors. 2004. Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. Eur. J. Biochem. 271:2028-2036. [DOI] [PubMed] [Google Scholar]
- 59.Pichler, H., and H. Riezman. 2004. Where sterols are required for endocytosis. Biochim. Biophys. Acta 1666:51-61. [DOI] [PubMed] [Google Scholar]
- 60.Pike, L. J. 2003. Lipid rafts: bringing order to chaos. J. Lipid Res. 44:655-667. [DOI] [PubMed] [Google Scholar]
- 61.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 62.Pöhlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rawat, S. S., M. Viard, S. A. Gallo, A. Rein, R. Blumenthal, and A. Puri. 2003. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review). Mol. Membr. Biol. 20:243-254. [DOI] [PubMed] [Google Scholar]
- 64.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 65.Reyes-Del Valle, J., S. Chavez-Salinas, F. Medina, and R. M. Del Angel. 2005. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J. Virol. 79:4557-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhainds, D., P. Bourgeois, G. Bourret, K. Huard, L. Falstrault, and L. Brissette. 2004. Localization and regulation of SR-BI in membrane rafts of HepG2 cells. J. Cell Sci. 117:3095-3105. [DOI] [PubMed] [Google Scholar]
- 67.Saunier, B., M. Triyatni, L. Ulianich, P. Maruvada, P. Yen, and L. D. Kohn. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seetharam, D., C. Mineo, A. K. Gormley, L. L. Gibson, W. Vongpatanasin, K. L. Chambliss, L. D. Hahner, M. L. Cummings, R. L. Kitchens, Y. L. Marcel, D. J. Rader, and P. W. Shaul. 2006. High-density lipoprotein promotes endothelial cell migration and reendothelialization via scavenger receptor-B type I. Circ. Res. 98:63-72. [DOI] [PubMed] [Google Scholar]
- 70.Shah, W. A., H. Peng, and S. Carbonetto. 2006. Role of non-raft cholesterol in lymphocytic choriomeningitis virus infection via alpha-dystroglycan. J. Gen. Virol. 87:673-678. [DOI] [PubMed] [Google Scholar]
- 71.Soldaini, E., A. Wack, U. D'Oro, S. Nuti, C. Ulivieri, C. T. Baldari, and S. Abrignani. 2003. T cell costimulation by the hepatitis C virus envelope protein E2 binding to CD81 is mediated by Lck. Eur. J. Immunol. 33:455-464. [DOI] [PubMed] [Google Scholar]
- 72.Stiasny, K., C. Koessl, and F. X. Heinz. 2003. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 77:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun, X., and G. R. Whittaker. 2003. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 77:12543-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun, Y., N. Wang, and A. R. Tall. 1999. Regulation of adrenal scavenger receptor-BI expression by ACTH and cellular cholesterol pools. J. Lipid Res. 40:1799-1805. [PubMed] [Google Scholar]
- 75.Thomssen, R., S. Bonk, C. Propfe, K. H. Heermann, H. G. Kochel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. (Berlin) 181:293-300. [DOI] [PubMed] [Google Scholar]
- 76.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. (Berlin) 182:329-334. [DOI] [PubMed] [Google Scholar]
- 77.Trigatti, B. L., A. Rigotti, and A. Braun. 2000. Cellular and physiological roles of SR-BI, a lipoprotein receptor which mediates selective lipid uptake. Biochim. Biophys. Acta 1529:276-286. [DOI] [PubMed] [Google Scholar]
- 78.Trowbridge, R., and E. J. Gowans. 1998. Identification of novel sequences at the 5′ terminus of the hepatitis C virus genome. J. Viral Hepat. 5:95-98. [DOI] [PubMed] [Google Scholar]
- 79.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vial, C., and R. J. Evans. 2005. Disruption of lipid rafts inhibits P2X1 receptor-mediated currents and arterial vasoconstriction. J. Biol. Chem. 280:30705-30711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 82.Voisset, C., A. O. de Beeck, P. Horellou, M. Dreux, T. Gustot, G. Duverlie, F. L. Cosset, N. Vu-Dac, and J. Dubuisson. 2006. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J. Gen. Virol. 87:2577-2581. [DOI] [PubMed] [Google Scholar]
- 83.Voisset, C., and J. Dubuisson. 2004. Functional hepatitis C virus envelope glycoproteins. Biol. Cell 96:413-420. [DOI] [PubMed] [Google Scholar]
- 84.von Hahn, T., B. D. Lindenbach, A. Boullier, O. Quehenberger, M. Paulson, C. M. Rice, and J. A. McKeating. 2006. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology 43:932-942. [DOI] [PubMed] [Google Scholar]
- 85.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wielgosz, M. M., D. A. Rauch, K. S. Jones, F. W. Ruscetti, and L. Ratner. 2005. Cholesterol dependence of HTLV-I infection. AIDS Res. Hum. Retrovir. 21:43-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu, L., G. Cao, J. Repa, and H. Stangl. 2004. Sterol regulation of scavenger receptor class B type I in macrophages. J. Lipid Res. 45:889-899. [DOI] [PubMed] [Google Scholar]
- 91.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhong, J., G. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]